Abstract
Plasmodiophora brassicae infection leads to hypertrophy of host roots and subsequent formation of galls, causing huge economic losses to agricultural producers of Cruciferae plants. Ethylene (ET) has been reported to play a vital role against necrotrophic pathogens in the classic immunity system. More clues suggested that the defense to pathogens in roots may be different from the acrial. The ET pathway may play a positive role in the infection of P. brassicae, as shown by recent transcriptome profiling. However, the molecular basis of ET remains poorly understood. In this study, we investigated the potential role of ethylene against P. brassicae infection in an ein3/eil1 double-mutant of Arabidopsis thaliana (A. thaliana). After infection, ein3/eil1 (Disease Index/DI: 93) showed more susceptibility compared with wild type (DI: 75). Then, we inoculated A. thaliana Columbia-0 (Col-0) with P. brassicae by 1-aminocyclopropane-1-carboxylic acid (ACC) and pyrazinamide (PZA), respectively. It was found that the symptoms of infected roots with ACC were more serious than those with PZA at 20 dpi (day post infection). However, the DI were almost the same in different treatments at 30 dpi. WRKY75 can be directly regulated by ET and was upregulated at 7 dpi with ACC, as shown by qRT-PCR. The wrky75-c mutant of A. thaliana (DI: 93.75) was more susceptible than the wild type in Arabidopsis. Thus, our work reveals the dual roles of ET in infection of P. brassicae and provides evidence of ET in root defense against pathogens.
1. Introduction
Clubroot is caused by the infection of a soilborne obligate biotrophic protist Plasmodiophora brassicae Woron, which is one of the most destructive diseases of Cruciferous crops [1], with billions of dollars lost. Clubroot is widely spreading in Brazil, South Africa, Australia, New Zealand, China, and Russia [2]. The model plant Arabidopsis thaliana and 16 Brassicaceae crops (about 3.2–4.0 million ha), including Brassica rapa pekinensis, Brassica napus, and Brassica juncea, are the hosts of P. brassicae [3]. The infection process of P. brassicae can be divided into two stages. Under proper conditions, primary zoospores, delivered by resting spores, penetrate root hairs and epidermal cells, and subsequently, secondary zoospores, which are produced in infected root hairs and epidermal cells, are released into the soil, This stage is known as the primary infection. The secondary infection stage occurs in cortical cells through the released zoospores, resulting in gall formation [4]. These galls are sucrose sinks that hijack the sugar partitioning of host plants via promoting phloem differentiation and phloem-specific expression of sugar transporters [5], thus blocking the uptake of nutrition and water in susceptible plants.
Plants have developed sophisticated immune systems against diverse pathogens that may affect the yield and quality of crops. In recent years, two-layered innate immune systems are widely accepted. One is triggered by the recognition of pathogen/damage-associated molecular patterns (P/DAMPs). The other one is triggered by recognition of the pathogen effectors via receptors known as surface pattern-recognition receptors (PRRs) and intracellular nucleotide-binding domain leucine-rich repeat containing receptors (NLRs). The recognition of two receptors results in pattern-triggered immunity (PTI) and effector-triggered immunity, respectively [6]. PTI and ETI share some overlapping downstream outputs, and the phytohormone signaling pathway plays a vital role in the plant immune signaling pathway network. Among them, salicylic acid (SA) and jasmonate (JA)/ethylene (ET) are the critical major defense players against biotrophic pathogens and necrotrophic pathogens, respectively [7]. However, our understanding of plant defense is mostly from the aerial part of the plant. Recently, considerable progress has been made in understanding the defense mechanisms of the root systems, and some differences have been highlighted between roots and shoots in response to pathogens [8]. Of note, both SA and JA pathways have a positive effect against the biotrophic root pathogen P. brassicae [9]. Although transcriptome analysis has indicated the ethylene signaling pathway may be involved in resistance to P. brassicae, experimental studies have yet to be conducted to support this hypothesis.
Ethylene, a gas phytohormone, has been shown to play a significant role in plant development and response to abiotic and biotic stresses, including salinity stress, heat stress, heavy metal stress, flooding stress, etc. [10]. Ethylene is produced from L-methionine, and the last step of the synthesis is catalyzed by ACC oxidases (ACO) with 1-aminocyclopropane-1-carboxylic acid (ACC) as the substrate. EIN3/EIL1 transcription factors, targeted by EIN2, are the key regulators of ethylene signaling pathway that maintain multiple responses to ethylene [11]. Many transcription factors, such as AP2/ERF, WRKY, and NAC, have been shown to be regulated by ethylene through EIN3/EIL1. WRKY75, involved in various biological processes including leaf senescence, phosphate deficiency, oxalic acid stress, flowering time, defense responses, and root hair development, can be directly targeted by EIN3 [12,13,14,15,16].
Ethylene may participate in inhibiting or stimulating cell division and expansion in different organs. For example, ethylene can promote cell division through several downstream AP2/ERF transcription factors in vascular tissue [17], yet it inhibits cell division by multiple mechanisms in leaves that are exposed to different environmental stresses [18]. In addition, ethylene exhibits positive and negative effects on cell expansion in petioles and leaves, respectively [19,20]. The RALFs–LRXs–FER pathway is thought to regulate plant immune and cell expansion. FERONIA (FER), a member of receptor-like kinases, has been found to participate in a variety of cellular processes, such as ethylene responses in Arabidopsis hypocotyls [21] and function as the receptor of RALF peptide in plant immune system [22]. RALFs and LRXs are involved in plant growth via cell wall signal transduction [23]. Rapid alkalinization factor (RALF) peptides have been reported to be involved in various processes, such as plant immune response [24], through regulating cell expansion in most cases.
