Identification of Antitumor miR-30e-5p Controlled Genes; Diagnostic and Prognostic Biomarkers for Head and Neck Squamous Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Analysis of miRNAs and miRNA Target Genes in HNSCC Patients
2.2. HNSCC Cell Lines
2.3. RNA Extraction and Quantitative Real-Time Reverse-Transcription PCR (qRT-PCR)
2.4. Transfection of miRNAs and siRNAs into HNSCC Cells
2.5. RIP Assay
2.6. Functional Assays of HNSCC Cells (Cell Proliferation, Migration, and Invasion Assays)
2.7. Plasmid Construction and Dual-Luciferase Reporter Assays
2.8. Immunohistochemistry
2.9. Gene Set Enrichment Analysis (GSEA)
2.10. Statistical Analysis
3. Results
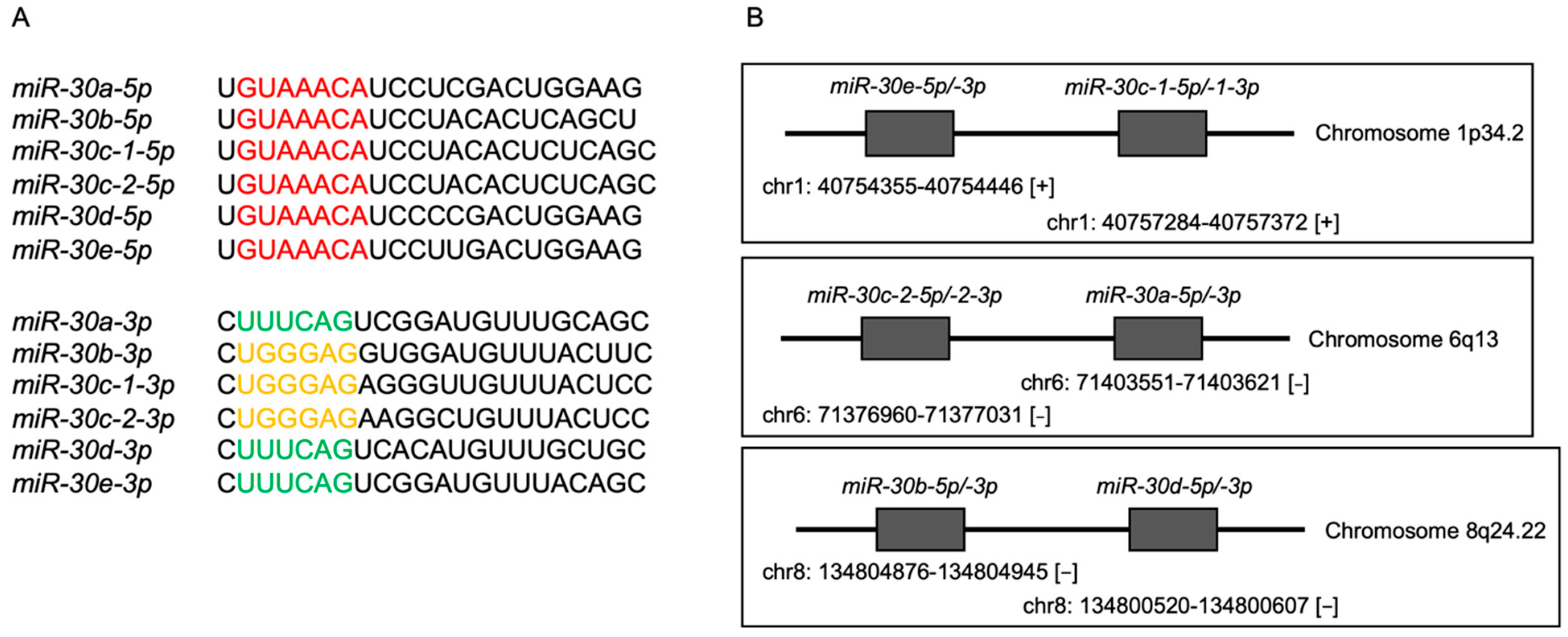
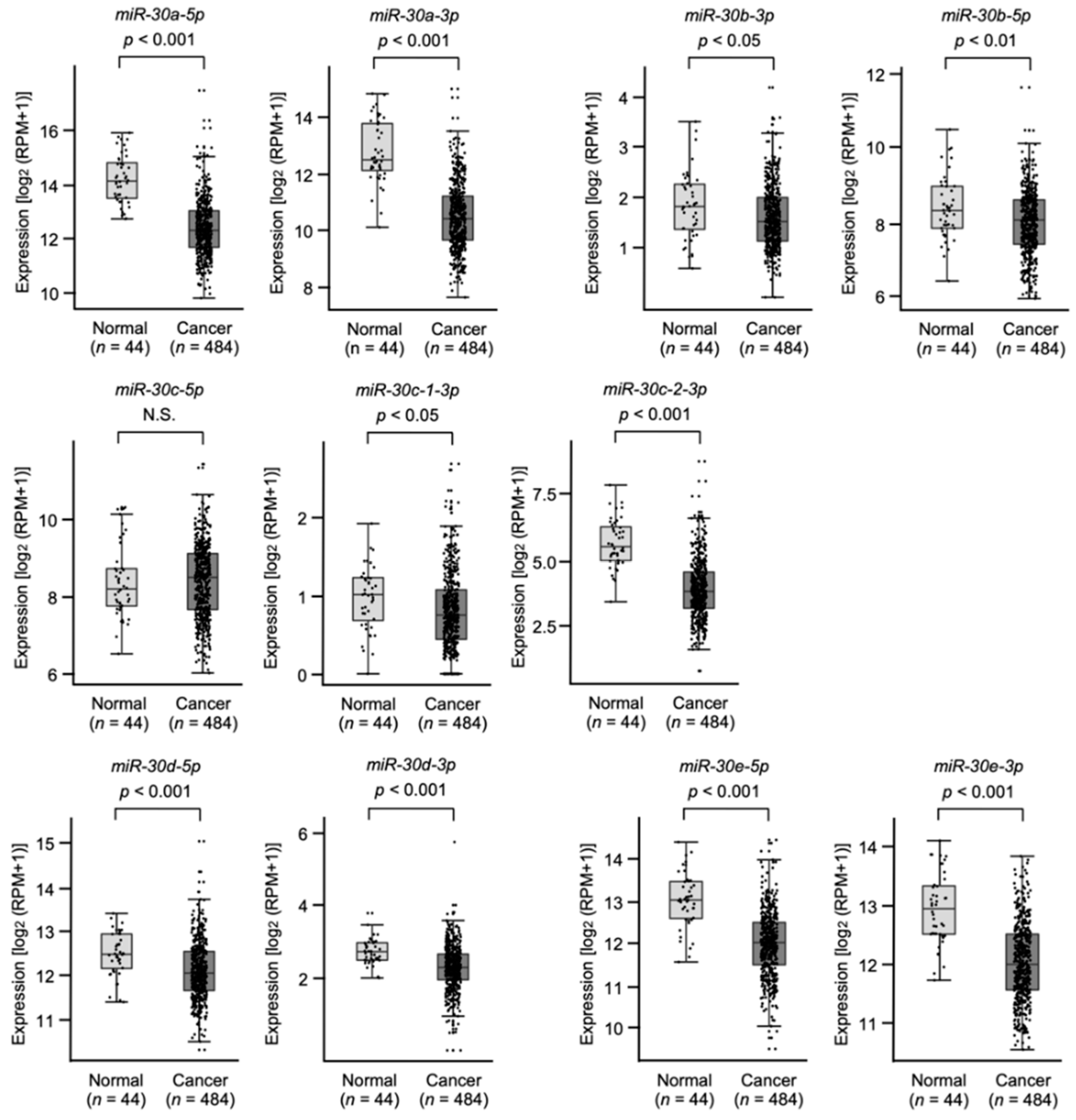
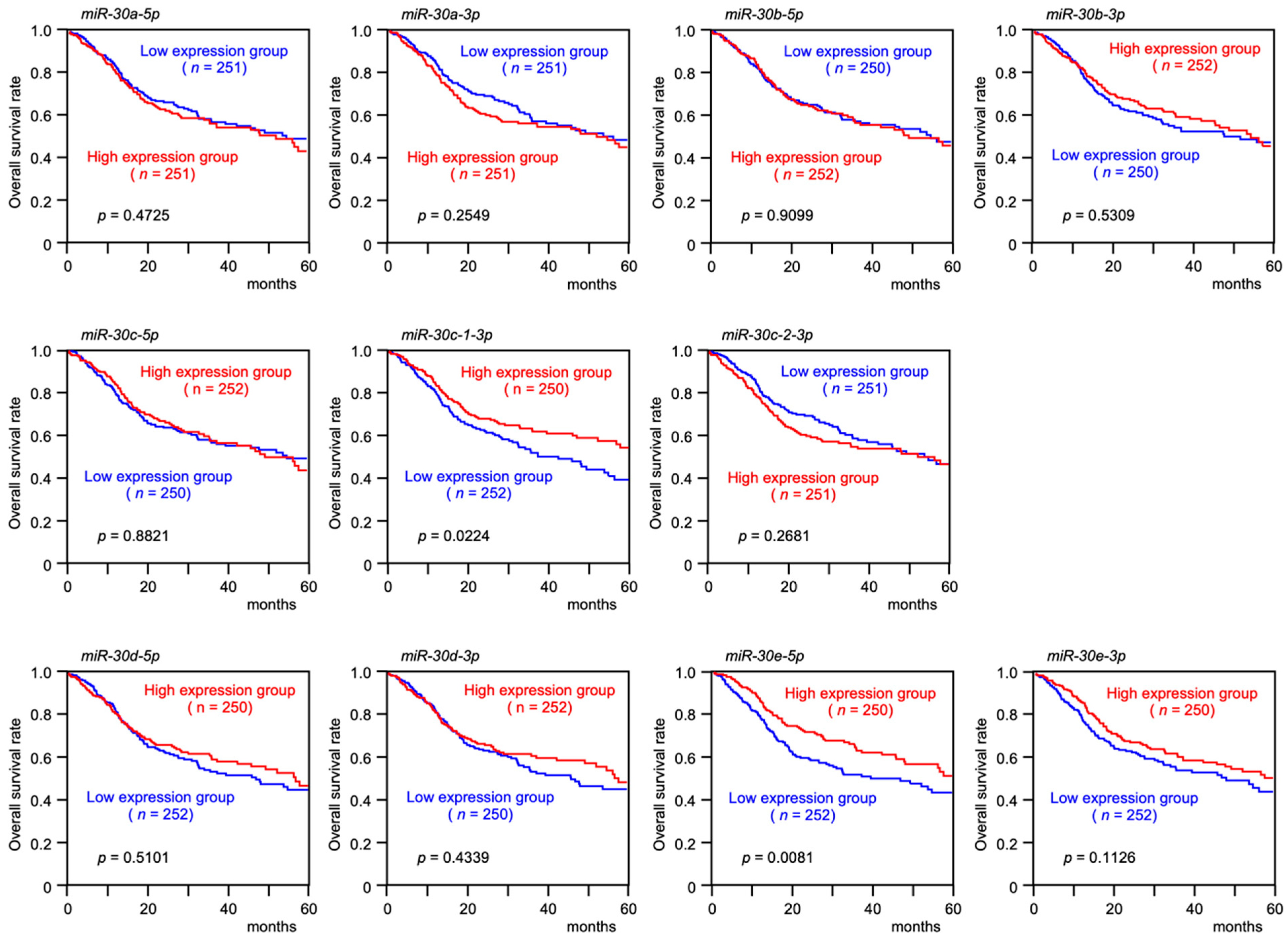
3.1. Expression Levels and the Clinical Significance of the miR-30 Family in HNSCC Clinical Specimens Assessed by TCGA Analysis
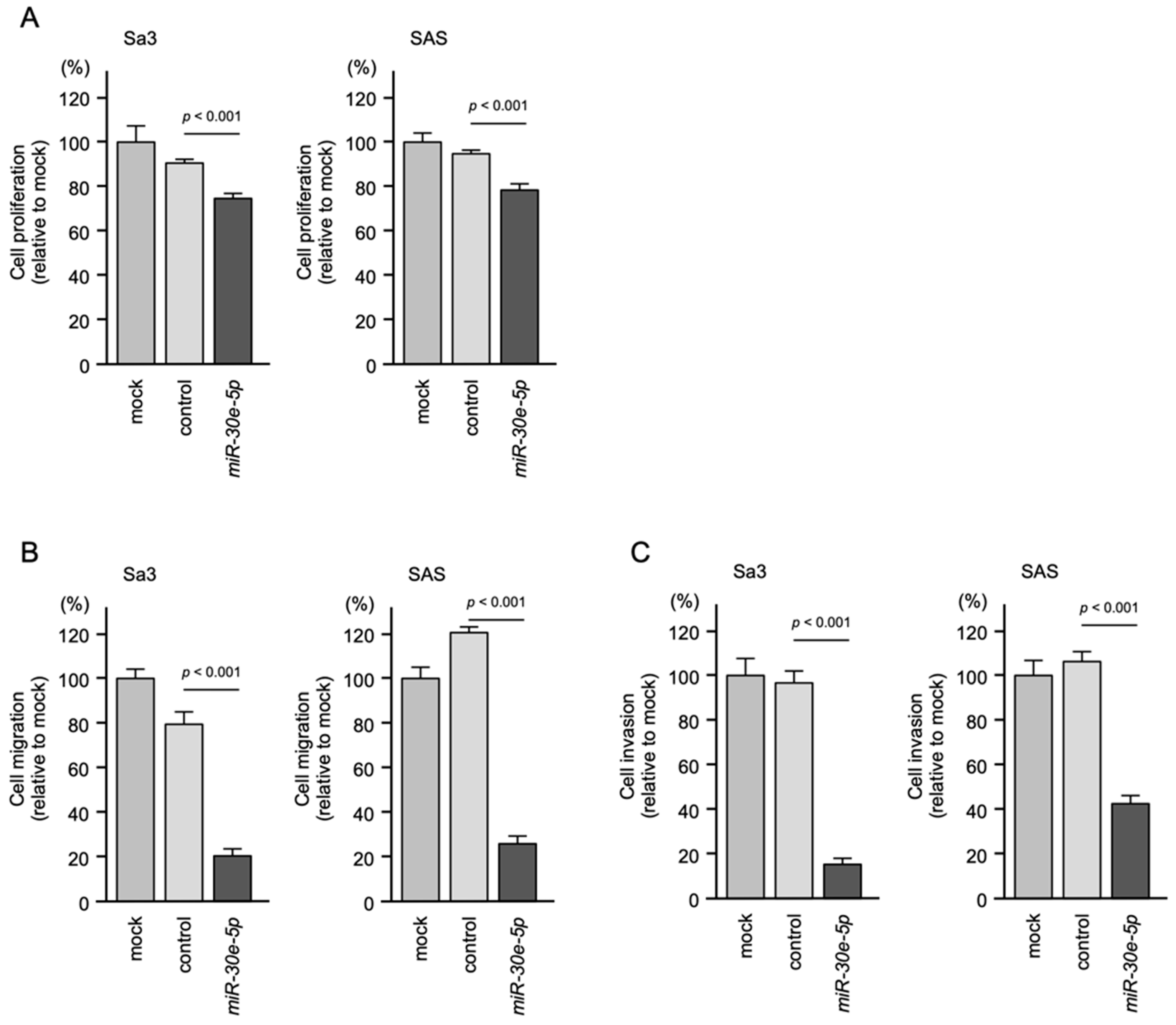
3.2. Effect of Transient Transfection of miR-30e-5p on HNSCC Cell Proliferation, Migration and Invasion
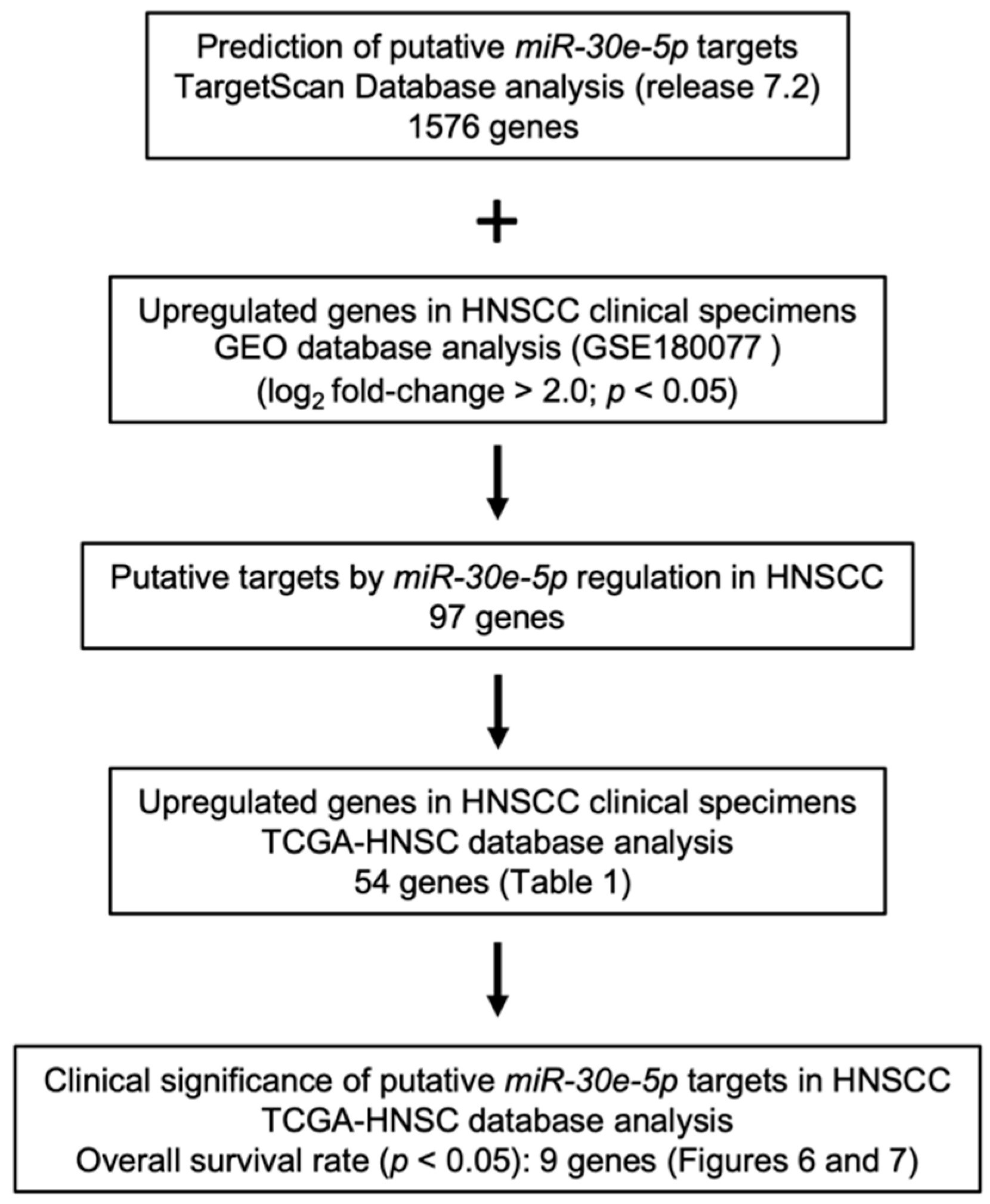
3.3. Screening for Oncogenic Targets of miR-30e-5p in HNSCC
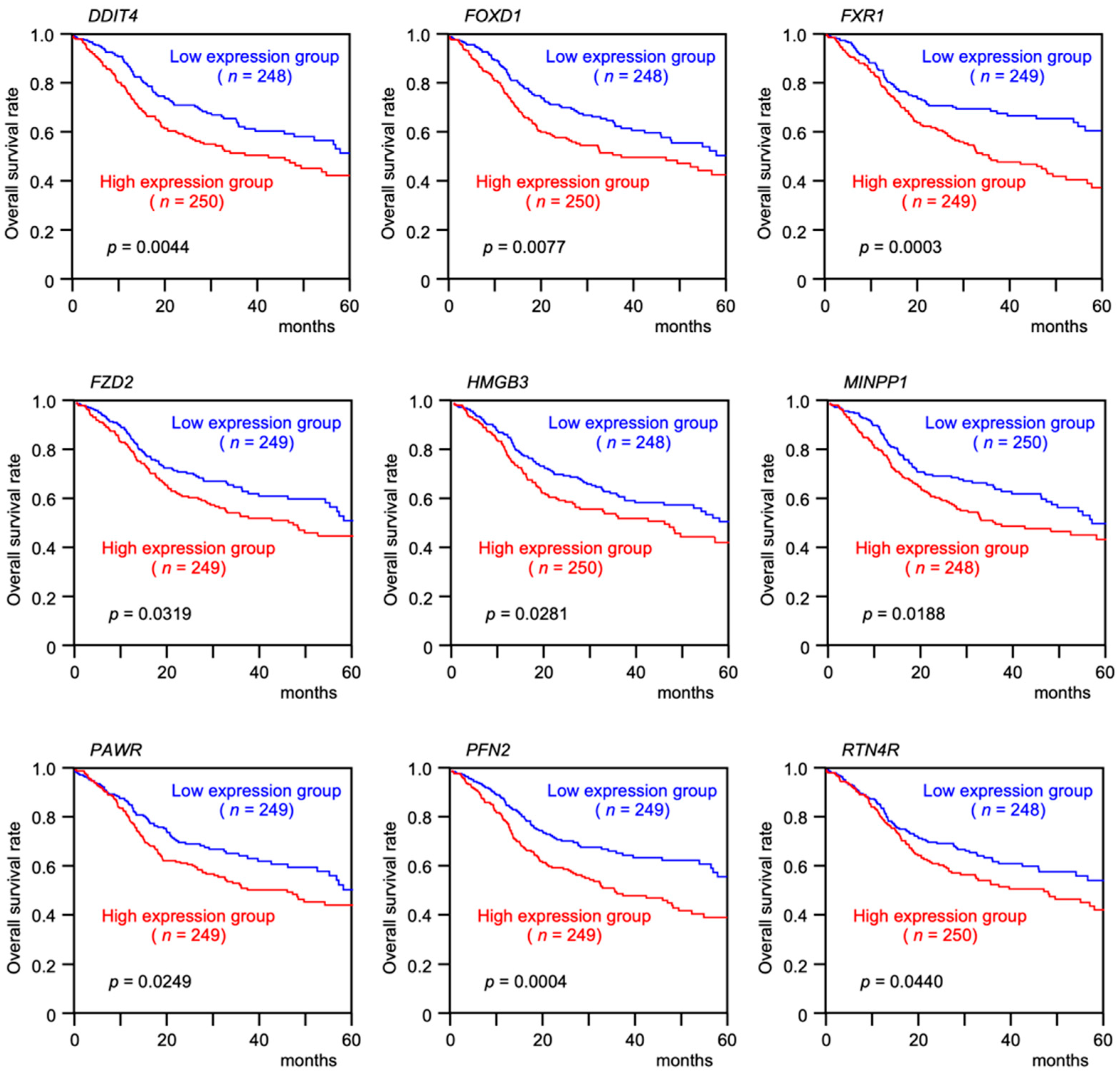
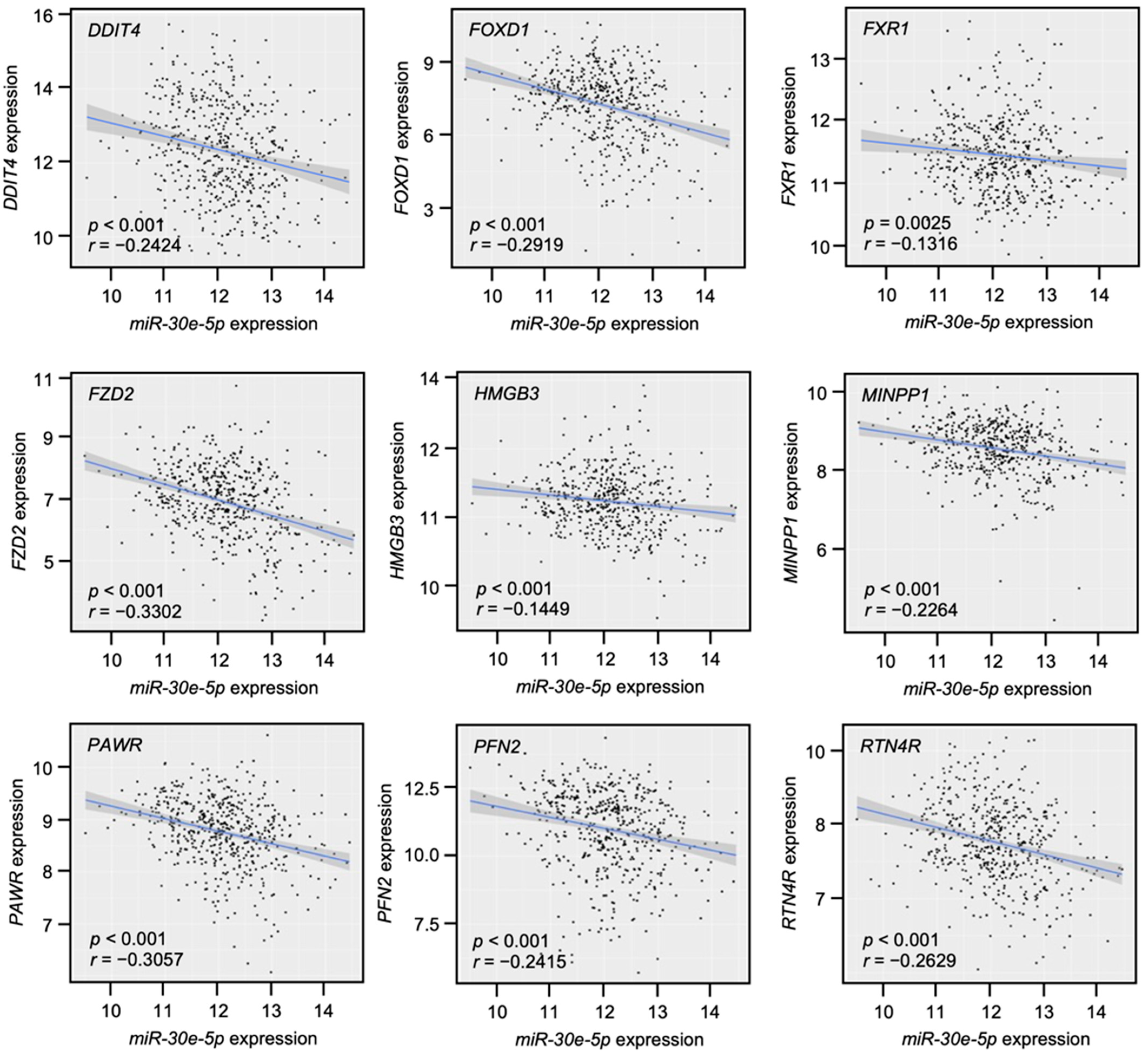
3.4. Clinical Significance of miR-30e-5p Targets in Patients with HNSCC Determined by TCGA Analysis
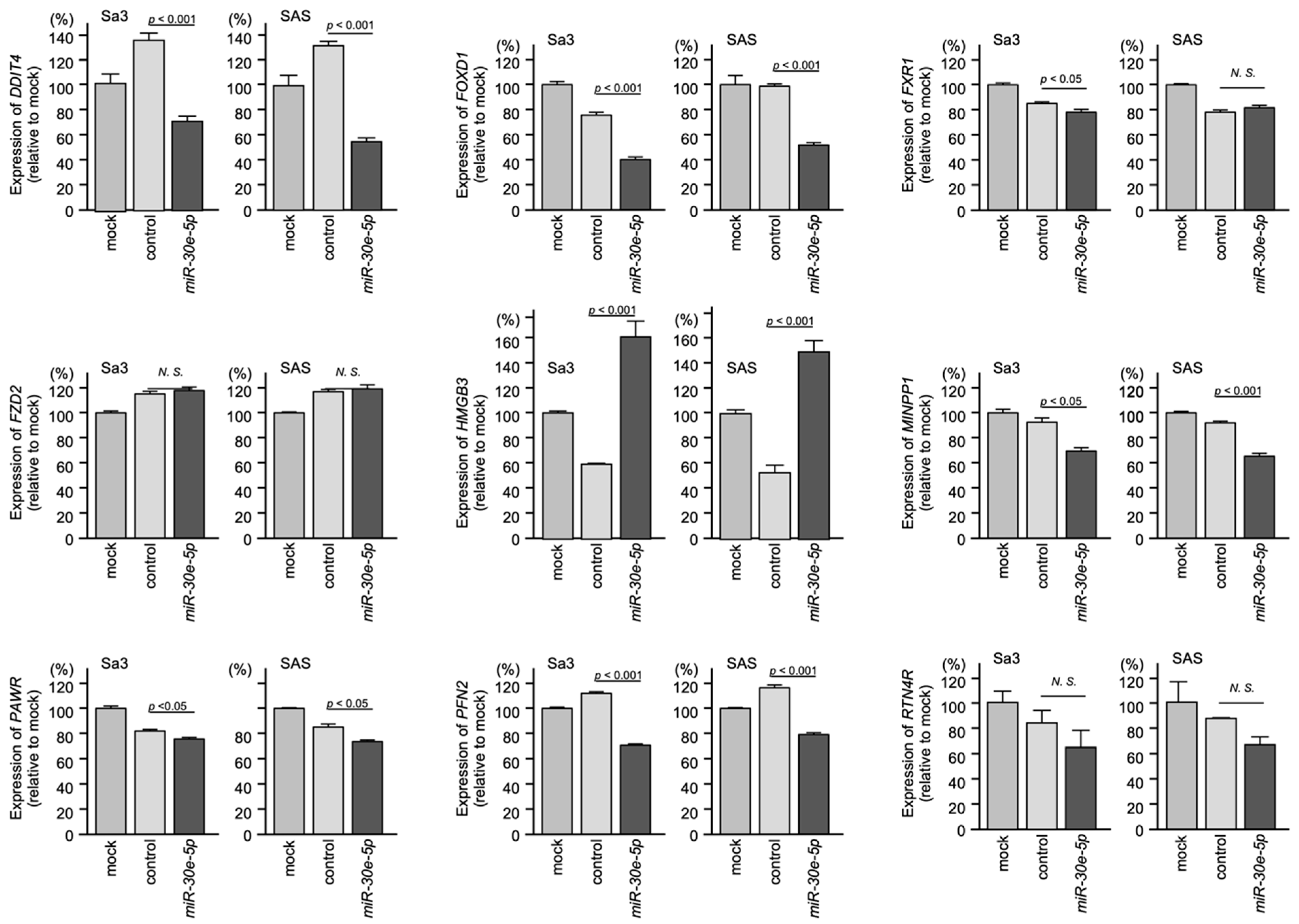
3.5. Regulated Expression of the Nine Identified Genes by miR-30e-5p in HNSCC Cells
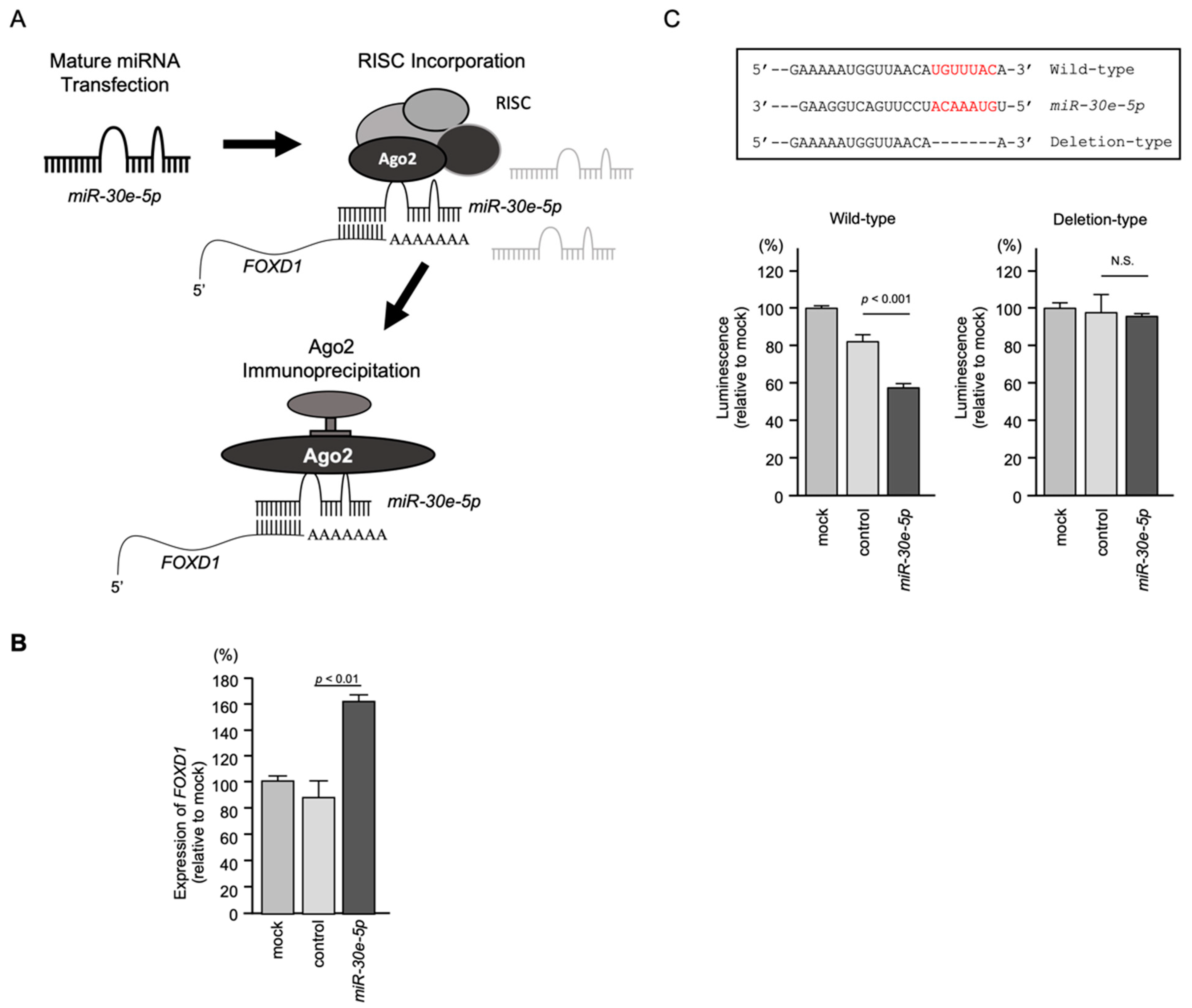
3.6. Incorporation of FOXD1 mRNA into the RNA-Induced Silencing Complex (RISC) and Direct Control of FOXD1 Expression by miR-30e-5p in HNSCC Cells
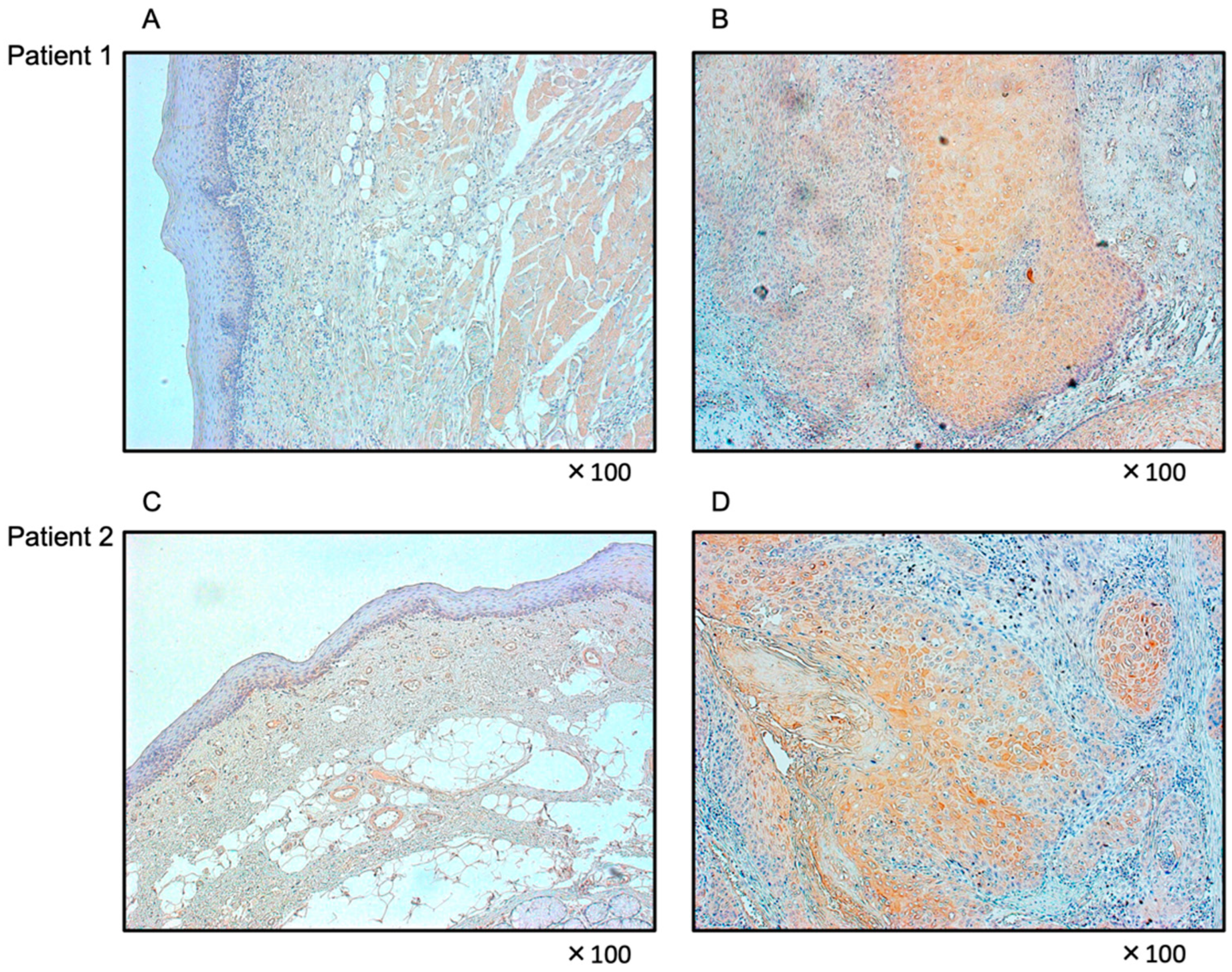
3.7. Expression of FOXD1 in HNSCC Clinical Specimens
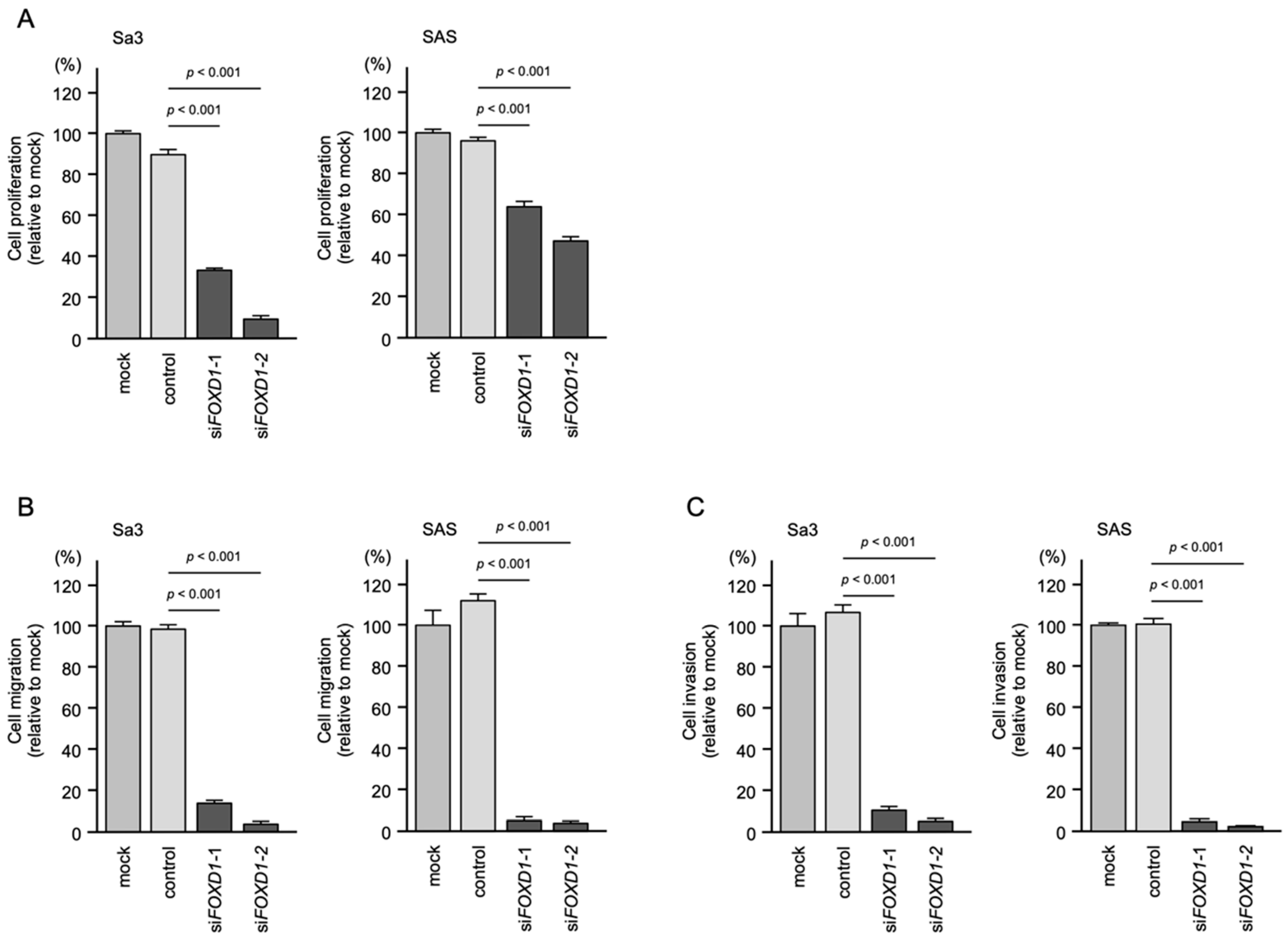
3.8. Effects of FOXD1 Knockdown on the Proliferation, Migration, and Invasion of HNSCC Cells
3.9. FOXD1-Mediated Molecular Pathways in HNSCC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.E.W.; Bell, R.B.; Bifulco, C.B.; Burtness, B.; Gillison, M.L.; Harrington, K.J.; Le, Q.T.; Lee, N.Y.; Leidner, R.; Lewis, R.L.; et al. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC). J. Immunother. Cancer 2019, 7, 184. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef]
- Bonner, J.A.; Harari, P.M.; Giralt, J.; Cohen, R.B.; Jones, C.U.; Sur, R.K.; Raben, D.; Baselga, J.; Spencer, S.A.; Zhu, J.; et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010, 11, 21–28. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Krek, A.; Grun, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef]
