Abstract
Temperature-dependent sex determination (TSD) decides the sex fate of an individual based on incubation temperature. However, other environmental factors, such as pollutants, could derail TSD sexual development. Cadmium is one such contaminant of soils and water bodies known to affect DNA methylation, an epigenetic DNA modification with a key role in sexual development of TSD vertebrate embryos. Yet, whether cadmium alters DNA methylation of genes underlying gonadal formation in turtles remains unknown. Here, we investigated the effects of cadmium on the expression of two gene regulators of TSD in the painted turtle, Chrysemys picta, incubated at male-producing and female-producing temperatures using qPCR. Results revealed that cadmium alters transcription of Dmrt1 and aromatase, overriding the normal thermal effects during embryogenesis, which could potentially disrupt the sexual development of TSD turtles. Results from a preliminary DNA methylation-sensitive PCR assay implicate changes in DNA methylation of Dmrt1 as a potential cause that requires further testing (aromatase methylation assays were precluded).
1. Introduction
Concerns over pollution and contamination of water bodies and soil have emerged in the past decades, and attention is growing due to the potential harm they can cause to living organisms. For instance, disruption of sex ratios in natural populations of vertebrates by such contaminants can cause population decline or extinction [1,2]. This occurs because some pollutants can alter sex determination, the biological process that sets the sexual fate of developing embryos and which ranges between two ends of a continuum [3,4,5]. On one end is genotypic sex determination (GSD), in which a genetic component triggers the molecular and hormonal cascade that commits the gonad to become a testis or an ovary. On the other end is environmental sex determination (ESD), in which an environmental factor (such as temperature—TSD, pH, population density, among others) is responsible for initiating the sexual development cascade [3,4,6]. In TSD, the incubation temperature experienced during a specific embryonic development period is the triggering factor [7]. TSD is predominant among turtle species, while GSD in the form of sex chromosomes evolved independently multiple times [8]. The painted turtle (Chrysemys picta) is a TSD species lacking sex chromosomes [9].
Sex determination in C. picta may be mediated in part by DNA methylation [10], an epigenetic modification that alters gene expression patterns without changing the DNA sequence, by substituting the 5′carbon of a cytosine with a methyl group. Depending on its location and context, DNA methylation may repress, activate, or modulate gene expression [11]. More often, methylated cytosines physically block access of the transcription machinery to the DNA [12], silencing methylated genes. In general, DNA methylation is an important molecular mechanism that organisms use to respond to environmental stimuli [13,14], and it is critical for sex determination in turtles and other reptiles with TSD [15,16,17,18]. For instance, in Alligator mississipiensis, the promoter region of Sox9, a gene involved in male sexual development, is methylated at female-producing temperatures, repressing its expression in developing ovaries [15]. Another example of a male development gene regulated by methylation is Dmrt1, which exhibits hypermethylation (and thus is downregulated) in developing females compared to males in TSD and GSD fish [19,20,21,22]. Importantly, Dmrt1 is under additional epigenetic control via histone methylation in the TSD turtle Trachemys scripta, where the histone demethylase KDM6B removes trimethylation from H3K27, a histone close to the Dmrt1 promoter [23]. Third, aromatase, a key gene for female development, is regulated in T. scripta by DNA methylation at specific CpG sites on its promoter region. Specifically, CpG sites located close to binding sites for Foxl2, a known aromatase regulator, and close to the TATA box, an RNA-polymerase recruiter site, are hypomethylated at female-producing compared to male-producing temperatures [24]. Furthermore, recent studies demonstrated a global sexually dimorphic DNA methylation during gonadogenesis of Lepidochelys olivacea sea turtles [17], a pattern observed in gonads of C. picta hatchlings incubated in different temperatures, including at gene regulators of sexual development [10]. Yet, how contaminants such as cadmium might alter DNA methylation of these important genes during embryogenesis in developing male and female turtles remains unknown.
Contaminants present in water or soil during the incubation period may affect sex determination, mainly by disrupting the hormonal balance during sexual development [1] or altering DNA methylation of relevant sexual development genes. For instance, cadmium that is present in multiple industrial processes [25] can disrupt DNA methylation patterns and potentially affect gonadal development [26,27]. Here we investigated the effects of cadmium exposure on DNA methylation of C. picta turtles during three different embryonic stages and the consequent effects on gene expression patterns of a gene important for testicular formation (Dmrt1) and a key regulator of ovarian formation (aromatase). In the absence of cadmium effects, we expect Dmrt1 to be hypermethylated (and thus downregulated) in developing females (at 31 °C) compared to males (at 26 °C), and aromatase to be hypermethylated (and thus downregulated) in developing males (at 26 °C) compared to females (at 31 °C), opposite transcriptional patterns to those observed typically at those temperatures in C. picta embryos [28,29].