Based on previous studies, it requires a deeper understanding of ET in the interaction between plant and P. brassicae. In this study, we firstly confirmed the positive role of ET by the inoculation process in the ein3/eil1 double mutant. To further investigate ET-mediated responses in different stages during clubroot infection, exogenous ACC and ACC oxidase inhibitors were applied during P. brassicae infection in Arabidopsis. The altered P. brassicae symptoms and transcriptional regulation were investigated to decipher the sophisticated mechanism of ET. Our study provides a more detailed view of ET function in the interaction with P. brassicae.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
A. thaliana Columbia-0 (Col-0) and three mutants, ein3 and eil1 single mutants and ein3/eil1 double mutant, were provided by Dr Guo (Southern University of Science and Technology). The wrky75-c mutant was obtained using the CRISPR/Cas9 genome editing system in the Arabidopsis Col-0 background through the floral dip method mediated by Agrobacterium.
Dried seeds were surface-sterilized with 70% ethyl alcohol for 3 min, stratified in 1/2 MS (Murashige and Skoog) medium at 4 °C for 3 d, and then placed in the growth chamber at 22 °C under 16 h light/8 h dark for 1 week. The healthy seedlings with a similar growth rate were selected to transfer into the soil (mix matrix soil, vermiculite, and perlite with 3:1:1) and grow at the same conditions in the growth chamber.
2.2. Vector Construction
To obtain the wrky75-c mutant lines through the CRISPR/Cas9 system, two sgRNAs (19 bp) in the WRKY75 genome, G1: 5′-GGAGTCGTCGAAAAAGAAG-3′ and G2: 5′-TTAACAGTGGACCAAGAAG-3′, were designed by CRISPR-P 2.0 (http://crispr.hzau.edu.cn/CRISPR2/ (accessed on 6 July 2020)) [25]. Then, the sgRNA’s fragment was cloned with the template pCBCT1T2 and the primes containing homologous arms near BSAI F: 5′-TCGAAGTAGTGATTGAGCGACAGAGGTAACCCAAGTTTTAGAGCTAGAAATAGC-3′ and R: 5′-TTCTAGCTCTAAAACTGGTGCTTGCTGGGCTTATCAATCTCTTAGTCGACTCTAC-3′. Finally, the CRISPR/Cas9 expression vector AtU6-G1-Ter1-AtU9-G2-Ter2 (U6:G1-U9:G2) was obtained via homologous recombination connecting the sgRNA’s fragment and PBSE401, and cut by the restriction enzyme BSAI. A 35S:DsRED expression cassette was transferred into U6:G1-U9:G2 through restriction-ligase reaction via EcoRI to simplify the screening of positive mutant lines.
2.3. RNA Isolation and Quantitative PCR Analyses
Total RNA was isolated from the treated roots with a RNAprep Pure plant kit (TIANGEN), and the reverse transcription of RNA was performed using a Fastking RT kit (with gDNase, TIANGEN). For each RNA sample, the concentration was measured with a NanoDrop 2000 spectrophotometer, and the integrity of RNA was confirmed using RNase-free agarose gel electrophoresis. Quantitative RT-PCR was performed on the ABI QuantStudio 7 Flex Real-Time PCR system using 2× realstar green fast mixture with ROX Ⅱ (SenStar). The PCR cycling reaction was performed according to the following program: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s, and 60 °C for 1 min. The relative gene expression was calculated using the 2–ΔΔCt method [26]. Three biological replicates were carried out for each experiment, and the results were normalized using the reference gene ACTIN2 and UBQ10. The amounts of RNA and cDNA used were 1μg and 200 ng, respectively. Data were analyzed in Excel Office. Significant differences in the expression levels were evaluated with the t-test (* p < 0.05; ** p < 0.01). All of the qRT-PCR primers are listed in Supplementary Table S1, and the most qRT-PCR primers in Arabidopsis were obtained at qPrimerDB (https://biodb.swu.edu.cn/qprimerdb/ (accessed on 23 March 2021)) [27]. To further understand the ET-mediated pathway in response to P. brassicae infection, a number of genes were detected, including SA and JA marker genes (PR2, THI2 and ARGAH2), some transcription factors (WRKY75, WRKY45, SHN1, ERF105 and At2G20350), and members of the RALFs–LRXs–FER pathway.
2.4. Treatment in the Inoculation Process
Healthy and consistent two-week-old seedlings were inoculated with 2 mL 1 × 107 spores/mL of P. brassicae pathotype 4 (P4) of China. The controls were treated with H2O in the same stage. For the ACC and PZA treatments, ACC and PZA were added into clubroot fluid, and the final concentrations were 100 mM, 500 mM, and 1000 mM for ACC and 50 mM, 250 mM, and 500 mM for PZA, and the inoculation process was the same as the others. After that, the phenotype of roots was photographed at 20 dpi and 30 dpi. The disease index was calculated using the following 0–4 scoring system [28] with DI = 100 × (1N1 + 2N2 + 3N3 + 4N4)/4Nt. Each treatment or lines contained at least 40 plants, and the data were analyzed in Excel 2019 using t-test (* p < 0.05; ** p < 0.01).