- Mitra, R.; Adams, C.M.; Jiang, W.; Greenawalt, E.; Eischen, C.M. Pan-cancer analysis reveals cooperativity of both strands of microRNA that regulate tumorigenesis and patient survival. Nat. Commun. 2020, 11, 968. [Google Scholar] [CrossRef]
- Koshizuka, K.; Nohata, N.; Hanazawa, T.; Kikkawa, N.; Arai, T.; Okato, A.; Fukumoto, I.; Katada, K.; Okamoto, Y.; Seki, N. Deep sequencing-based microRNA expression signatures in head and neck squamous cell carcinoma: Dual strands of pre-miR-150 as antitumor miRNAs. Oncotarget 2017, 8, 30288–30304. [Google Scholar] [CrossRef]
- Yonemori, K.; Seki, N.; Idichi, T.; Kurahara, H.; Osako, Y.; Koshizuka, K.; Arai, T.; Okato, A.; Kita, Y.; Arigami, T.; et al. The microRNA expression signature of pancreatic ductal adenocarcinoma by RNA sequencing: Anti-tumour functions of the microRNA-216 cluster. Oncotarget 2017, 8, 70097–70115. [Google Scholar] [CrossRef]
- Yang, S.J.; Yang, S.Y.; Wang, D.D.; Chen, X.; Shen, H.Y.; Zhang, X.H.; Zhong, S.L.; Tang, J.H.; Zhao, J.H. The miR-30 family: Versatile players in breast cancer. Tumor Biol. 2017, 39, 1010428317692204. [Google Scholar] [CrossRef]
- Saleh, A.D.; Cheng, H.; Martin, S.E.; Si, H.; Ormanoglu, P.; Carlson, S.; Clavijo, P.E.; Yang, X.; Das, R.; Cornelius, S.; et al. Integrated Genomic and Functional microRNA Analysis Identifies miR-30-5p as a Tumor Suppressor and Potential Therapeutic Nanomedicine in Head and Neck Cancer. Clin. Cancer Res. 2019, 25, 2860–2873. [Google Scholar] [CrossRef]
- Mao, L.; Liu, S.; Hu, L.; Jia, L.; Wang, H.; Guo, M.; Chen, C.; Liu, Y.; Xu, L. miR-30 Family: A Promising Regulator in Development and Disease. BioMed Res. Int. 2018, 2018, 9623412. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Anaya, J. Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Koshizuka, K.; Hanazawa, T.; Kikkawa, N.; Arai, T.; Okato, A.; Kurozumi, A.; Kato, M.; Katada, K.; Okamoto, Y.; Seki, N. Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 2017, 108, 1681–1692. [Google Scholar] [CrossRef]
- Koma, A.; Asai, S.; Minemura, C.; Oshima, S.; Kinoshita, T.; Kikkawa, N.; Koshizuka, K.; Moriya, S.; Kasamatsu, A.; Hanazawa, T.; et al. Impact of Oncogenic Targets by Tumor-Suppressive miR-139-5p and miR-139-3p Regulation in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 9947. [Google Scholar] [CrossRef]