2. Materials and Methods
2.1. Egg Collection, Incubation, and Cadmium (Cd) Treatment
Freshly laid eggs were collected from a turtle farm in Iowa and transported in moist vermiculite to the laboratory for incubation following our standard protocols [30], with some modifications. Briefly, eggs were cleaned, marked with unique IDs, and randomly assigned to incubation boxes with moist sand. Before the onset of incubation, 1 mL of a solution of 1 μg/mL of CdCl2 diluted in water was applied topically to each egg. This dose was chosen because previous studies at female-producing temperature using ecologically relevant cadmium doses found that 1 μg/g egg reduced germ cells in neonate gonads of Trachemys scripta turtles, whereas the observed germ cell reduction using 0.1 μg/g was not statistically significant, but importantly, that C. picta appears much more susceptible to cadmium than T. scripta [27]. Thus, given a C. picta egg mass of ~6 g [31,32,33], a dose of 1 μg per egg (~0.17 μg/g egg) fell within the ecologically relevant range [27]. Water was chosen as solvent because CdCl2 is hygroscopic and highly soluble in water, the turtle eggshell absorbs water and water-soluble contaminants readily [34,35], and to avoid unknown effects of other solvents. Eggs were incubated at 26 °C (male-producing temperature, MPT) (n = 80) and 31 °C (female-producing temperature—FPT) (n = 88), of which 40 were intended for hatching to test cadmium effects on sex ratios and the remainder were intended for assessment of embryonic gene expression and DNA methylation. The position of the boxes inside the incubator was rotated daily between left/right side and top/bottom shelfs to account for any thermal gradient that might exist, following standard procedures [30,36]. Moisture was maintained by replacing evaporated water (determined by weight reduction of the box) weekly. Embryonic development and viability were assessed by egg candling weekly. Embryos and tissues were dissected at Yntema’s stages 09 (before the thermosensitive period—TSP), 15 (just prior to the onset of TSP), and 22 (end of the TSP), and stored in RNA later (Invitrogen) at −20 °C until processing. Because the gonadal primordium cannot be separated in stage 9 embryos, trunks were sampled at that early stage. Adrenal-kidney-gonad (AKG) complexes were sampled at stage 15 because the gonads could not be separated from the AK. Stage 15 gonads are bipotential in Chrysemys [37]. Lastly, separated gonads were sampled from stage 22 embryos. While some of the differences among stages are likely attributable to differences in the tissues sampled, our approach permits testing for differences in gene expression and methylation between FPT and MPT at each developmental period, as well as between embryos exposed to cadmium (this study) and embryos incubated in the absence of cadmium under otherwise identical conditions, which serve as negative controls. Hatchlings obtained after 45 days of incubation were kept in water tanks at 25 °C and fed commercial turtle food for three months before autopsy to ensure gonadal differentiation was complete before sexing by gonadal inspection. All procedures were done in accordance with guidelines and regulations approved by the IACUC of Iowa State University.
2.2. DNA and RNA Extractions, cDNA Conversion
DNA and RNA were extracted from embryo trunks (stage 09, n = 15 per temperature), AKG complexes (stage 15, n = 12 per temperature), and gonads (stage 22, n = 10 per temperature) using Qiagen AllPrep DNA/RNA ™ Mini (stages 9 and 15) and Micro (stage 22) kits, following the manufacturer’s instructions. Differences in the sample size per stage were due to embryonic mortality at later stages of development due to cadmium exposure. RNA and DNA were quantified using a NanoDrop Spectrophotometer. DNA was stored at −20 °C until further use.
The RNA extraction protocol from the extraction kits included a treatment with DNAse to reduce gDNA. RNA quality was assessed by the presence of ribosomal bands in 1% agarose gels. RNA was stored at −80 °C until 500 ng of RNA was converted to cDNA using Invitrogen SuperScript™ VILO™ Synthesis kit following the manufacturer’s instructions; cDNA was stored at −20 °C until further use.
2.3. qPCR Assay and Data Analysis
To assess whether cadmium exposure altered gene expression patterns of Dmrt1, we carried out qPCR reactions in an Mx3000P real-time PCR thermal cycler (Stratagene), using the cDNA produced as described above. For Dmrt1, we used a TaqMan probe-based multiplex qPCR to amplify a fragment of only the canonical isoform of this gene (147 bp) and a fragment of the ß-actin transcript as a reference gene (138 bp), following previous Dmrt1 profiling in this species [38]. All primers used in this study are listed in Table 1. We used the IDT Prime Time Gene Expression Master Mix, which contains the DNA polymerase, dNTPs, MgCl2, enhancers, and stabilizers, in concentrations undisclosed by the manufacturer. Multiplex optimization reactions with primers concentration of 500 nM and probes concentration of 500 nM (Dmrt1) and 250 nM (ß-actin) were carried out, with 5 μL of commercial master mix, ß-actin and Dmrt1 forward and reverse primers, ß-actin and Dmrt1 Taqman probe, 2 μL of cDNA (50 ng), and water up to 15 μL.

Table 1.
Primers and probes used in this study (see text for details).
For aromatase, a gene with no known alternative spliceoforms, we carried out SYBR Green reactions to amplify a 168 bp transcript fragment of aromatase, using GAPDH as a reference gene (127 bp amplicon), following [39,40,41]. Reactions were prepared using Brilliant® SYBR® Green qPCR Master Mix with aromatase and GAPDH primers in separate qPCR reactions.