2.5. Mutant Identification
A. thaliana mutants ein3, eil1, and ein3/eil1 were confirmed by sequencing the products of PCR. The wrky75-c mutant positive lines were screened by fluorescent tag DsRed and confirmed by sequencing the target region (all of the primers are listed in Supplementary Table S1).
3. Results
3.1. Phenotype Analysis of Mutants in Ethylene Signaling Pathway
To explore the potential role of ethylene in plant responses to P. brassicae, Col-0, ein3/eil1 double mutant, and ein3 and eil1 single mutants were planted and inoculated with P4 of ten-day-old seedlings. The symptoms of the roots were observed in treated and untreated plants after 30 d (Figure 1). Compared to Col-0 (DI = 75), ein3/eil1 (DI = 93) was more susceptible, as indicated by the bigger galls and few or no lateral roots (Figure 2). However, ein3 and eil1 single mutants showed smaller galls compared with ein3/eil1, suggesting the redundancy function between EIN3 and EIL1 in the ET pathway. Ethylene may be needed in resistance to P. brassicae in Arabidopsis, but the details remain to be explored.
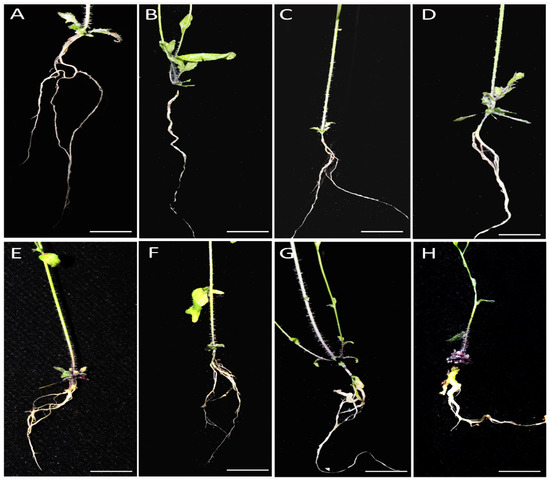
Figure 1.
Phenotypes of infected and uninfected roots of wild type and mutant lines after 30 days of inoculation. (A–D) were control lines while (E–H) were infected lines with the same order: Col-0, ein3, eil1, and ein3/eil1.
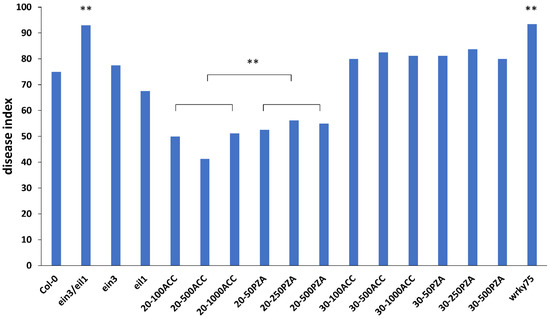
Figure 2.
Disease index of different materials and treatments. Col-0, ein3/eil1, ein3, eil1, and Col-0 treated with 100 mM ACC, 500 mM ACC, 1000 mM ACC, 50 mM PZA, 250 mM PZA, and 500 mM PZA at 20dai and 30dai lines were evaluated with the 0–4 scoring system. For each line, 40 plants were analyzed. Student’s t test, ** p < 0.01.
3.2. Exogenous Application of ACC Revealed a Dual Role during the Infection of P. brassicae
Previous studies have shown that exogenous application of plant immune hormones, including SA and JA, reduces clubroot symptoms in Col-0 [9]. To confirm the function of ethylene, the same strategy was adopted to evaluate the role of ethylene’s response to clubroot. The clubroot symptoms were identified at 20 dpi and 30 dpi. In general, the treatment with ACC decreased the clubroot symptoms compared with PZA at 20 dpi (Figure 2), and the treatment with PZA led to bigger galls in the lateral root, especially in 250 mM PZA (Figure 3). In addition, the effect of ET and its inhibitor may be dose-dependent when no more than 500 mM ACC and 250 mM PZA were applied separately (Figure 2), which may delay or accelerate gall formation with different patterns between ACC and PZA under high concentrations. Although inoculation with 1000 mM ACC proved more severe than in those with 100 mM and 500 mM ACC, it was better than those treated with PZA. In the case of 500 mM PZA, it showed similar symptoms with those inoculated by 250 mM PZA. However, the treatment with ACC and PZA displayed similar results at 30 dpi, while the clubroots treated with ACC exhibited similar DI to those with PZA (Figure 2). Of note, the galls of the plants treated with 250 mM and 50 mM PZA displayed slight root at 30 dpi, which is consistent with previous results that ethylene delayed the invasion of P. brassicae (Figure 4). The development of galls may be induced by ethylene from 20 dpi to 30 dpi. Taken together, we conclude that exogenous ethylene may delay the infection of P. brassicae. However, once the invasion is established, ethylene may promote the development of galls. The proper concentration for ACC and PZA is 500 mM and 250 mM, respectively.

Figure 3.