- Asai, S.; Koma, A.; Nohata, N.; Kinoshita, T.; Kikkawa, N.; Kato, M.; Minemura, C.; Uzawa, K.; Hanazawa, T.; Seki, N. Impact of miR-1/miR-133 Clustered miRNAs: PFN2 Facilitates Malignant Phenotypes in Head and Neck Squamous Cell Carcinoma. Biomedicines 2022, 10, 663. [Google Scholar] [CrossRef]
- Minemura, C.; Asai, S.; Koma, A.; Kase-Kato, I.; Tanaka, N.; Kikkawa, N.; Kasamatsu, A.; Yokoe, H.; Hanazawa, T.; Uzawa, K.; et al. Identification of Tumor-Suppressive miR-30e-3p Targets: Involvement of SERPINE1 in the Molecular Pathogenesis of Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 3808. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Yamada, Y.; Sugawara, S.; Arai, T.; Kojima, S.; Kato, M.; Okato, A.; Yamazaki, K.; Naya, Y.; Ichikawa, T.; Seki, N. Molecular pathogenesis of renal cell carcinoma: Impact of the anti-tumor miR-29 family on gene regulation. Int. J. Urol. 2018, 25, 953–965. [Google Scholar] [CrossRef]
- Kanthaje, S.; Baikunje, N.; Kandal, I.; Ratnacaram, C.K. Repertoires of MicroRNA-30 family as gate-keepers in lung cancer. Front. Biosci. 2021, 13, 141–156. [Google Scholar] [CrossRef]
- Xu, G.; Cai, J.; Wang, L.; Jiang, L.; Huang, J.; Hu, R.; Ding, F. MicroRNA-30e-5p suppresses non-small cell lung cancer tumorigenesis by regulating USP22-mediated Sirt1/JAK/STAT3 signaling. Exp. Cell Res. 2018, 362, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.X.; Zhang, H.; Li, X.H.; Liu, Y.H. MiR-30e-5p inhibits proliferation and metastasis of nasopharyngeal carcinoma cells by target-ing USP22. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6342–6349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qin, H.; Jiang, B.; Chen, W.; Cao, W.; Zhao, X.; Yuan, H.; Qi, W.; Zhuo, D.; Guo, H. miR-30e-5p suppresses cell proliferation and migration in bladder cancer through regulating metadherin. J. Cell. Biochem. 2019, 120, 15924–15932. [Google Scholar] [CrossRef]
- Zhang, S.; Li, G.; Liu, C.; Lu, S.; Jing, Q.; Chen, X.; Zheng, H.; Ma, H.; Zhang, D.; Ren, S.; et al. miR-30e-5p represses angiogenesis and metastasis by directly targeting AEG-1 in squamous cell carcinoma of the head and neck. Cancer Sci. 2020, 111, 356–368. [Google Scholar] [CrossRef]
- Laudato, S.; Patil, N.; Abba, M.L.; Leupold, J.H.; Benner, A.; Gaiser, T.; Marx, A.; Allgayer, H. P53-induced miR-30e-5p inhibits colorectal cancer invasion and metastasis by targeting ITGA6 and ITGB1. Int. J. Cancer 2017, 141, 1879–1890. [Google Scholar] [CrossRef]