For all genes, qPCR conditions included an initial denaturing step at 95 °C for 3 min followed by 50 cycles of 95 °C for 30 s and 60 °C for 1 min. All reactions were run in duplicate. We generated standard curves by pooling cDNA from all samples (100 ng per individual) and then serially diluted the pooled cDNA using a 1:5 ratio to obtain a total of eight standards. Standards were included in duplicate in each qPCR plate to generate standard curves to calculate qPCR efficiency and R2 values.
All samples showed a coefficient of variation <10% between technical replicates; thus, they were all included in the analysis. ß-actin and GAPDH displayed constant expression across stages and temperatures in our study. We performed a Student’s t-test in R [42] to test for differences in the normalized ratio of Dmrt1 to ß-actin, and of aromatase to GAPDH, between temperatures at each stage and assessed significance at an α of 0.05. As negative controls, we leveraged previous qPCR profiling of Dmrt1 and aromatase expression of stage 9, 15, and 22 C. picta in embryos incubated at identical thermal conditions but in the absence of cadmium exposure as reported elsewhere [38].
2.4. PCR Test of DNA Methylation
In order to assess DNA methylation status, we first digested the DNA with a DNA-methylation-sensitive restriction enzyme, then amplified digested and undigested DNA via PCR using a combination of primers designed specifically to produce differential amplification if the DNA is methylated (and thus digested) or not [43]. We used this method in previous studies of DNA methylation in C. picta hatchlings (not embryos) for the gene Fezf2 [10]. This primer design included one forward primer (F1) and two reverse primers (R1 and R2), one of which (R1) binds closer to F1, and one (R2) which binds further away from F1 (Figure 1). Primers F1 + R1 amplify a smaller region nested within the larger region amplified by F1 + R2. Primers F1 and R1 flank a non-methylated area, which should be amplified irrespective of DNA methylation status and thus serve as a positive control. In contrast, primers F1 and R2 flank a region that encompasses the differentially methylated CpG island and amplify depending on the methylation status of this area. Methylated regions are protected from the restriction enzyme and thus are not digested, resulting in two amplicon bands in the gel electrophoresis, while unmethylated regions are digested and result in a single PCR amplicon band. We designed primers at the promoter regions of Dmrt1, focusing on sequences that contained the restriction site of HpaII (CCGG).
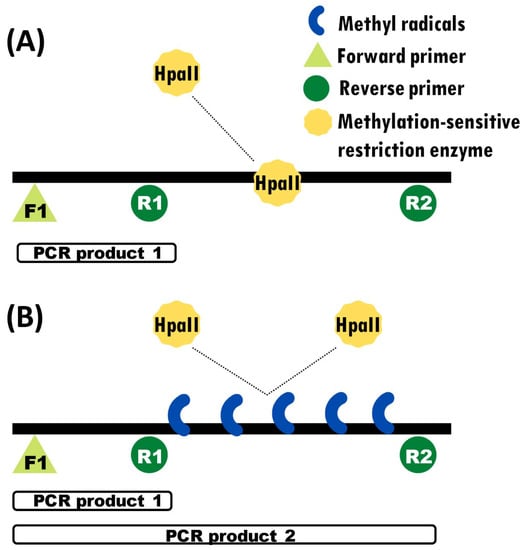
Figure 1.
Experimental design of the methylation-sensitive PCR assay. Black bars denote the target DNA sequence, onto which the relative position of the primers, restriction sites, and methylation marks are illustrated. White bars denote the relative size of fragments amplified by PCR. (A) Amplification of only one PCR product due to the DNA digestion by HpaII. (B) Amplification of two PCR products thanks to the protection conferred by the DNA methyl groups, which prevents DNA digestion by the restriction enzyme.
Before PCR amplification, the DNA from embryos of the three stages examined was digested with HpaII (Thermo Fisher, Waltham, MA, USA) following the manufacturer’s instructions. Primers for aromatase and Dmrt1 were designed at their promoter regions using the design described above. PCR was carried out separately using DNA subjected to the restriction enzyme and undigested (control) samples in 15 μL reactions with 1× Taq buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.4 μM of each primer (F1, R1 and R2), 0.4 U Taq polymerase, and 10.5 μL water, with 50 ng of DNA as template. PCR conditions included an initial denaturing step at 94 °C for 3 min, followed by 35 cycles of denaturing at 94 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 1 min. PCR products were visualized in a 1% agarose gel stained with EtBr, and their size was estimated by a 1 kb plus ladder (Invitrogen). DNA methylation status was determined qualitatively by the intensity of the amplicon bands in the agarose gel.
