Exploring the Mutational Landscape of Isolated Congenital Heart Defects: An Exome Sequencing Study Using Cardiac DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. DNA Extraction
2.3. Exome Sequencing
2.4. Raw Data Processing
2.5. GnomAD and CADD
2.6. Mosaic Variant Calling
2.7. Trio Analysis
- (1)
- A heterozygous single nucleotide variant (SNV) with GATK’s single sample quality score (QUAL) > 300 or a heterozygous small insertion or deletion (indel) with QUAL > 1000;
- (2)
- Both parents were homozygous for the reference allele with a corresponding genotype quality (GQ) ≥ 30;
- (3)
- The variant was observed only once or twice in our case cohort of 73 ICHD patients.Inframe indels were excluded from further analysis because of the high risk of false-positive calls for this variant type. From the remaining HQ DNV, only LOF variants and missense variants with gnomAD AF ≤ 0.1% were retained for further interpretation.
- (1)
- A SNV with QUAL > 300 or a small indel with QUAL > 1000;
- (2)
- The variant was also present in one or both parents;
- (3)
- The variant was observed only once or twice in our case cohort of 73 ICHD patients.
2.8. Transmission Disequilibrium Testing
- (1)
- SNV with QUAL > 300 or a small indel with QUAL > 1000;
- (2)
- The gnomAD AF was ≤ 0.1%.
2.9. Association Testing
2.10. Gene Panels and Gene Expression in Human Heart during Embryonic Development
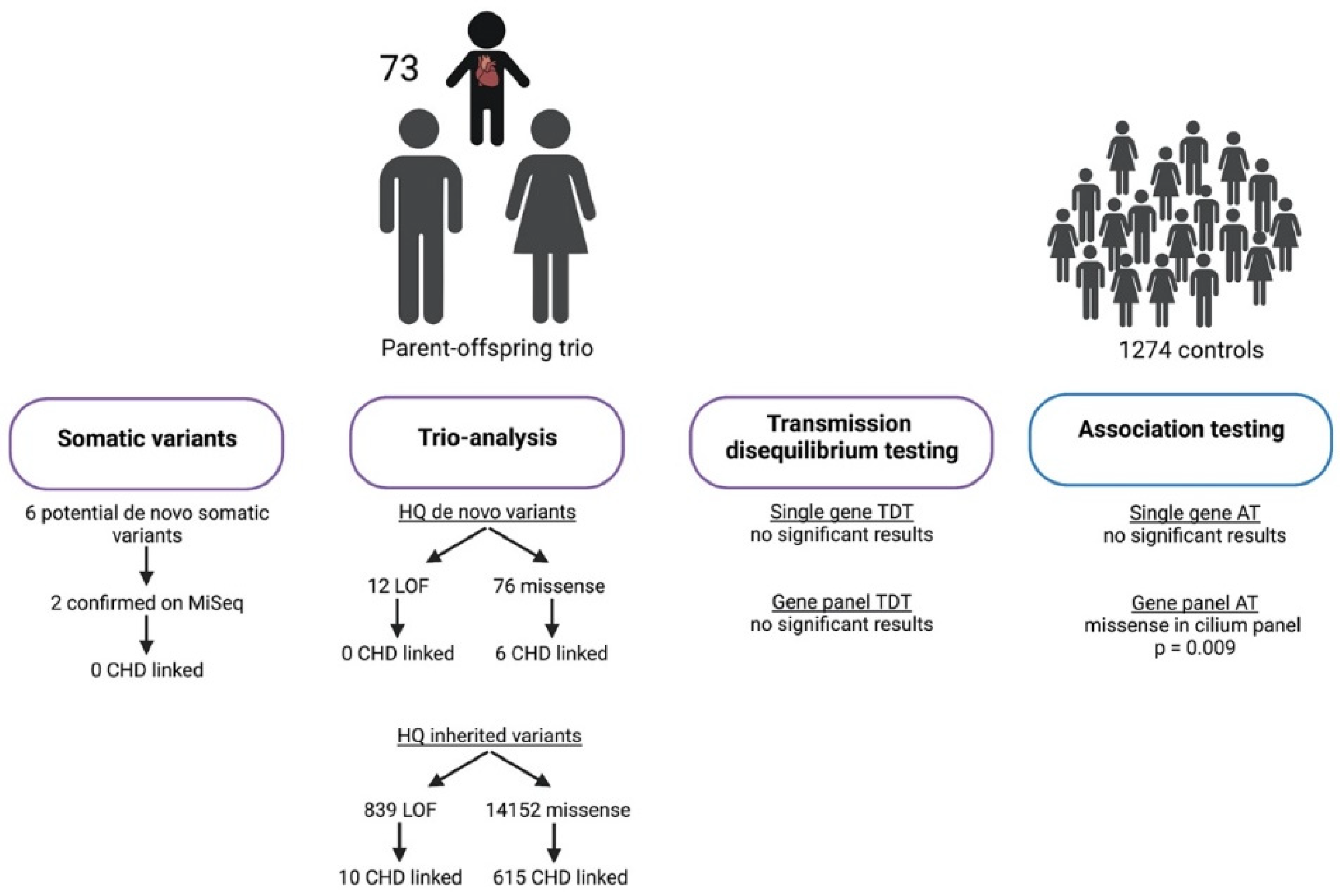
3. Results
3.1. Study Cohort
3.2. Mosaic Variant Calling
3.3. Trio Analysis
3.3.1. High-Quality de Novo Variants
3.3.2. High-Quality Rare Inherited Variants
3.4. Transmission Disequilibrium Testing