- Majumder, M.; Johnson, R.H.; Palanisamy, V. Fragile X-related protein family: A double-edged sword in neurodevelopmental disorders and cancer. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 409–424. [Google Scholar] [CrossRef]
- Qian, J.; Hassanein, M.; Hoeksema, M.D.; Harris, B.K.; Zou, Y.; Chen, H.; Lu, P.; Eisenberg, R.; Wang, J.; Espinosa, A.; et al. The RNA binding protein FXR1 is a new driver in the 3q26-29 amplicon and predicts poor prognosis in human cancers. Proc. Natl. Acad. Sci. USA 2015, 112, 3469–3474. [Google Scholar] [CrossRef]
- Majumder, M.; Palanisamy, V. RNA binding protein FXR1-miR301a-3p axis contributes to p21WAF1 degradation in oral cancer. PLoS Genet. 2020, 16, e1008580. [Google Scholar] [CrossRef]
- Qie, S.; Majumder, M.; Mackiewicz, K.; Howley, B.V.; Peterson, Y.K.; Howe, P.H.; Palanisamy, V.; Diehl, J.A. Fbxo4-mediated degradation of Fxr1 suppresses tumorigenesis in head and neck squamous cell carcinoma. Nat. Commun. 2017, 8, 1534. [Google Scholar] [CrossRef]
- Pinto-Costa, R.; Sousa, M.M. Profilin as a dual regulator of actin and microtubule dynamics. Cytoskeleton 2020, 77, 76–83. [Google Scholar] [CrossRef]
- Zhou, K.; Chen, J.; Wu, J.; Xu, Y.; Wu, Q.; Yue, J.; Song, Y.; Li, S.; Zhou, P.; Tu, W.; et al. Profilin 2 Promotes Proliferation and Metastasis of Head and Neck Cancer Cells by Regulating PI3K/AKT/β-Catenin Signaling Pathway. Oncol. Res. 2019, 27, 1079–1088. [Google Scholar] [CrossRef]
- Cao, Q.; Liu, Y.; Wu, Y.; Hu, C.; Sun, L.; Wang, J.; Li, C.; Guo, M.; Liu, X.; Lv, J.; et al. Profilin 2 promotes growth, metastasis, and angiogenesis of small cell lung cancer through cancer-derived exosomes. Aging 2020, 12, 25981–25999. [Google Scholar] [CrossRef]
- Yan, J.; Ma, C.; Gao, Y. MicroRNA-30a-5p suppresses epithelial-mesenchymal transition by targeting profilin-2 in high invasive non-small cell lung cancer cell lines. Oncol. Rep. 2017, 37, 3146–3154. [Google Scholar] [CrossRef][Green Version]
- Katoh, M.; Katoh, M. Human FOX gene family (Review). Int. J. Oncol. 2004, 25, 1495–1500. [Google Scholar] [CrossRef]
- Quintero-Ronderos, P.; Laissue, P. The multisystemic functions of FOXD1 in development and disease. J. Mol. Med. 2018, 96, 725–739. [Google Scholar] [CrossRef]
- Lin, C.H.; Lee, H.H.; Chang, W.M.; Lee, F.P.; Chen, L.C.; Lu, L.S.; Lin, Y.F. FOXD1 Repression Potentiates Radiation Effectiveness by Downregulating G3BP2 Expression and Promoting the Activation of TXNIP-Related Pathways in Oral Cancer. Cancers 2020, 12, 2690. [Google Scholar] [CrossRef]
- Sun, Q.; Novak, D.; Hüser, L.; Poelchen, J.; Wu, H.; Granados, K.; Federico, A.; Liu, K.; Steinfass, T.; Vierthaler, M.; et al. FOXD1 promotes dedifferentiation and targeted therapy resistance in melanoma by regulating the expression of connective tissue growth factor. Int. J. Cancer 2021, 149, 657–674. [Google Scholar] [CrossRef]
- Wu, Q.; Ma, J.; Wei, J.; Meng, W.; Wang, Y.; Shi, M. FOXD1-AS1 regulates FOXD1 translation and promotes gastric cancer progression and chemoresistance by activating the PI3K/AKT/mTOR pathway. Mol. Oncol. 2021, 15, 299–316. [Google Scholar] [CrossRef]
- Chen, C.; Tang, J.; Xu, S.; Zhang, W.; Jiang, H. miR-30a-5p Inhibits Proliferation and Migration of Lung Squamous Cell Carcinoma Cells by Targeting FOXD1. BioMed Res. Int. 2020, 2020, 2547902. [Google Scholar] [CrossRef]
- Zhou, L.; Jia, S.; Ding, G.; Zhang, M.; Yu, W.; Wu, Z.; Cao, L. Down-regulation of miR-30a-5p is Associated with Poor Prognosis and Promotes Chemoresistance of Gemcitabine in Pancreatic Ductal Adenocarcinoma. J. Cancer 2019, 10, 5031–5040. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Cong, H.; Wang, H.; Zhang, D.; Liu, C.; Chu, H.; Qing, Q.; Wang, K. MiR-30a-5p inhibits osteosarcoma cell proliferation and migration by targeting FOXD1. Biochem. Biophys. Res. Commun. 2018, 503, 1092–1097. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qiu, C.; Lu, N.; Liu, Z.; Jin, C.; Sun, C.; Bu, H.; Yu, H.; Dongol, S.; Kong, B. FOXD1 is targeted by miR-30a-5p and miR-200a-5p and suppresses the proliferation of human ovarian carcinoma cells by promoting p21 expression in a p53-independent manner. Int. J. Oncol. 2018, 52, 2130–2142. [Google Scholar] [CrossRef] [PubMed]
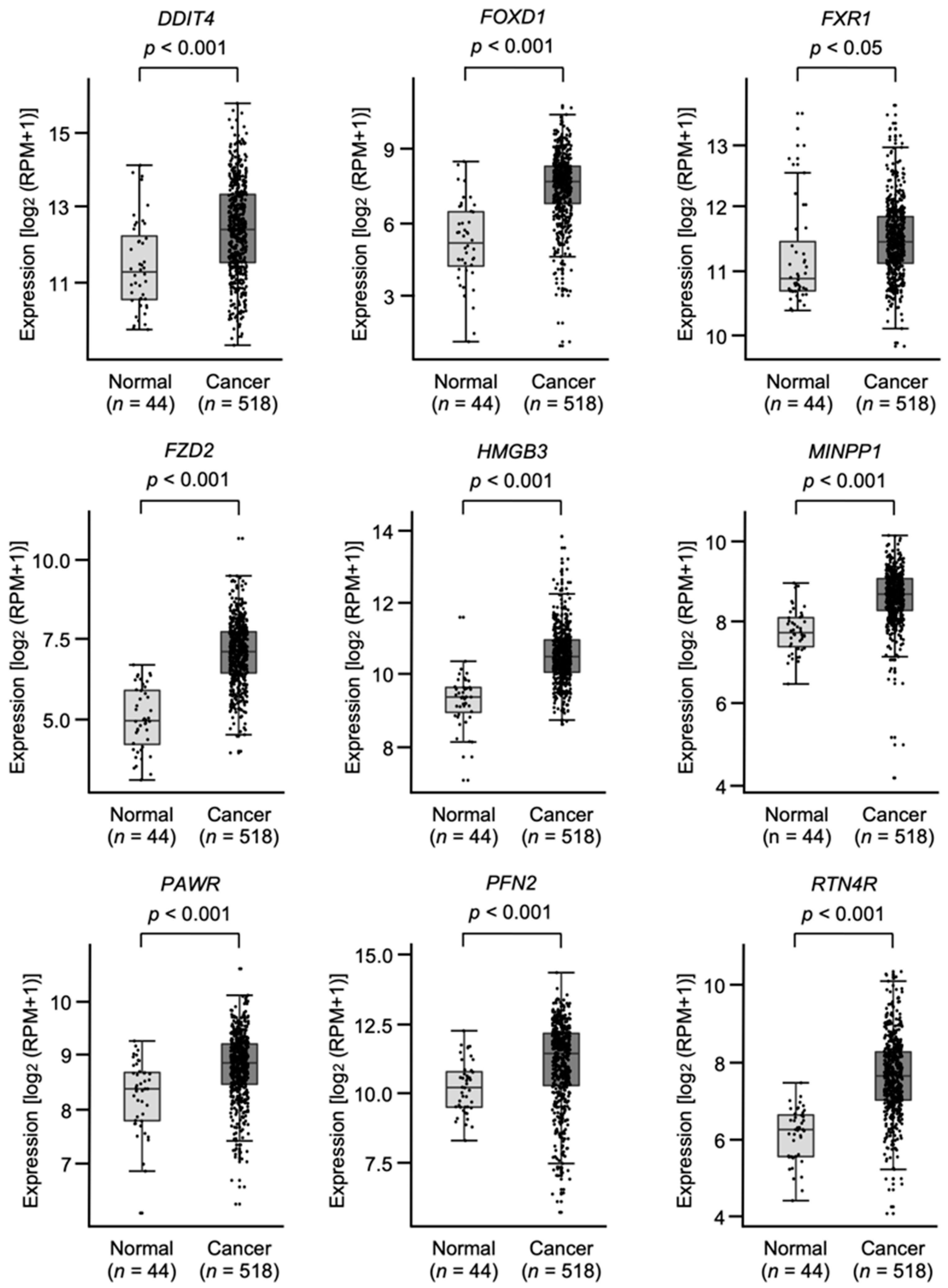
| Entrez Gene ID | Gene Symbol | Gene Name | Total Binding Sites | GEO 1 p Value | GEO log2(FC 2) | 5y OS 3 p Value |
|---|---|---|---|---|---|---|
| 8087 | FXR1 | Fragile X mental retardation, autosomal homolog 1 | 1 | 0.014 | 3.06 | <0.001 |
| 5217 | PFN2 | Profilin 2 | 1 | 0.004 | 2.67 | <0.001 |
| 54541 | DDIT4 | DNA-damage-inducible transcript 4 | 1 | 0.017 | 2.70 | 0.004 |
| 2297 | FOXD1 | Forkhead box D1 | 1 | 0.008 | 3.79 | 0.008 |
| 9562 | MINPP1 | Multiple inositol-polyphosphate phosphatase 1 | 1 | 0.013 | 2.02 | 0.019 |
| 5074 | PAWR | PRKC, apoptosis, WT1, regulator | 2 | 0.003 | 2.04 | 0.025 |
| 3149 | HMGB3 | High mobility group box 3 | 1 | 0.004 | 2.17 | 0.028 |
| 2535 | FZD2 | Frizzled class receptor 2 | 1 | 0.006 | 2.36 | 0.032 |
| 65078 | RTN4R | Reticulon 4 receptor | 1 | 0.008 | 2.20 | 0.044 |
| 115908 | CTHRC1 | Collagen triple helix repeat containing 1 | 1 | 0.010 | 2.96 | 0.059 |
| 3218 | HOXB8 | Homeobox B8 | 1 | 0.046 | 3.29 | 0.077 |
| 9143 | SYNGR3 | Synaptogyrin 3 | 1 | 0.042 | 2.12 | 0.077 |
| 1012 | CDH13 | Cadherin 13 | 1 | 0.013 | 2.13 | 0.091 |
| 6683 | SPAST | Spastin | 2 | 0.012 | 2.10 | 0.092 |
| 79718 | TBL1XR1 | Transducin (β)-like 1 X-linked receptor 1 | 2 | 0.002 | 2.63 | 0.104 |
| 114088 | TRIM9 | Tripartite motif containing 9 | 1 | 0.005 | 4.27 | 0.113 |
| 84733 | CBX2 | Chromobox homolog 2 | 1 | 0.005 | 3.00 | 0.133 |
| 27 | ABL2 | ABL proto-oncogene 2, non-receptor tyrosine kinase | 1 | 0.008 | 2.19 | 0.155 |
| 23657 | SLC7A11 | solute carrier family 7 (Anionic amino acid transporter light chain, xc- system), member 11 | 1 | 0.007 | 4.00 | 0.160 |
| 154214 | RNF217 | Ring finger protein 217 | 1 | 0.016 | 2.36 | 0.186 |
| 79712 | GTDC1 | Glycosyltransferase-like domain containing 1 | 1 | 0.004 | 4.31 | 0.193 |
| 26059 | ERC2 | ELKS/RAB6-interacting/CAST family member 2 | 1 | 0.027 | 3.54 | 0.210 |
| 3237 | HOXD11 | Homeobox D11 | 1 | 0.033 | 4.24 | 0.214 |
| 89796 | NAV1 | Neuron navigator 1 | 1 | 0.007 | 2.88 | 0.234 |
| 6659 | SOX4 | SRY (sex determining region Y)-box 4 | 1 | 0.005 | 2.30 | 0.258 |
| 54434 | SSH1 | Slingshot protein phosphatase 1 | 1 | 0.016 | 2.05 | 0.280 |
| 2048 | EPHB2 | EPH receptor B2 | 1 | 0.013 | 2.53 | 0.303 |
| 9258 | MFHAS1 | Malignant fibrous histiocytoma amplified sequence 1 | 1 | 0.005 | 2.27 | 0.311 |
| 54566 | EPB41L4B | Erythrocyte membrane protein band 4.1 like 4B | 1 | 0.004 | 3.06 | 0.413 |
| 8448 | DOC2A | Double C2-like domains, α | 2 | 0.021 | 3.00 | 0.485 |
| 28982 | FLVCR1 | Feline leukemia virus subgroup C cellular receptor 1 | 1 | 0.005 | 2.43 | 0.492 |
| 55785 | FGD6 | FYVE, RhoGEF and PH domain containing 6 | 1 | 0.014 | 2.21 | 0.530 |
| 490 | ATP2B1 | ATPase, Ca++ transporting, plasma membrane 1 | 1 | 0.007 | 2.93 | 0.556 |
| 4644 | MYO5A | Myosin VA (heavy chain 12, myoxin) | 1 | 0.003 | 2.06 | 0.605 |
| 4015 | LOX | Lysyl oxidase | 1 | 0.006 | 4.46 | 0.613 |
| 50805 | IRX4 | Iroquois homeobox 4 | 1 | 0.039 | 2.67 | 0.652 |
| 23432 | GPR161 | G protein-coupled receptor 161 | 1 | 0.008 | 2.50 | 0.662 |
| 2729 | GCLC | Glutamate-cysteine ligase, catalytic subunit | 1 | 0.004 | 3.14 | 0.710 |
| 8038 | ADAM12 | ADAM metallopeptidase domain 12 | 2 | 0.003 | 4.29 | 0.721 |
| 3631 | INPP4A | Inositol polyphosphate-4-phosphatase, type I, 107kDa | 1 | 0.003 | 2.12 | 0.747 |
| 9832 | JAKMIP2 | Janus kinase and microtubule interacting protein 2 | 1 | 0.010 | 4.19 | 0.769 |
| 121268 | RHEBL1 | Ras homolog enriched in brain like 1 | 1 | 0.004 | 2.92 | 0.775 |
| 144455 | E2F7 | E2F transcription factor 7 | 3 | 0.021 | 2.47 | 0.786 |
| 94032 | CAMK2N2 | Calcium/calmodulin-dependent protein kinase II inhibitor 2 | 1 | 0.003 | 2.95 | 0.823 |
| 84206 | MEX3B | Mex-3 RNA binding family member B | 2 | 0.016 | 2.75 | 0.880 |
| 2887 | GRB10 | Growth factor receptor-bound protein 10 | 1 | 0.008 | 2.07 | 0.890 |
| 23333 | DPY19L1 | Dpy-19-like 1 (C. elegans) | 1 | 0.013 | 3.07 | 0.893 |
| 9435 | CHST2 | Carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 | 1 | 0.006 | 3.30 | 0.917 |
| 54165 | DCUN1D1 | DCN1, defective in cullin neddylation 1, domain containing 1 | 2 | 0.009 | 2.50 | 0.938 |
| 54714 | CNGB3 | Cyclic nucleotide gated channel β 3 | 1 | 0.008 | 2.98 | 0.968 |
| 221002 | RASGEF1A | RasGEF domain family, member 1A | 1 | 0.005 | 2.81 | 0.968 |
| 55144 | LRRC8D | Leucine rich repeat containing 8 family, member D | 1 | 0.002 | 2.60 | 0.978 |
| 8534 | CHST1 | Carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 1 | 0.005 | 4.41 | 0.980 |
| 8632 | DNAH17 | Dynein, axonemal, heavy chain 17 | 1 | 0.006 | 5.26 | 0.999 |
| Monovariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | p-Value | HR | 95% CI | p-Value |
| DDIT4 (High vs. Low expression) | 1.506 | 1.138–1.994 | 0.004 | 1.292 | 0.939–1.776 | 0.115 |
| FOXD1 (High vs. Low expression) | 1.526 | 1.153–2.019 | 0.003 | 1.374 | 1.002–1.890 | 0.049 |
| FXR1 (High vs. Low expression) | 1.651 | 1.242–2.193 | 0.001 | 1.303 | 0.930–1.825 | 0.124 |
| FZD2 (High vs. Low expression) | 1.337 | 1.011–1.768 | 0.042 | 1.069 | 0.776–1.473 | 0.684 |
| HMGB3 (High vs. Low expression) | 1.413 | 1.069–1.867 | 0.015 | 1.186 | 0.861–1.633 | 0.297 |
| MINPP1 (High vs. Low expression) | 1.451 | 1.096–1.921 | 0.009 | 1.308 | 0.940–1.818 | 0.111 |
| PAWR (High vs. Low expression) | 1.438 | 1.084–1.908 | 0.012 | 1.255 | 0.913–1.727 | 0.162 |
| PFN2 (High vs. Low expression) | 1.642 | 1.238–2.178 | 0.001 | 1.238 | 0.885–1.731 | 0.213 |
| RTN4R (High vs. Low expression) | 1.326 | 1.003–1.752 | 0.048 | 0.962 | 0.699–1.323 | 0.810 |