In the case of genes that are hypermethylated in males, we expected to see a single band in females (corresponding to the small amplicon produced by F1 + R1 primers in the nested non-methylated region) and two bands in males (the common smaller band from F1 + R1 primers and a larger band from F1 + R2 primers). The opposite is expected in cases when genes are hypermethylated in females: one band in males and two bands in females. Because the DNA-methylation-sensitive PCR assay was not conducted during the previous study [38], our experimental design for the methylation assessment lacks a direct negative control with embryos not exposed to cadmium, such that our PCR results are preliminary and should be taken as suggestive until further testing, and interpreted only in the context of the gene expression profiling.
3. Results
3.1. Cadmium Induces High Embryonic Mortality
Only 6.67% of the 40 eggs incubated until hatching to assess sex ratios when exposed to cadmium survived. While the mortality observed in parallel incubation experiments conducted that same year was particularly high (60–65%) for unknown reasons [44], the mortality of cadmium-treated embryos exceeded that baseline by >30%. Early embryo mortality under cadmium exposure (before stage 09 of development) affected 24% of the eggs (n = 11), while 49% of the eggs (n = 22) died during late development (between stages 23 and 26), and 8% (n = 4) of the eggs died after they pipped or hatched. Consequently, only three eggs hatched and survived to three months of age. These three hatchlings were not sex-reversed (n = 2 males from 26 °C and n = 1 female from 31 °C). Other than the increased embryonic mortality, no obvious effect of cadmium exposure was observed in the morphology and anatomy of the three surviving hatchlings. However, the small number of surviving individuals precluded testing for the effect of cadmium on sex reversal as originally intended, such that further studies are warranted using alternative cadmium doses. Of the eggs incubated for embryonic methylation and gene expression analysis, 15, 12, and 10 embryos were collected from each temperature at stages 9, 15, and 22, respectively.
3.2. Cadmium Exposure Alters Gene Expression
All standard curves for the qPCR reactions exhibited a R2 > 0.98, and negative controls showed no amplification. Our quantitative results indicate that both Dmrt1 and aromatase transcriptional patterns were altered by exposure to cadmium. Specifically, Dmrt1 transcription was higher, but not significantly so (Table 2), in presumptive developing females (i.e., at FPT—31 °C) at stages 15 (TSP-onset) and 22 (late-TSP), rather than being upregulated in presumptive males (i.e., at MPT—26 °C) at stage 22 as occurs under uncontaminated conditions (Figure 2A,B). Aromatase transcription at MPT was also affected, as expression peaked at stage 15 and dropped at stage 22, with no significant upregulation (Table 2) at FPT by stage 22, as occurs under normal conditions [28,29] (Figure 2C,D). Thus, cadmium exposure eliminated the differential transcription characteristic of Dmrt1 and aromatase expression (p > 0.05 in all cases) as expected during typical development [28,29,38].

Table 2.
Results of the Student’s t-test statistical analysis for Dmrt1 and Aromatase between temperatures at each stage examined.
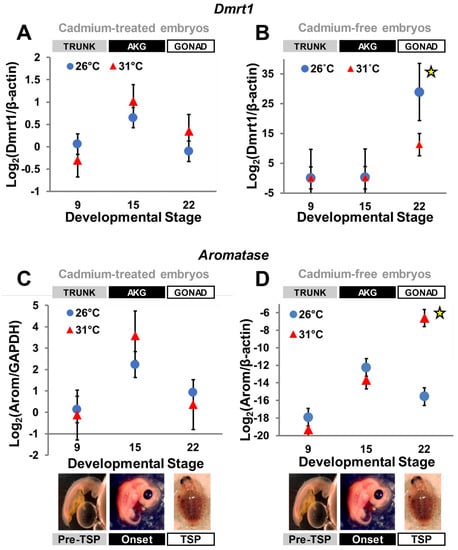
Figure 2.
Dmrt1 and aromatase transcription under and without cadmium exposure. Data derived from this study (A,C), and from Mizoguchi and Valenzuela 2020 [38] (B) and Valenzuela et al. 2013 [28] (D), which serve as negative control. Bars represent standard deviations. In embryos exposed to cadmium (A,C), differences in expression between temperatures were not significant at any of the stages examined, whereas in the absence of cadmium exposure, Dmrt1 (B) and aromatase (D) are normally upregulated at stage 22 at MPT or FPT, respectively (indicated by the yellow star symbol).
3.3. Cadmium Exposure Induces Similar DNA Methylation Profiles of Dmrt1, a Testicular Development Regulator
Results from the methylation-sensitive PCR assay showed similar levels of DNA methylation between temperatures for Dmrt1 at all stages (Figure 3). This was assessed qualitatively by the reduction of PCR amplification (fainter bands) of the larger amplicon in the digested DNA samples compared to the undigested controls to a similar level at both incubation temperatures, indicating that DNA was equally unprotected at 26 °C and 31 °C from digestion by DNA methylation. This similarity between MPT and FPT reflects either demethylation or inhibition of DNA methylation due to cadmium exposure, which should lead to similar levels of transcription in developing males and females. Unfortunately, a freezer malfunction ruined the samples before PCR amplification of aromatase, precluding aromatase methylation tests.
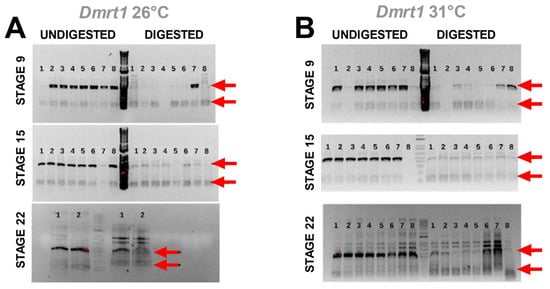
Figure 3.
Agarose gels of DNA methylation-sensitive PCR assay for Dmrt1 at MPT (A) and FPT (B). Invitrogen’s 1 kb Plus DNA ladder was used. Arrows show the size of the two amplicons expected (120 bp and 438 bp) when DNA is methylated and thus protected from digestion by the restriction enzyme, as detailed in the text.
4. Discussion
In this study, we investigated for the first time the effects of cadmium exposure on the expression of two gene members of the sexual development network, Dmrt1 and aromatase (Figure 2). Dmrt1 encodes a transcription factor involved in male-specific differentiation. Aromatase is a cytochrome P450 enzyme responsible for aromatizing testosterone into estrogen and is key for ovarian formation [24,33]. We observed that cadmium exposure during embryogenesis alters gene expression patterns in ways consistent with a potential to disrupt sexual development.
Cadmium is known to affect DNA methylation by interacting with the DNA-binding domain of DNA methyltransferase, the enzyme responsible for establishing DNA methylation marks, leading to hypomethylation [26]. Concordantly, we observed quantitative changes in gene expression that match this mode of action for cadmium. For instance, cadmium exposure can alter gene expression when gene promoters are regulated by DNA methylation, as is the case for Dmrt1 in fish [13,20,21,22], aromatase in TSD turtles and fish [19,24], and Sox9 in alligators [15]. These genes are differentially transcribed in the developing embryo of C. picta turtles [28,29,38]. Yet, cadmium obliterated the significant differential expression of Dmrt1 and aromatase typically observed between MPT and FPT in this species (Figure 2). Indeed, no significant differential expression was detected at any stage examined here, whereas in previous studies, Dmrt1 and aromatase were significantly upregulated at stages 19 and 22 at MPT (Dmrt1) or FPT (aromatase), respectively [30,31,34]. This is a notable finding, given that such abnormal expression patterns could greatly affect downstream genetic pathways and, consequently, sexual development. Furthermore, the alteration of the transcription of both a male- and a female-inducing gene during our experiment indicates that cadmium can interfere in sexual development by directly altering gene expression patterns of relevant sex-related genes. Previous studies demonstrated that cadmium can affect germ cell abundance in ovaries of neonatal TSD turtles, but no sex reversal was reported [27]. Whether cadmium induces sex reversal remains unknown, particularly at MPT, because high mortality precluded sex ratio analyses here, and the previous work only tested FPTs [27].
For instance, upregulation of aromatase at MPT by any factor could lead to higher estrogen production, which could directly interfere with the male sexual development pathway. Indeed, estrogen affects the expression of steroidogenic factor-1 (Sf-1, NR5A1), an activator of the anti-Mullerian hormone (AMH) [45], critical for testis development. On the other hand, the downregulation of aromatase in females, as occurs by aromatase inhibitors, leads to sex reversal or masculinization of gonads [36,37]. Further, Dmrt1 expression responds rapidly to aromatase inhibition [18], revealing Dmrt1’s early role in initiating male differentiation in some TSD turtles. Although we did not observe any sex reversal in our study due to the increased mortality induced by cadmium, previous studies in C. picta observed oocyte apoptosis in C. picta neonates after cadmium exposure, perhaps due to the disruption of aromatase expression [27]. Similarly, aromatase inhibition was linked to oocyte apoptosis in juvenile and adult zebrafish [38,39]. Furthermore, downregulation of Dmrt1 at MPT induces the upregulation of Foxl2, an important gene in the female sexual development pathway, and a known inhibitor of Sf-1 [46]. Foxl2 also inhibits the expression of Sox9, another key male development gene [18]. Moreover, experimental knock-down of Dmrt1 feminizes embryos, inducing elongated gonads and the development of oviducts or even ovotestis, whereas Dmrt1 overexpression at FPT leads to masculinization or complete sex reversal [18].
Our transcription profiling results for Dmrt1 (lack of significant differential expression) were associated with the qualitative patterns of DNA methylation assessed by the DNA methylation-sensitive PCR assay. Specifically, results from the PCR test are consistent with cadmium having altered DNA methylation, as they revealed a reduction of PCR amplification of the larger amplicon (this larger amplicon was expected if DNA was methylated) in the digested DNA samples compared to the undigested controls for both genes, to similar levels at both temperatures. The observation of similar methylation-sensitive PCR amplification at MPT and FPT suggests that DNA methylation levels were equivalent at both temperatures, perhaps because cadmium caused de-methylation at FPT. However, instances where chronic exposure to cadmium causes hypermethylation exist [26], such that further studies are warranted to determine the exact mode of action of cadmium in C. picta.
If DNA-methylation was indeed altered, and given that the transcription of both Dmrt1 and aromatase was affected by cadmium exposure, a female-inducing gene that is normally upregulated at warm temperatures and a male-inducing gene that is normally upregulated at colder temperatures, we hypothesize that cadmium affects DNA methylation at a global genome level in turtles, rather than just targeting specific genes. Such effect would agree with previous studies that identified changes in global methylation in the European eel, human embryo fibroblast, and hens, associated with cadmium exposure [47,48,49]. If true for turtles, then it would be expected that other genes regulated by DNA methylation were likely affected, consistent with the increased mortality observed in our study.
In conclusion, we detected for the first time that embryos exposed to cadmium suffer increased mortality and experienced altered expression of gene regulators of sexual development. Our preliminary data suggest that this modified Dmrt1 transcription may be associated with changes in DNA methylation, but strong inferences are precluded in the absence of methylation-sensitive PCR data from embryos not exposed to cadmium. Despite these caveats, our study contributes to the growing body of work highlighting the detrimental consequences of exposure to chemicals, especially endocrine-disrupting contaminants (EDCs), on gene expression, gonadal development, and sex reversal (reviewed in [1]). Because cadmium is a metal that accumulates in organisms, including the painted turtle, and in yolk [50], cadmium is an environmental contaminant that poses substantial dangers to sexual development [27,47,48,49]. Because no contaminant is found alone in nature, but instead in mixtures, their effects can be seen even at smaller doses [51]. Such exposure can lead to higher mortality rates, abnormal sexual development, decreased fertility, and generally lower fitness [1]. Natural populations are vulnerable to soil and water contamination, and our findings help illuminate some of the mechanism by which contaminants interfere in embryonic development. Our work should help guide broader research to aid managers in better preserving turtle populations and determining safe thresholds for human exposure to similar contaminants.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes13081318/s1, Supplementary Table S1: Normalized gene expression data used in this study.
Author Contributions
Conceptualization, B.M. and N.V.; methodology, B.M. and N.V.; investigation, B.M., A.M.L. and N.E.T.; formal analysis, B.M., A.M.L. and N.E.T.; resources, N.V.; writing—original draft preparation, B.M.; writing—review and editing, N.V., A.M.L. and N.E.T.; visualization, B.M.; supervision, B.M. and N.V.; funding acquisition, N.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by the National Science Foundation of USA (NSF) grant number IOS 1555999 to N.V., and the APC was funded in part by NSF grant IOS 2127995 to N.V.
Institutional Review Board Statement
All experimental procedures were in accordance with the Institutional Animal Care and Use Committee (IACUC) of Iowa State University.
Informed Consent Statement
Not applicable.
Data Availability Statement
Quantitative data used in this study are provided in the Supplementary Information, Supplementary Table S1.
Acknowledgments
The authors would like to thank Thea Gessler and Basanta Bista for their contribution in collecting eggs for this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mizoguchi, B.A.; Valenzuela, N. Ecotoxicological Perspectives of Sex Determination. Sex. Dev. 2016, 10, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Kohno, S. Can Xenobiotics Alter the Sex Ratio of Crocodilians in the Wild? Sex. Dev. 2021, 15, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, N.; Adams, D.C.; Janzen, F.J. Pattern does not equal process: Exactly when is sex environmentally determined? Am. Nat. 2003, 161, 676–683. [Google Scholar] [CrossRef] [Green Version]
- Stöck, M.; Kratochvíl, L.; Kuhl, H.; Rovatsos, M.; Evans, B.J.; Suh, A.; Valenzuela, N.; Veyrunes, F.; Zhou, Q.; Gamble, T.; et al. A brief review of vertebrate sex evolution with a pledge for integrative research: Towards ‘sexomics’. Philos. Trans. R. Soc. Lond. Ser. B 2021, 376. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, S.L.; Castelli, M.A.; Dissanayake, D.S.B.; Holleley, C.E.; Georges, A. Temperature-induced sex reversal in reptiles: Prevalence, discovery, and evolutionary implications. Sex. Dev. 2021, 15, 148–156. [Google Scholar] [CrossRef]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Sex Determination: Why So Many Ways of Doing It? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef] [Green Version]
- Valenzuela, N.; Lance, V.A. (Eds.) Temperature Dependent Sex Determination in Vertebrates; Smithsonian Books: Washington, DC, USA, 2004. [Google Scholar]
- Bista, B.; Valenzuela, N. Turtle insights into the evolution of the reptilian karyotype and the genomic architecture of sex determination. Genes 2020, 11, 416. [Google Scholar] [CrossRef] [Green Version]
- Valenzuela, N.; Badenhorst, D.; Montiel, E.E.; Literman, R. Molecular cytogenetic search for cryptic sex chromosomes in painted turtles Chrysemys picta. Cytogenet. Genome Res. 2014, 144, 39–46. [Google Scholar] [CrossRef]
- Radhakrishnan, S.; Literman, R.; Mizoguchi, B.; Valenzuela, N. MeDIP-seq and nCpG analyses illuminate sexually dimorphic methylation of gonadal development genes with high historic methylation in turtle hatchlings with temperature-dependent sex determination. Epigenet. Chromatin 2017, 10, 28. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Piferrer, F. Epigenetics of sex determination and gonadogenesis. Dev. Dyn. 2013, 242, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Head, J.A. Patterns of DNA Methylation in Animals: An Ecotoxicological Perspective. Integr. Comp. Biol. 2014, 54, 77–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrott, B.B.; Kohno, S.; Cloy-McCoy, J.A.; Guillette, L.J. Differential Incubation Temperatures Result in Dimorphic DNA Methylation Patterning of the SOX9 and Aromatase Promoters in Gonads of Alligator (Alligator mississippiensis) Embryos1. Biol. Reprod. 2014, 90, 1–11. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Hannigan, B.; Crews, D. Temperature shift alters DNA methylation and histone modification patterns in gonadal aromatase (cyp19a1) gene in species with temperature-dependent sex determination. PLoS ONE 2016, 11, e0167362. [Google Scholar] [CrossRef] [Green Version]
- Venegas, D.; Marmolejo-Valencia, A.; Valdes-Quezada, C.; Govenzensky, T.; Recillas-Targa, F.; Merchant-Larios, H. Dimorphic DNA methylation during temperature-dependent sex determination in the sea turtle Lepidochelys olivacea. Gen. Comp. Endocrinol. 2016, 236, 35–41. [Google Scholar] [CrossRef]
- Ge, C.; Ye, J.; Zhang, H.; Zhang, Y.; Sun, W.; Sang, Y.; Capel, B.; Qian, G. Dmrt1 induces the male pathway in a turtle species with temperature-dependent sex determination. Development 2017, 144, 2222–2233. [Google Scholar] [CrossRef] [Green Version]
- Piferrer, F.; Anastasiadi, D.; Valdivieso, A.; Sánchez-baizán, N.; Moraleda-Prados, J.; Ribas, L. The Model of the Conserved Epigenetic Regulation of Sex. Front. Genet. 2019, 10, 857. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Zheng, J.; Chi, M.; Liu, S.; Jiang, W.; Cheng, S.; Gu, Z.; Chen, L. Molecular identification of dmrt1 and its promoter CpG methylation in correlation with gene expression during gonad development in Culter alburnus. Fish Physiol. Biochem. 2019, 45, 245–252. [Google Scholar] [CrossRef]
- He, Y.; Wu, X.; Zhu, Y.; Yang, D. Expression Profiles of dmrt1 in Schizothorax kozlovi, and Their Relation to CpG Methylation of Its Promoter and Temperature Relation to CpG Methylation of Its Promoter and Temperature. Zool. Sci. 2020, 37, 140–147. [Google Scholar] [CrossRef]
- Ou, M.; Chen, K.; Gao, D.; Wu, Y.; Luo, Q.; Liu, H.; Zhao, J. Characterization, expression and CpG methylation analysis of Dmrt1 and its response to steroid hormone in blotched snakehead (Channa maculata). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2022, 257, 110672. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Ye, J.; Weber, C.; Sun, W.; Zhang, H.; Zhou, Y.; Cai, C.; Qian, G.; Capel, B. The histone demethylase KDM6B regulates temperature-dependent sex determination in a turtle species. Science 2018, 360, 645–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, Y.; Buemio, A.; Chu, R.; Vafaee, M.; Crews, D. Epigenetic Control of Gonadal Aromatase (cyp19a1) in Temperature-Dependent Sex Determination of Red-Eared Slider Turtles. PLoS ONE 2013, 8, e63599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutton, M. Sources of cadmium in the environment. Ecotoxicol. Environ. Saf. 1983, 7, 9–24. [Google Scholar] [CrossRef]
- Takiguchi, M.; Achanzar, W.E.; Qu, W.; Li, G.; Waalkes, M.P. Effects of cadmium on DNA- (Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp. Cell Res. 2003, 286, 355–365. [Google Scholar] [CrossRef]
- Kitana, N.; Callard, I.P. Effect of cadmium on gonadal development in freshwater turtle (Trachemys scripta, Chrysemys picta) embryos. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2008, 43, 262–271. [Google Scholar] [CrossRef]
- Valenzuela, N.; Neuwald, J.L.; Literman, R. Transcriptional evolution underlying vertebrate sexual development. Dev. Dyn. 2013, 242, 307–319. [Google Scholar] [CrossRef]