3.5. Association Testing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van der Linde, D.; Konings, E.E.; Slager, M.A.; Witsenburg, M.; Helbing, W.A.; Takkenberg, J.J.; Roos-Hesselink, J.W. Birth prevalence of congenital heart disease worldwide: A systematic review and meta-analysis. J. Am. Coll. Cardiol. 2011, 58, 2241–2247. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S.; Zühlke, L.; Black, G.C.; Choy, M.K.; Li, N.; Keavney, B.D. Global birth prevalence of congenital heart defects 1970–2017: Updated systematic review and meta-analysis of 260 studies. Int. J. Epidemiol. 2019, 48, 455–463. [Google Scholar] [CrossRef]
- Stoll, C.; Dott, B.; Alembik, Y.; Roth, M.P. Associated noncardiac congenital anomalies among cases with congenital heart defects. Eur. J. Med. Genet. 2015, 58, 75–85. [Google Scholar] [CrossRef]
- Fahed, A.C.; Gelb, B.D.; Seidman, J.G.; Seidman, C.E. Genetics of congenital heart disease: The glass half empty. Circ. Res. 2013, 112, 707–720. [Google Scholar] [CrossRef]
- Andersen, T.A.; Troelsen Kde, L.; Larsen, L.A. Of mice and men: Molecular genetics of congenital heart disease. Cell Mol. Life Sci. 2014, 71, 1327–1352. [Google Scholar] [CrossRef]
- Cerrone, M.; Remme, C.A.; Tadros, R.; Bezzina, C.R.; Delmar, M. Beyond the One Gene-One Disease Paradigm: Complex Genetics and Pleiotropy in Inheritable Cardiac Disorders. Circulation 2019, 140, 595–610. [Google Scholar] [CrossRef]
- Reamon-Buettner, S.M.; Borlak, J. TBX5 mutations in non-Holt-Oram syndrome (HOS) malformed hearts. Hum. Mutat. 2004, 24, 104. [Google Scholar] [CrossRef]
- Reamon-Buettner, S.M.; Borlak, J. Somatic NKX2-5 mutations as a novel mechanism of disease in complex congenital heart disease. J. Med. Genet. 2004, 41, 684–690. [Google Scholar] [CrossRef]
- Reamon-Buettner, S.M.; Hecker, H.; Spanel-Borowski, K.; Craatz, S.; Kuenzel, E.; Borlak, J. Novel NKX2-5 mutations in diseased heart tissues of patients with cardiac malformations. Am. J. Pathol. 2004, 164, 2117–2125. [Google Scholar] [CrossRef]
- Draus, J.M., Jr.; Hauck, M.A.; Goetsch, M.; Austin, E.H., 3rd; Tomita-Mitchell, A.; