Circulating microRNAs as the Potential Diagnostic and Prognostic Biomarkers for Nasopharyngeal Carcinoma
Abstract
1. Introduction
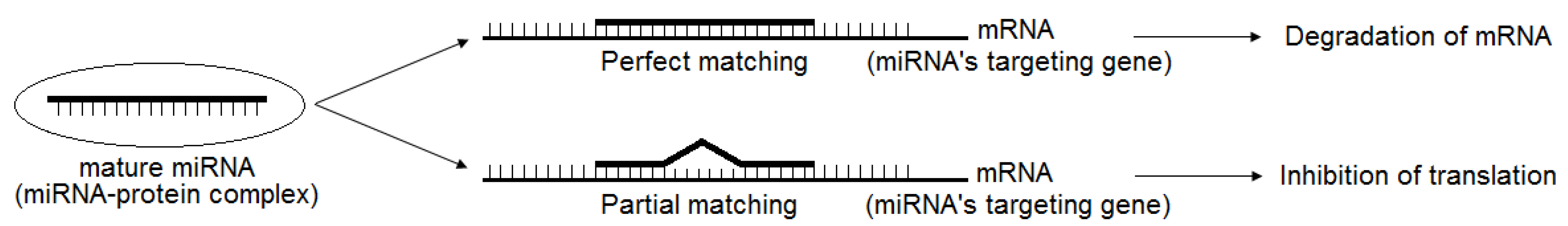
2. Brief Introduction of miRNAs and Their Regulation
3. Circulating miRNAs (CIR-miRNAs), Their Origin and Unique Characteristics
4. CIR-miRNAs as Diagnosis and Prognosis Biomarkers for NPC
5. Challenges of Circulating miRNAs as Indicators for Diagnosis and Prognosis
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ear, E.N.S.; Irekeola, A.A.; Yean Yean, C. Diagnostic and Prognostic Indications of Nasopharyngeal Carcinoma. Diagnostics 2020, 10, 611. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.H.; Austin, S.A.; Chen, S.H.; Sonne, D.C.; Gurushanthaiah, D. Nasopharyngeal Carcinoma Diagnostic Challenge in a Nonendemic Setting: Our Experience with 101 Patients. Perm. J. 2017, 21, 16–180. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.F.; Huang, M.; Ono, N.; Morita, A.; Kanaya, S.; Altaf-Ul-Amin, M. Development of a biomarker database toward performing disease classification and finding disease interrelations. Database 2021, 2021, baab011. [Google Scholar] [CrossRef]
- Tan, L.P.; Tan, G.W.; Sivanesan, V.M.; Goh, S.L.; Ng, X.J.; Lim, C.S.; Kim, W.R.; Mohidin, T.B.B.M.; Dali, N.S.M.; Ong, S.H.; et al. Systematic comparison of plasma EBV DNA, anti-EBV antibodies and miRNA levels for early detection and prognosis of nasopharyngeal carcinoma. Int. J. Cancer 2020, 146, 2336–2347. [Google Scholar] [CrossRef]
- Liu, W.; Chen, G.; Gong, X.; Wang, Y.; Zheng, Y.; Liao, X.; Liao, W.; Song, L.; Xu, J.; Zhang, X. The diagnostic value of EBV-DNA and EBV-related antibodies detection for nasopharyngeal carcinoma: A meta-analysis. Cancer Cell Int. 2021, 21, 164. [Google Scholar] [CrossRef]
- Yang, S.; Song, C. A meta-analysis on the EBV DNA and VCA-IgA in diagnosis of Nasopharyngeal Carcinoma. Pakistan J. Med. Sci. 2013, 29, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Middeldorp, J.; Yu, K.J.; Juwana, H.; Hsu, W.-L.; Lou, P.-J.; Wang, C.-P.; Chen, J.-Y.; Liu, M.-Y.; Pfeiffer, R.M.; et al. Characterization of ELISA detection of broad-spectrum anti-Epstein-Barr virus antibodies associated with nasopharyngeal carcinoma. J. Med. Virol. 2013, 85, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Lao, T.D.; Nguyen, T.A.H.; Ngo, K.D.; Thieu, H.H.; Nguyen, M.T.; Nguyen, D.H.; Le, T.A.H. Molecular Screening of Nasopharyngeal Carcinoma: Detection of LMP-1, LMP-2 Gene Expression in Vietnamese Nasopharyngeal Swab Samples. Asian Pac. J. Cancer Prev. 2019, 20, 2757–2761. [Google Scholar] [CrossRef]
- Lan, Y.-Y.; Hsiao, J.-R.; Chang, K.-C.; Chang, J.S.-M.; Chen, C.-W.; Lai, H.-C.; Wu, S.-Y.; Yeh, T.-H.; Chang, F.-H.; Lin, W.-H.; et al. Epstein-Barr Virus Latent Membrane Protein 2A Promotes Invasion of Nasopharyngeal Carcinoma Cells through ERK/Fra-1-Mediated Induction of Matrix Metalloproteinase 9. J. Virol. 2012, 86, 6656–6667. [Google Scholar] [CrossRef]
- Morris, M.; Laverick, L.; Wei, W.; Davis, A.; O’Neill, S.; Wood, L.; Wright, J.; Dawson, C.; Young, L. The EBV-Encoded Oncoprotein, LMP1, Induces an Epithelial-to-Mesenchymal Transition (EMT) via Its CTAR1 Domain through Integrin-Mediated ERK-MAPK Signalling. Cancers 2018, 10, 130. [Google Scholar] [CrossRef]
- Siak, P.Y.; Khoo, A.S.-B.; Leong, C.O.; Hoh, B.-P.; Cheah, S.-C. Current Status and Future Perspectives about Molecular Biomarkers of Nasopharyngeal Carcinoma. Cancers 2021, 13, 3490. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.P.; Kurzrock, R. Epstein-Barr Virus and Cancer. Clin. Cancer Res. 2004, 10, 803–821. [Google Scholar] [CrossRef] [PubMed]
- Dowd, J.B.; Palermo, T.; Brite, J.; McDade, T.W.; Aiello, A. Seroprevalence of Epstein-Barr Virus Infection in U.S. Children Ages 6–19, 2003–2010. PLoS ONE 2013, 8, e64921. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Xiang, J.; Wu, M.; Xiong, W.; Tang, H.; Deng, M.; Li, X.; Liao, Q.; Su, B.; Luo, Z.; et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 Combined as Non-Invasive Biomarkers in Nasopharyngeal Carcinoma. PLoS ONE 2012, 7, e46367. [Google Scholar] [CrossRef]
- Sun, K.; Jia, K.; Lv, H.; Wang, S.-Q.; Wu, Y.; Lei, H.; Chen, X. EBV-Positive Gastric Cancer: Current Knowledge and Future Perspectives. Front. Oncol. 2020, 10, 2096. [Google Scholar] [CrossRef]
- Feng, Y.; Xia, W.; He, G.; Ke, R.; Liu, L.; Xie, M.; Tang, A.; Yi, X. Accuracy Evaluation and Comparison of 14 Diagnostic Markers for Nasopharyngeal Carcinoma: A Meta-Analysis. Front. Oncol. 2020, 10, 1779. [Google Scholar] [CrossRef]
- Wang, W.-Y.; Twu, C.-W.; Liu, Y.-C.; Lin, H.-H.; Chen, C.-J.; Lin, J.-C. Fibronectin promotes nasopharyngeal cancer cell motility and proliferation. Biomed. Pharmacother. 2019, 109, 1772–1784. [Google Scholar] [CrossRef]
- Cobanoglu, U.; Mergan, D.; Dülger, A.C.; Celik, S.; Kemik, O.; Sayir, F. Are Serum Mac 2-Binding Protein Levels Elevated in Esophageal Cancer? A Control Study of Esophageal Squamous Cell Carcinoma Patients. Dis. Markers 2018, 2018, 3610239. [Google Scholar] [CrossRef]
- Sang, Y.; Chen, M.; Luo, D.; Zhang, R.-H.; Wang, L.; Li, M.; Luo, R.; Qian, C.-N.; Shao, J.-Y.; Zeng, Y.-X.; et al. TEL2 suppresses metastasis by down-regulating SERPINE1 in nasopharyngeal carcinoma. Oncotarget 2015, 6, 29240–29253. [Google Scholar] [CrossRef][Green Version]
- Mustikaningtyas, E.; Juniati, S.H.; Romdhoni, A.C. Intracell Heat Shock Protein 70 Expression and Nasopharyngeal Carcinoma Stage. Indian J. Otolaryngol. Head Neck Surg. 2019, 71, 321–326. [Google Scholar] [CrossRef]
- Janvilisri, T. Omics-Based Identification of Biomarkers for Nasopharyngeal Carcinoma. Dis. Markers 2015, 2015, 762128. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli, K.; Qian, P.-Y. Proteomics: Challenges, Techniques and Possibilities to Overcome Biological Sample Complexity. Hum. Genom. Proteom. 2009, 1. [Google Scholar] [CrossRef] [PubMed]
- De Haan, N.; Pučić-Baković, M.; Novokmet, M.; Falck, D.; Lageveen-Kammeijer, G.; Razdorov, G.; Vučković, F.; Trbojević-Akmačić, I.; Gornik, O.; Hanić, M.; et al. Developments and perspectives in high-throughput protein glycomics: Enabling the analysis of thousands of samples. Glycobiology 2022. [Google Scholar] [CrossRef]
- Lao, T.D.; Le, T.A.H. MicroRNAs: Biogenesis, Functions and Potential Biomarkers for Early Screening, Prognosis and Therapeutic Molecular Monitoring of Nasopharyngeal Carcinoma. Processes 2020, 8, 966. [Google Scholar] [CrossRef]
- Spence, T.; Bruce, J.; Yip, K.W.; Liu, F.-F. MicroRNAs in nasopharyngeal carcinoma. Chin. Clin. Oncol. 2016, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Claret, F.X.; Wu, W. MicroRNAs as therapeutic targets in nasopharyngeal carcinoma. Front. Oncol. 2019, 9, 756. [Google Scholar] [CrossRef]