Clubroot symptoms of infected roots with exogenous ACC and PZA in Arabidopsis. (A): 100 mM ACC; (B): 500 mM ACC; (C): 1000 mM ACC; (D): 50 mM PZA; (E): 250 mM PZA; (F): 500 mM PZA. Roots were photographed at 20 dpi.

Figure 4.
Clubroot symptoms of infected roots with exogenous ACC and PZA in Arabidopsis. (A): 100 mM ACC; (B): 500 mM ACC; (C): 1000 mM ACC; (D): 50 mM PZA; (E): 250 mM PZA; (F): 500 mM PZA. Roots were photographed at 30 dpi.
3.3. Clubroot Induced ET-Response Genes during Infection
To explore the molecular mechanism of A. thaliana responses to P. brassicae with exogenous ethylene and inhibitor, gene expression of treated and untreated roots in different stages was detected at 7 dpi (the primary infection in root hairs and epidermal cells), 14 dpi (the secondary infection in cortical cells), and 20 dpi (the development of galls) by qRT-PCR. We focused on the gene expression by the treatment with 500 mM ACC and 250 mM PZA due to distinct clubroot symptoms (Figure 5). Firstly, the expression of EIN3 and EIL1 was detected in different stages. The expression level of EIN3 and EIL1 had almost no change at 7 dpi and 14 dpi (Figure 5A). Then, a number of transcription factors and genes related to ET and plant defense were tested. Previous studies have shown that both JA and SA contribute to resistance to clubroot [9]. PR2, an SA-responsive gene, was suppressed in infected Col-0 but significantly induced by 500 mM ACC at 7 dpi and 20 dpi, while the expression level of PR2 was induced in all treatments at 14 dpi (Figure 5B). However, RGAH2 and THI2, two JA marker genes, were not induced by 500 mM ACC in all stages. In contrast, RGAH2 was induced in infected Col-0 at 14 dpi and 20 dpi, and THI2 was induced 70-fold higher compared with Col-0 at 20 dpi (Figure 5B). These results indicated that ET-regulated resistance was independent of JA, and SA might be induced by ET in response to clubroot disease at the early infection stage.
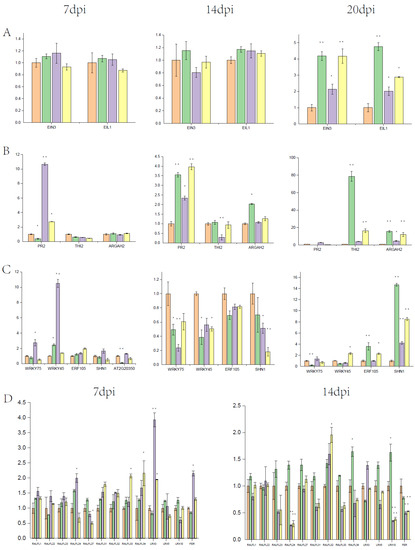
Figure 5.
Relative expression of ET- and clubroot-related genes at 7 dpi, 14 dpi, and 20 dpi, respectively. (A): The relative expression of EIN3 and EIL1; (B): The relative expression of SA and JA marker genes; (C): The relative expression of ET-regulated transcription factors; (D): The relative expression of genes in the RALFs–LRXs–FER pathway. The samples were untreated Col-0, treated Col-0, treated Col-0-500 mM ACC, and treated Col-0-250 mM PZA from the left. Data are means (±SD) (n = 3). Student’s t-test, * p < 0.05, ** p < 0.01.
WRKY and ERF transcription factors are commonly found in the downstream of ET signaling. Although WRKY45 was induced at 7 dpi and suppressed at 14 dpi by both clubroot and ACC, the effect of ACC was more significant compared with that of clubroot (Figure 5C). WRKY75 was induced by 500 mM ACC at 7 dpi and suppressed at 14 dpi (Figure 5C). AT2G20350, a member of the B-6 subfamily of ERF/AP2 transcription factors, was intensively suppressed by about ten folds in infected Col-0 at 7 dpi, but the relative expression was recovered by 500 mM ACC and suppressed by about two folds by 250 mM PZA compared with untreated Col-0 (Figure 5C). Another B-6 subfamily of ERF/AP2 transcription factor SHN1 is involved in wax and cutin biosynthesis. It was found that SHN1 was induced by 500 mM ACC and suppressed by 250 mM PZA at 7 dpi, and the expression was suppressed in all of the other treatments (Figure 5C).
FER was induced by 500 mM ACC and suppressed by 250 mM PZA at 7 dpi. The expression of RALFs and LRXs was detected, as shown in Figure 5D. At 7 dpi, RALFs were induced in infected Col-0, but they displayed higher expression by 500 mM ACC. The expression of RALFL27 was decreased both in ACC and PZA treated roots, and the trend was more notable in PZA-treated roots. RALFL24 showed the opposite expression pattern in the treatment between ACC and PZA. At 14 dpi, RALFs were upregulated in infected Col-0, and possessed similar expression patterns between the treatments with ACC and PZA. These results indicate that RALFs’ function in response to clubroot and ethylene may be involved in this process in the infection of clubroot. LRX3 was induced by 500 mM ACC and suppressed by 250 mM PZA at 7 dpi, which shared the same expression pattern with FER.
3.4. Positive Effect of WRKY75 on the Infection of Clubroot in an ET-Dependent Manner
Since WRKY75 can be induced by ethylene and SA, which is a direct target gene of EIN3, it is necessary to understand whether it participates in response to clubroot. The wrky75-c mutant, obtained by gene editing through the CRISPR/Cas9 system, was investigated via the inoculation test. A homozygous mutant with a G insertion at the 144th position of the coding region from the initiation codon was gained. wrky75-c displayed more susceptibility than Col-0, and wrky75-c showed delayed leaf senescence compared to Col-0 (Figure 6), consistent with previously reported T-DNA insertion line N121525 (wrky75-25) [15]. However, the infection of wrky75-c revealed early-senescence in leaves compared to both Col-0 and untreated wrky75-c. Of note, the clubroot symptoms in wrky75-c were similar to the mutants, such as ein3/eil1, ein2, and etr1-1 [29], which represented extremely short roots and barely lateral roots. It is speculated that WRKY75 may have a positive role in response to clubroot through the ethylene pathway during the early infection.
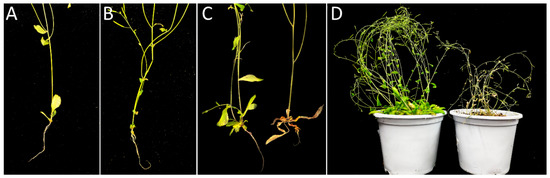
Figure 6.
Phenotype of infected and uninfected roots in wild type and wrky75-c after 30 days of inoculation. Inoculation P. brassicae of two-week-old seedlings. Roots were photographed at 30 dpi. (A): Control, (B): infected Col-0, (C,D): starting from the left: uninfected and infected wrky75-c.
4. Discussion
Previous studies have shown that ET is a positive regulator against P. brassicae. A number of mutants related to ET, such as ein2, ein3-1, ein4, and etr1-1, have been reported to be more susceptible than the wild-type in Arabidopsis [29]. In our study, ein3/eil1 double mutant and ein3 and eil1 single mutants were tested in response to clubroot. ein3/eil1 and eil1 represented increased clubroot symptoms compared to Col-0 (Figure 1), which is consistent with previous studies and provides further proof that ET is needed against P. brassicae. However, the clubroot symptoms of ein3 were opposite from previous studies. Several studies have shown that some phytohormone-impaired/insensitive lines in Arabidopsis have a positive or negative effect on different root knot nematodes that may be due to various root exudates [30]. For example, the ET-regulated pathway reduces the root attractiveness of the soybean cyst nematode but increases the attractiveness of the sugar beet cyst nematode Heterodera schachtii. We assume that the differences between the isolation e3 and P4 may be the cause of clubroot symptoms of ein3. Meanwhile, the transcription level of PR2 and THI2 was different in infected Col-0 compared to untreated Col-0 (Figure 5B) [9], which may contribute to the special properties between eH (belongs to P1) and P4 of P. brassicae. In addition, recent studies have shown that root galls caused by P. brassicae are a mixture of multiple strains [31]. The pathogens of P. brassicae in our study (P4) were obtained from the field, although as identified as P4 by Professor Zhongyun Piao, they may contain more than one strain, while e3 and eH are both single strains. Taken together, our results indicate that the mechanism of clubroot resistance may be different among different pathotypes of P. brassicae.
Although ethylene has a positive role in resistance to clubroot in Cruciferae, detailed studies have yet to be conducted. The effect of ethylene on clubroot is mostly focused on the transcriptional level between clubroot-resistant and clubroot-susceptible roots. Inoculation of 5 × 105 spores at mL−1 for the ethylene signaling pathway and biosynthesis mutants leads to more susceptibility than Col-0, displaying almost no clubroot symptoms [29]. However, eto-2, an ET-overproducing mutant, also showed more susceptibility compared with Col-0. Yet the clubroot symptoms of eto-2 were reduced compared with Col-0 and ET signaling pathway and biosynthesis mutants when higher concentrations of spores (1 × 106 mL−1 to 1 × 107 mL−1) were used. All of these results revealed that a sophisticated process was governed by ET in response to clubroot. The application of the exogenous ethylene precursor ACC and ACC oxidase inhibitor PZA changed the clubroot symptoms under the 1 × 107 mL−1 pathogen. The clubroot symptoms between 20 dpi and 30 dpi showed a remarkable difference, and the clubroot symptoms treated with 500 mM ACC were reduced at 20 dpi but increased at 30 dpi compared with those treated with 250 mM PZA. Although the galls of roots treated with 500 mM ACC were bigger than those by 250 mM PZA at 30 dpi, the roots by 250 mM PZA had rot symptoms (Figure 4 and Figure 5). Therefore, it is hypothesized that ET has a dual role in response to clubroot by delaying the establishment of infection but promoting the development of galls.
Plant hormones are thought to play a vital role in plant response to various pathogens. Thus far, the defense mechanisms in leaves have been studied adequately, but the knowledge about root response to pathogens remains elusive. JA and SA, two antagonistic phytohormones in the classical plant immune systems in leaves, have been reported to have a positive effect on resistance to P. brassicae. However, the regulation process is still elusive. SA is thought to have a positive role in clubroot resistance because the cpr5-2 mutant (which constitutively promotes the SA signaling pathway is more resistant to clubroot disease than the wild type [9]. However, the ein3/eil1 double mutant, which constitutively accumulates SA, is more susceptible compared with the wild type [32]. It seems that the signaling pathway mediated by SA is blocked by P. brassicae, which has been confirmed with the discovery of methyltransferase (PbBSMT) secreted by P. brassicae. PbBSMT can inactivate SA by converting SA to methyl salicylate (MeSA) [33]. Herein, the SA signaling pathway was promoted by exogenous ACC treatment because PR2, a member of the SA signaling pathway, was significantly induced by 500 mM ACC compared with the wild type and the treatment by 250 mM PZA at 7 dpi. Moreover, the robust resistance to P. brassicae in the bik1 mutant may also be related to SA accumulation due to the high PR1 levels in bik1 compared with the wild type [34]. ET is usually thought to act synergistically with JA in the defense response against necrotrophic pathogens, but interestingly, two JA marker genes, THI2 and ARGAH2, were not induced by 500 mM ACC at 7 dpi and 14 dpi. Instead, THI2 and ARGAH2 were significantly induced in the infected Col-0 at 14 dpi and 20 dpi compared to the untreated Col-0 (Figure 5B). Although the expression levels of THI2 and ARGAH2 were induced by 500 mM ACC compared with Col-0 at 20 dpi, the relative expression was lower in contrast to the infected Col-0 and the treatment with 250 mM PZA. ARGAH2 can enhance the expression of NATA1, thereby decreasing the clubroot symptoms [9]. Based on these results, we conclude that ET may contribute to clubroot resistance through activating the SA signaling pathway during early infection, but may promote greater susceptibility through inhibiting the JA signaling pathway during the second infection of clubroot.
Based on the above results, we investigated the regulation mechanism of ET response to clubroot during the early infection. WRKY75, WRKY45, and SHN1 shared similar expression patterns, which were significantly induced by 500 mM ACC compared with the others (Figure 5C). Although AT2G20350 was not significantly induced by 500 mM ACC, the expression was about ten times lower in the infected Col-0 and by the treatment with 250 mM PZA compared with the untreated Col-0 at 7 dpi. It indicates that these transcription factors may affect the resistance to clubroot, which was partially supported by the results of the inoculation of P4 for the mutant wrky75-c. It has been reported that WRKY75 is ethylene-induced and directly targeted by EIN3 [15]. WRKY75 can promote SA accumulation by inducing SA synthesis gene SID2 during leaf senescence [15], and WRKY75 participates in a variety of other biological processes such as phosphorus stress, root hair patterning, and flowering. Likewise, WRKY45 is involved in leaf senescence and phosphorus stress, which is directly regulated by WRKY75 during leaf senescence [35,36,37]. WRKY45 may be mediated by WRKY75 during the ET treatments at the early infection of clubroot, while the relationship between WRKY45 and SA remains unknown. Both SHN1 and AT2G20350 belong to B-6 of the ERF/AP2 transcription factor family, and most of their functions remain unclear. SHN1 is involved in positively regulating defense to Botrytis cinerea [38,39]. It is reported that the ethylene signaling pathway can limit the pathogen to outer cell layers. For example, the ethylene pathway can prevent the invasion of oomycete Pythium irregular into the epidermis and outer and inner cortex of the tap roots [40]. SHN1 regulates the contents of the cuticle, which consist of wax, and is the first barrier to prevent plants from invading pathogens. SHN1 may contribute to the delaying of infection during the early infection. However, studies on AT2G20350 have not been documented to date, and hence the functions remain unknown.
Membrane-localized receptors have vital roles in response to distinguishing nonbeneficial components during the infection of pathogens [41]. FER, a sensor of cell wall integrity, together with RALFs and LRXs, regulates the immune signaling pathway during host–pathogen interactions [42]. It is reported that the function of FER in plant defense differs due to the lifestyle of pathogens [43]. In our study, the RALFs–LRXs–FER pathway was induced by 500 mM ACC at 7 dpi, which may have a positive effect on clubroot invasion. In addition, WRKY75 and WRKY45 can be upregulated in FER mutant leaves [43]. P. brassicae prefers acidic soils with a pH value of less than 5.5, so it may hinder the uptake of Ca, K, Mg, and P [44]. Taken together, the RALFs–LRXs–FER signaling pathway induced by ET may function through two pathways, i.e., by regulating WRKY75 and WRKY45 and by mediating the soil alkalization.
5. Conclusions
Our study found that ET was required in the plant–P. brassicae interaction, and ET had a dual role in the infection of P. brassicae in A. thaliana. ET activated the SA signaling pathway and a number of transcription factors including WRKY75, WRKY45, SHN1, and At2G20350, hence delaying the establishment of infection during the primary infection. In contrast, the JA pathway was suppressed, thereby promoting the development of galls during the secondary infection.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13081299/s1, Table S1: Primers used for mutant identification and qRT-PCR.
Author Contributions
Data curation, K.W., Y.S., Q.S., M.L., L.Z. and B.A.; Formal analysis, K.W.; Supervision, C.Y., F.Y., A.X. and Z.H.; Writing—original draft, K.W.; Writing—review and editing, A.X. and Z.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31771835), the National Key Research and Development Program (2018YFE0108000), Key R&D of Yangling Seed Industry Innovation Center (Ylzy-yc2021-01), and Tang Scholar.
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Acknowledgments
We thank Hongwei Guo for providing the mutants of ein3, eil1 and ein3/eil1.
Conflicts of Interest
The authors declare that they have no competing interests.
Abbreviations
| ET | ethylene |
| JA | jasmonate |
| SA | salicylic acid |
| PCR | polymerase chain reaction |
| P/DAMPs | pathogen/damage-associated molecular patterns |
| PRRs | pattern-recognition receptors |
| NLRs | intracellular nucleotide-binding domain leucine-rich repeat containing receptors |
| PTI | pattern-triggered immunity |
| ETI | effector-triggered immunity |
| RAFL | rapid alkalinization factor |
References
- Chai, A.L.; Xie, X.W.; Shi, Y.X.; Li, B.J. Special Issue: Research status of clubroot (Plasmodiophora brassicae) on cruciferous crops in China. Can. J. Plant Pathol. 2014, 36, 142–153. [Google Scholar] [CrossRef]
- Hwang, S.F.; Strelkov, S.E.; Feng, J.; Gossen, B.D.; Howard, R.J. Plasmodiophora brassicae: A review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol. Plant Pathol. 2012, 13, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Xu, L.; Liu, F.; Chen, K.; Sun, C.; Li, J.; Fang, X. Host Range of Plasmodiophora brassicae on Cruciferous Crops and Weeds in China. Plant Dis. 2016, 100, 933–939. [Google Scholar] [CrossRef]
- Liu, L.; Qin, L.; Zhou, Z.; Hendriks, W.; Liu, S.; Wei, Y. Refining the Life Cycle of Plasmodiophora brassicae. Phytopathology 2020, 110, 1704–1712. [Google Scholar] [CrossRef]
- Walerowski, P.; Gündel, A.; Yahaya, N.; Truman, W.; Sobczak, M.; Olszak, M.; Rolfe, S.; Borisjuk, L.; Malinowski, R. Clubroot Disease Stimulates Early Steps of Phloem Differentiation and Recruits SWEET Sucrose Transporters within Developing Galls. Plant Cell 2018, 30, 3058–3073. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Chuberre, C.; Plancot, B.; Driouich, A.; Moore, J.P.; Bardor, M.; Gügi, B.; Vicré, M. Plant Immunity Is Compartmentalized and Specialized in Roots. Front. Plant Sci. 2018, 9, 1692. [Google Scholar] [CrossRef]
- Lemarié, S.; Robert-Seilaniantz, A.; Lariagon, C.; Lemoine, J.; Marnet, N.; Jubault, M.; Manzanares-Dauleux, M.J.; Gravot, A. Both the Jasmonic Acid and the Salicylic Acid Pathways Contribute to Resistance to the Biotrophic Clubroot Agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol. 2015, 56, 2158–2168. [Google Scholar]
- Müller, M.; Munné-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Larsen, P.B. Mechanisms of ethylene biosynthesis and response in plants. Essays Biochem. 2015, 58, 61–70. [Google Scholar] [PubMed]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, J.; Lin, G.; Wang, A.; Wang, Z.; Lu, G. Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Rep. 2013, 32, 1589–1599. [Google Scholar] [CrossRef]
- Rishmawi, L.; Pesch, M.; Juengst, C.; Schauss, A.C.; Schrader, A.; Hülskamp, M. Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol. 2014, 165, 186–195. [Google Scholar] [CrossRef]
- Guo, P.; Li, Z.; Huang, P.; Li, B.; Fang, S.; Chu, J.; Guo, H. A Tripartite Amplification Loop Involving the Transcription Factor WRKY75, Salicylic Acid, and Reactive Oxygen Species Accelerates Leaf Senescence. Plant Cell 2017, 29, 2854–2870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.P.; Chen, L.G.; Yu, D.Q. Transcription Factor WRKY75 Interacts with DELLA Proteins to Affect Flowering. Plant Physiol. 2018, 176, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Etchells, J.P.; Provost, C.M.; Turner, S.R. Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet. 2012, 8, e1002997. [Google Scholar] [CrossRef]
- Dubois, M.; Van den Broeck, L.; Inzé, D. The Pivotal Role of Ethylene in Plant Growth. Trends Plant Sci 2018, 23, 311–323. [Google Scholar] [CrossRef]
- Polko, J.K.; van Zanten, M.; van Rooij, J.A.; Marée, A.F.; Voesenek, L.A.; Peeters, A.J.; Pierik, R. Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol. 2012, 193, 339–348. [Google Scholar] [CrossRef]
- Plett, J.M.; Williams, M.; LeClair, G.; Regan, S.; Beardmore, T. Heterologous over-expression of ACC SYNTHASE8 (ACS8) in Populus tremula x P. alba clone 717-1B4 results in elevated levels of ethylene and induces stem dwarfism and reduced leaf size through separate genetic pathways. Front. Plant Sci. 2014, 5, 514. [Google Scholar] [CrossRef][Green Version]
- Deslauriers, S.D.; Larsen, P.B. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Mol. Plant 2010, 3, 626–640. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.C.; Zhang, L.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, Z.; Wu, D.; Yu, F. RALF-FERONIA Signaling: Linking Plant Immune Response with Cell Growth. Plant Commun. 2020, 1, 100084. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ding, Y.; Zhou, Y.; Jin, W.; Xie, K.; Chen, L.L. CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Mol. Plant 2017, 10, 530–532. [Google Scholar] [CrossRef]