- Radhakrishnan, S.; Literman, R.; Neuwald, J.; Severin, A.; Valenzuela, N. Transcriptomic responses to environmental temperature by turtles with temperaturedependent and genotypic sex determination assessed by RNAseq inform the genetic architecture of embryonic gonadal development. PLoS ONE 2017, 12, e0172044. [Google Scholar] [CrossRef]
- Valenzuela, N. Egg incubation and collection of painted turtle embryos. Cold Spring Harb. Protoc. 2009, 4, 10–12. [Google Scholar] [CrossRef] [Green Version]
- Rollinson, N.; Brooks, R.J. Optimal offspring provisioning when egg size is “constrained”: A case study with the painted turtle Chrysemys picta. Oikos 2008, 117, 144–151. [Google Scholar] [CrossRef]
- Rowe, J.W. Egg Size and Shape Variation within and among Nebraskan Painted Turtle (Chrysemys picta bellii) Populations: Relationships to Clutch and Maternal Body Size. Copeia 1994, 1994, 1034–1040. [Google Scholar] [CrossRef]
- Janzen, F.J.; Warner, D.A. Parent–offspring conflict and selection on egg size in turtles. J. Evol. Biol. 2009, 22, 2222–2230. [Google Scholar] [CrossRef] [PubMed]
- de Solla, S.R.; Martin, P.A. Absorption of current use pesticides by snapping turtle (Chelydra serpentina) eggs in treated soil. Chemosphere 2011, 85, 820–825. [Google Scholar] [CrossRef]
- Booth, D. Incubation of rigid-shelled turtle eggs: Do hydric conditions matter? J. Comp. Physiol. B 2022, 172, 627–633. [Google Scholar] [CrossRef]
- Bowden, R.M.; Ewert, M.A.; Nelson, C.E. Environmental sex determination in a reptile varies seasonally and with yolk hormones. Proc. Biol. Sci. 2000, 267. [Google Scholar] [CrossRef] [Green Version]
- Bull, J.J.; Vogt, R.C. Temperature-sensitive periods of sex determination in Emydid turtles. J. Exp. Zool. 1981, 218, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, B.; Valenzuela, N. Alternative splicing and thermosensitive expression of Dmrt1 during urogenital development in the painted turtle, Chrysemys picta. PeerJ 2020, 8, e8639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishikawa, H.; Reierstad, S.; Demura, M.; Rademaker, A.W.; Kasai, T.; Inoue, M.; Usui, H.; Shozu, M.; Bulun, S.E. High Aromatase Expression in Uterine Leiomyoma Tissues of African-American Women. J. Clin. Endocrinol. Metab. 2009, 94, 1752–1756. [Google Scholar] [CrossRef]
- Lambard, S.; Galeraud-Denis, I.; Bouraïma, H.; Bourguiba, S.; Chocat, A.; Carreau, S. Expression of aromatase in human ejaculated spermatozoa: A putative marker of motility. Mol. Hum. Reprod. 2003, 9, 117–124. [Google Scholar] [CrossRef] [Green Version]
- Carreau, S.; Wolczynski, S.; Galeraud-Denis, I. Aromatase, oestrogens and human male reproduction. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Caetano, L.C.; Gennaro, F.G.O.; Coelho, K.; Araújo, F.M.; Vila, R.A.; Araújo, A.; de Melo Bernardo, A.; Marcondes, C.R.; Chuva de Sousa Lopes, S.M.; Ramos, E.S. Differential expression of the MHM region and of sex-determining-related genes during gonadal development in chicken embryos. Genet. Mol. Res. 2014, 13, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, V.; Literman, R.; Neuwald, J.L.; Mizoguchi, B.; Iverson, J.B.; Riley, J.L.; Litzgus, J.D. Extreme thermal fluctuations from climate change unexpectedly accelerate demographic collapse of vertebrates with temperature-dependent sex determination. Sci. Rep. 2019, 9, 4254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elf, P.K. Yolk steroid hormones and sex determination in reptiles with TSD. Gen. Comp. Endocrinol. 2003, 132, 349–355. [Google Scholar] [CrossRef]
- Eggers, S.; Ohnesorg, T.; Sinclair, A. Genetic regulation of mammalian gonad development. Nat. Rev. Endocrinol. 2014, 10, 673–683. [Google Scholar] [CrossRef]
- Jiang, G.; Xu, L.; Song, S.; Zhu, C.; Wu, Q.; Zhang, L.; Wu, L. Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology 2008, 244, 49–55. [Google Scholar] [CrossRef]
- Pierron, F.; Baillon, L.; Sow, M.; Gotreau, S.; Gonzalez, P. Effect of low-dose cadmium exposure on DNA methylation in the endangered European eel. Environ. Sci. Technol. 2014, 48, 797–803. [Google Scholar] [CrossRef]
- Zhang, J.; Fu, Y.; Li, J.; Wang, J.; He, B.; Xu, S. Effects of subchronic cadmium poisoning on DNA methylation in hens. Environ. Toxicol. Pharmacol. 2009, 27, 345–349. [Google Scholar] [CrossRef]
- Rie, M.T.; Lendas, K.A.; Callard, I.P. Cadmium: Tissue distribution and binding protein induction in the painted turtle, Chrysemys picta. Comp. Biochem. Physiology. Toxicol. Pharmacol. 2001, 130, 41–51. [Google Scholar] [CrossRef]
- Willingham, E. Endocrine-disrupting compounds and mixtures: Unexpected dose-response. Arch. Environ. Contam. Toxicol. 2004, 46, 265–269. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).