- Wen, W.; Mai, S.J.; Lin, H.X.; Zhang, M.Y.; Huang, J.L.; Hua, X.; Lin, C.; Long, Z.Q.; Lu, Z.J.; Sun, X.Q.; et al. Identification of two microRNA signatures in whole blood as novel biomarkers for diagnosis of nasopharyngeal carcinoma. J. Transl. Med. 2019, 17, 186. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, Q.; Gu, M.; Zhang, K.; Xia, T.; Zhang, S.; Chen, W.; Yin, H.; Yao, H.; Fan, Y.; et al. MIR106A-5p upregulation suppresses autophagy and accelerates malignant phenotype in nasopharyngeal carcinoma. Autophagy 2021, 17, 1667–1683. [Google Scholar] [CrossRef]
- Xia, H.; Ng, S.S.; Jiang, S.; Cheung, W.K.C.; Sze, J.; Bian, X.-W.; Kung, H.; Lin, M.C. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem. Biophys. Res. Commun. 2010, 391, 535–541. [Google Scholar] [CrossRef]
- Zhang, L.; Deng, T.; Li, X.; Liu, H.; Zhou, H.; Ma, J.; Wu, M.; Zhou, M.; Shen, S.; Li, X.; et al. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis 2010, 31, 559–566. [Google Scholar] [CrossRef]
- Wu, L.; Zheng, K.; Yan, C.; Pan, X.; Liu, Y.; Liu, J.; Wang, F.; Guo, W.; He, X.; Li, J.; et al. Genome-wide study of salivary microRNAs as potential noninvasive biomarkers for detection of nasopharyngeal carcinoma. BMC Cancer 2019, 19, 843. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lu, J.; Wang, F.; Liu, X.; Peng, X.; Yu, B.; Zhao, F.; Li, X. Dynamic Changes in Plasma MicroRNAs Have Potential Predictive Values in Monitoring Recurrence and Metastasis of Nasopharyngeal Carcinoma. Biomed. Res. Int. 2018, 2018, 7329195. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Grimson, A.; Farh, K.K.-H.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA Targeting Specificity in Mammals: Determinants beyond Seed Pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef]
- Abdelfattah, A.M.; Park, C.; Choi, M.Y. Update on non-canonical microRNAs. Biomol. Concepts 2014, 5, 275–287. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Sheng, P.; Fields, C.; Aadland, K.; Wei, T.; Kolaczkowski, O.; Gu, T.; Kolaczkowski, B.; Xie, M. Dicer cleaves 5′-extended microRNA precursors originating from RNA polymerase II transcription start sites. Nucleic Acids Res. 2018, 46, 5737–5752. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, X.; Ma, Z.; Huo, Y.; Xiao, Z.; Li, Y.; Wang, Y. Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA 2011, 17, 1511–1528. [Google Scholar] [CrossRef] [PubMed]
- Ender, C.; Meister, G. Argonaute proteins at a glance. J. Cell Sci. 2010, 123, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Xu, J.; Qian, X.; Ding, R. MiR-24-3p Attenuates IL-1 β -Induced Chondrocyte Injury Associated With Osteoarthritis by Targeting BCL2L12. J. Orthop. Surg. Res. 2020, 16, 371. [Google Scholar] [CrossRef]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating MicroRNAs. Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Jiang, T.; Kang, X. Circulating microRNAs in cancer: Origin, function and application. J. Exp. Clin. Cancer Res. 2012, 31, 38. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zou, X.; Wu, L.; Zhang, S.; Wang, T.; Liu, P.; Zhu, W.; Zhu, J. Identification of a 7-microRNA signature in plasma as promising biomarker for nasopharyngeal carcinoma detection. Cancer Med. 2020, 9, 1230–1241. [Google Scholar] [CrossRef]
- Liu, N.; Cui, R.-X.; Sun, Y.; Guo, R.; Mao, Y.-P.; Tang, L.-L.; Jiang, W.; Liu, X.; Cheng, Y.-K.; He, Q.-M.; et al. A four-miRNA signature identified from genome-wide serum miRNA profiling predicts survival in patients with nasopharyngeal carcinoma. Int. J. Cancer 2014, 134, 1359–1368. [Google Scholar] [CrossRef]
- Zheng, X.-H.; Cui, C.; Ruan, H.-L.; Xue, W.-Q.; Zhang, S.-D.; Hu, Y.-Z.; Zhou, X.-X.; Jia, W.-H. Plasma microRNA profiles of nasopharyngeal carcinoma patients reveal miR-548q and miR-483-5p as potential biomarkers. Chin. J. Cancer 2014, 33, 330–338. [Google Scholar] [CrossRef]
- Liu, X.; Luo, H.-N.; Tian, W.-D.; Lu, J.; Li, G.; Wang, L.; Zhang, B.; Liang, B.-J.; Peng, X.-H.; Lin, S.-X.; et al. Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol. Ther. 2013, 14, 1133–1142. [Google Scholar] [CrossRef]
- Xie, Z.; Chen, G.; Zhang, X.; Li, D.; Huang, J.; Yang, C.; Zhang, P.; Qin, Y.; Duan, Y.; Gong, B.; et al. Salivary MicroRNAs as Promising Biomarkers for Detection of Esophageal Cancer. PLoS ONE 2013, 8, e57502. [Google Scholar] [CrossRef]
- Fan, C.; Tang, Y.; Wang, J.; Xiong, F.; Guo, C.; Wang, Y.; Xiang, B.; Zhou, M.; Li, X.; Wu, X.; et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J. Cancer 2018, 9, 2852–2864. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Huang, H.; Huang, L.; Sun, M.; Yan, Q.; Song, Y.; Wei, F.; Bo, H.; Gong, Z.; Zeng, Y.; et al. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci. China Life Sci. 2014, 57, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Židovec Lepej, S.; Matulić, M.; Gršković, P.; Pavlica, M.; Radmanić, L.; Korać, P. miRNAs: EBV Mechanism for Escaping Host’s Immune Response and Supporting Tumorigenesis. Pathogens 2020, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, F.; Wu, W.; Wang, Y.; Ding, H.; Qian, L. Epstein-Barr virus-encoded microRNAs as regulators in host immune responses. Int. J. Biol. Sci. 2018, 14, 565–576. [Google Scholar] [CrossRef]
- Lu, T.; Guo, Q.; Lin, K.; Chen, H.; Chen, Y.; Xu, Y.; Lin, C.; Su, Y.; Chen, Y.; Chen, M.; et al. Circulating Epstein-Barr virus microRNAs BART7-3p and BART13-3p as novel biomarkers in nasopharyngeal carcinoma. Cancer Sci. 2020, 111, 1711–1723. [Google Scholar] [CrossRef]
- Wardana, T.; Gunawan, L.; Herawati, C.; Oktriani, R.; Anwar, S.; Astuti, I.; Aryandono, T.; Mubarika, S. Circulation EBV Mir-Bart-7 Relating to Clinical Manifestation in Nasopharyngeal Carcinoma. Asian Pac. J. Cancer Prev. 2020, 21, 2777–2782. [Google Scholar] [CrossRef]
- Gao, W.; Wong, T.; Lv, K.; Zhang, M.; TSANG, R.K.; Chan, J.Y. Detection of Epstein–Barr virus (EBV)-encoded microRNAs in plasma of patients with nasopharyngeal carcinoma. Head Neck 2018, 41, 780–792. [Google Scholar] [CrossRef]
- Tay, J.K.; Lim, M.Y.; Kanagalingam, J. Screening in Nasopharyngeal Carcinoma: Current Strategies and Future Directions. Curr. Otorhinolaryngol. Rep. 2014, 2, 1–7. [Google Scholar] [CrossRef]
- Shannon-Lowe, C.; Rickinson, A. The Global Landscape of EBV-Associated Tumors. Front. Oncol. 2019, 9, 713. [Google Scholar] [CrossRef]
- Ayee, R.; Ofori, M.E.; Wright, E.; Quaye, O. Epstein Barr Virus Associated Lymphomas and Epithelia Cancers in Humans. J. Cancer 2020, 11, 1737–1750. [Google Scholar] [CrossRef]

| Year | CIR-miRNAs candidate | Description | Source | Reference |
|---|---|---|---|---|
| 2020 | hsa-miR-let-7b-5p, hsa-miR-140-3p, hsa-miR-144-3p, hsa-miR-17-5p, hsa-miR-20a-5p, hsa-miR-20b-5p, hsa-miR-205-5p | The panel of 7 miRNA, extracted from plasma, performed better in distinguishing NPC patients from healthy controls, the sensitivity and specificity being 0.74 and 0.76, respectively. | Plasma | [56] |
| 2019 | hsa-miR-188-5p, hsa-miR-1908, hsa-miR-3196, hsa-miR-3935, hsa-miR-4284, hsa-miR-4433-5p, hsa-miR-4665-3p, hsa-miR-513b | Theses 8 miRNA signatures diagnosed NPC with an accuracy of 97.14%, sensitivity of 96.43%, specificity of 100%, positive predictive value of 100%, and negative predictive value of 87.5% in a group of 84 NPC samples and 21 healthy samples. | Whole blood | [27] |
| 2019 | hsa-miR-937-5p, hsa-miR-650, hsa-miR-3612, hsa-miR-4478, hsa-miR-4259, hsa-miR-3714, hsa-miR-4730, hsa-miR-1203, hsa-miR-30b-3p, hsa-miR-1321, hsa-miR-1202, hsa-miR-575 | These CIR-miRNAs were significantly down-regulated in saliva of NPC patients compared to healthy controls, detected by miRNA microarray platform with the high accuracy (sensitivity = 100.00%, specificity = 96.00%). | Salivary | [31] |
| 2014 | hsa-miR-22, hsa-miR-miR-572, hsa-miR-638, hsa-miR-1234 | Different changes were observed in the serum of patients with NPC. The value of prognosis of the TNM staging system was reported. The patients of with high-risk scores had poorer overall survival and low metastasis-free survival than those with the patients with low-risk scores. | Serum | [57] |
| 2014 | hsa-miR-548q, hsa-miR-483-5p | miR-548q and miR-483-5p highly expressed in NPC cell lines and 31 plasma samples from NPC patients, compared with 19 non-cancerous controls. Combining these 2 CIR-miRNAs resulted in 67.1% sensitivity and 68.0% specificity. | Plasma | [58] |
| 2014 | hsa-miR-483-5p, hsa-miR-103, hsa-miR-29a | Differentially expressed CIR-miRNAs were identified as being effective biomarkers for predicting survival in NPC patients. | Plasma | [26] |
| 2013 | hsa-miR-16, miR-21, hsa-miR-24, hsa-miR-155, hsa-and miR-378 | Sensitivity and specificity reached 87.7% and 82.0%, respectively, when combining the panel of CIR-miRNAs, hsa-miR-16, miR-21, hsa-miR-24, hsa-miR-155, hsa-and miR-378. | Plasma | [59] |
| 2012 | hsa-miR-17, hsa-miR-20a, hsa-miR-29c, hsa-miR-223 | miRNAs were differentially expressed in the serum of 20 NPC patients compared with that of 20 non-cancerous controls. Using these 4 CIR-miRNAs, a diagnostic value with sensitivity of 97.3% and specificity of 96.5% was established. | Serum | [14] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, T.A.H.; Lao, T.D. Circulating microRNAs as the Potential Diagnostic and Prognostic Biomarkers for Nasopharyngeal Carcinoma. Genes 2022, 13, 1160. https://doi.org/10.3390/genes13071160
Le TAH, Lao TD. Circulating microRNAs as the Potential Diagnostic and Prognostic Biomarkers for Nasopharyngeal Carcinoma. Genes. 2022; 13(7):1160. https://doi.org/10.3390/genes13071160
Chicago/Turabian StyleLe, Thuy Ai Huyen, and Thuan Duc Lao. 2022. "Circulating microRNAs as the Potential Diagnostic and Prognostic Biomarkers for Nasopharyngeal Carcinoma" Genes 13, no. 7: 1160. https://doi.org/10.3390/genes13071160
APA StyleLe, T. A. H., & Lao, T. D. (2022). Circulating microRNAs as the Potential Diagnostic and Prognostic Biomarkers for Nasopharyngeal Carcinoma. Genes, 13(7), 1160. https://doi.org/10.3390/genes13071160