- Rao, X.; Lai, D.; Huang, X. A new method for quantitative real-time polymerase chain reaction data analysis. J. Comput. Biol. 2013, 20, 703–711. [Google Scholar] [CrossRef]
- Lu, K.; Li, T.; He, J.; Chang, W.; Zhang, R.; Liu, M.; Yu, M.; Fan, Y.; Ma, J.; Sun, W.; et al. qPrimerDB: A thermodynamics-based gene-specific qPCR primer database for 147 organisms. Nucleic Acids Res. 2018, 46, d1229–d1236. [Google Scholar] [CrossRef]
- Siemens, J.; Nagel, M.; Ludwig-Müller, J.; Sacristán, M. The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: Parameters for disease quantification and screening of mutant lines. J. Phytopathol. 2002, 150, 592–605. [Google Scholar] [CrossRef]
- Knaust, A.; Ludwig-Muller, J. The Ethylene Signaling Pathway is Needed to Restrict Root Gall Growth in Arabidopsis after Infection with the Obligate Biotrophic Protist Plasmodiophora brassicae. J. Plant Growth Regul. 2013, 32, 9–21. [Google Scholar] [CrossRef]
- Sikder, M.M.; Vestergård, M.; Kyndt, T.; Kudjordjie, E.N.; Nicolaisen, M. Phytohormones selectively affect plant parasitic nematodes associated with Arabidopsis roots. New Phytol. 2021, 232, 1272–1285. [Google Scholar] [CrossRef]
- Fu, H.; Yang, Y.; Mishra, V.; Zhou, Q.; Zuzak, K.; Feindel, D.; Harding, M.W.; Feng, J. Most Plasmodiophora brassicae Populations in Single Canola Root Galls from Alberta Fields are Mixtures of Multiple Strains. Plant Dis. 2020, 104, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X.; et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 2009, 21, 2527–2540. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Muller, J.; Julke, S.; Geiss, K.; Richter, F.; Mithofer, A.; Sola, I.; Rusak, G.; Keenan, S.; Bulman, S. A novel methyltransferase from the intracellular pathogen Plasmodiophora brassicae methylates salicylic acid. Mol. Plant Pathol. 2015, 16, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Bi, K.; He, Z.C.; Gao, Z.X.; Zhao, Y.; Fu, Y.P.; Cheng, J.S.; Xie, J.T.; Jiang, D.H. Arabidopsis Mutant bik1 Exhibits Strong Resistance to Plasmodiophora brassicae. Front. Physiol. 2016, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.G.; Xiang, S.Y.; Chen, Y.L.; Li, D.B.; Yu, D.Q. Arabidopsis WRKY45 Interacts with the DELLA Protein RGL1 to Positively Regulate Age-Triggered Leaf Senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef]
- Wang, H.; Xu, Q.; Kong, Y.H.; Chen, Y.; Duan, J.Y.; Wu, W.H.; Chen, Y.F. Arabidopsis WRKY45 Transcription Factor Activates PHOSPHATE TRANSPORTER1; 1 Expression in Response to Phosphate Starvation1 W OPEN. Plant Physiol. 2014, 164, 2020–2029. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, L.; Wu, S.; Chen, Y.; Yu, D.; Chen, L. AtWRKY75 positively regulates age-triggered leaf senescence through gibberellin pathway. Plant Divers. 2021, 43, 331–340. [Google Scholar] [CrossRef]
- Sajeevan, R.S.; Nataraja, K.N.; Shivashankara, K.S.; Pallavi, N.; Gurumurthy, D.S.; Shivanna, M.B. Expression of Arabidopsis SHN1 in Indian Mulberry (Morus indica L.) Increases Leaf Surface Wax Content and Reduces Post-harvest Water Loss. Front. Plant Sci. 2017, 8, 418. [Google Scholar] [CrossRef]
- Sela, D.; Buxdorf, K.; Shi, J.X.; Feldmesser, E.; Schreiber, L.; Aharoni, A.; Levy, M. Overexpression of AtSHN1/WIN1 provokes unique defense responses. PLoS ONE 2013, 8, e70146. [Google Scholar] [CrossRef]
- Blake, S.N.; Barry, K.M.; Gill, W.M.; Reid, J.B.; Foo, E. The role of strigolactones and ethylene in disease caused by Pythium irregulare. Mol. Plant Pathol 2016, 17, 680–690. [Google Scholar] [CrossRef]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Herger, A.; Dünser, K.; Kleine-Vehn, J.; Ringli, C. Leucine-Rich Repeat Extensin Proteins and Their Role in Cell Wall Sensing. Curr. Biol. 2019, 29, R851–R858. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Nolan, T.M.; Song, G.; Liu, S.; Xie, Z.; Chen, J.; Schnable, P.S.; Walley, J.W.; Yin, Y. FERONIA Receptor Kinase Contributes to Plant Immunity by Suppressing Jasmonic Acid Signaling in Arabidopsis thaliana. Curr. Biol. 2018, 28, 3316–3324. [Google Scholar] [CrossRef]
- Bhering, A.D.; do Carmo, M.G.; Matos, T.S.; Lima, E.S.; do Amaral Sobrinho, N.M. Soil Factors Related to the Severity of Clubroot in Rio de Janeiro, Brazil. Plant Dis. 2017, 101, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).