| Age (≥70 vs. <70) | 1.628 | 1.197–2.213 | 0.002 | 1.922 | 1.369–2.698 | <0.001 |
| Disease Stage (III, IV vs. I, II) | 1.746 | 1.158–2.633 | 0.008 | 1.774 | 1.159–2.716 | 0.008 |
| Pathological Grade (3, 4 vs. 1, 2) | 0.901 | 0.653–1.245 | 0.529 | - | - | - |
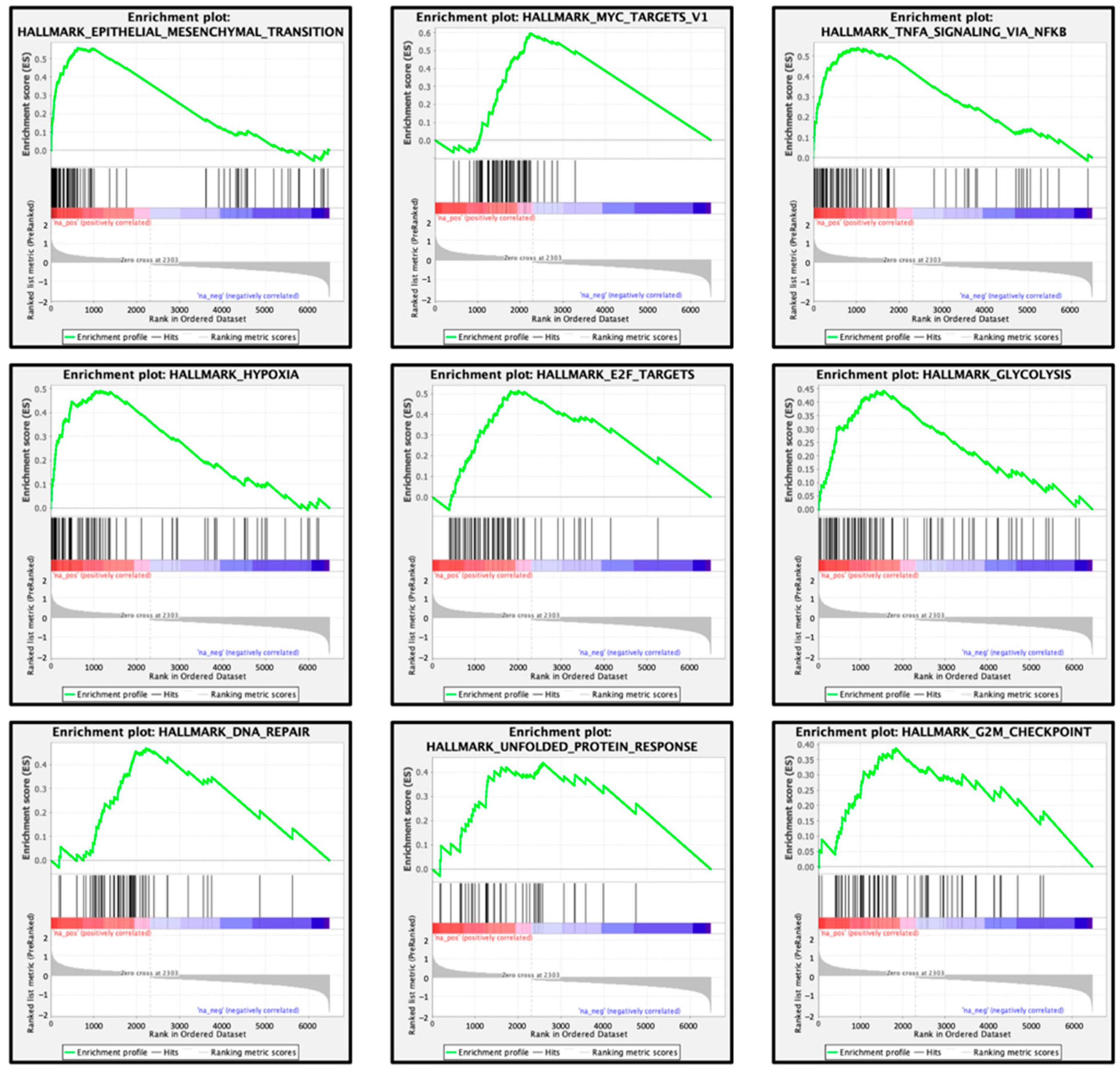
| Enriched Gene Sets in High FOXD1 Expression Group | ||
|---|---|---|
| Name | Normalized Enrichment Score | FDR q-Value |
| Epithelial mesenchymal transition | 3.674 | q < 0.001 |
| MYC targets V1 | 3.412 | q < 0.001 |
| TNFα signaling via NFκB | 3.262 | q < 0.001 |
| Hypoxia | 3.044 | q < 0.001 |
| E2F targets | 2.988 | q < 0.001 |
| Glycolysis | 2.797 | q < 0.001 |
| DNA repair | 2.602 | q < 0.001 |
| Unfolded protein response | 2.275 | 0.001 |
| G2M checkpoint | 2.248 | 0.001 |
| TGFβ signaling | 2.025 | 0.001 |
| KRAS signaling up | 1.946 | 0.005 |
| Coagulation | 1.848 | 0.010 |
| Inflammatory response | 1.753 | 0.014 |
| Apical junction | 1.739 | 0.015 |
| Oxidative phosphorylation | 1.713 | 0.015 |
| P53 pathway | 1.702 | 0.016 |
| IL6/JAK/STAT3 signaling | 1.617 | 0.027 |
| Apoptosis | 1.601 | 0.029 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minemura, C.; Asai, S.; Koma, A.; Kikkawa, N.; Kato, M.; Kasamatsu, A.; Uzawa, K.; Hanazawa, T.; Seki, N. Identification of Antitumor miR-30e-5p Controlled Genes; Diagnostic and Prognostic Biomarkers for Head and Neck Squamous Cell Carcinoma. Genes 2022, 13, 1225. https://doi.org/10.3390/genes13071225
Minemura C, Asai S, Koma A, Kikkawa N, Kato M, Kasamatsu A, Uzawa K, Hanazawa T, Seki N. Identification of Antitumor miR-30e-5p Controlled Genes; Diagnostic and Prognostic Biomarkers for Head and Neck Squamous Cell Carcinoma. Genes. 2022; 13(7):1225. https://doi.org/10.3390/genes13071225
Chicago/Turabian StyleMinemura, Chikashi, Shunichi Asai, Ayaka Koma, Naoko Kikkawa, Mayuko Kato, Atsushi Kasamatsu, Katsuhiro Uzawa, Toyoyuki Hanazawa, and Naohiko Seki. 2022. "Identification of Antitumor miR-30e-5p Controlled Genes; Diagnostic and Prognostic Biomarkers for Head and Neck Squamous Cell Carcinoma" Genes 13, no. 7: 1225. https://doi.org/10.3390/genes13071225
APA StyleMinemura, C., Asai, S., Koma, A., Kikkawa, N., Kato, M., Kasamatsu, A., Uzawa, K., Hanazawa, T., & Seki, N. (2022). Identification of Antitumor miR-30e-5p Controlled Genes; Diagnostic and Prognostic Biomarkers for Head and Neck Squamous Cell Carcinoma. Genes, 13(7), 1225. https://doi.org/10.3390/genes13071225






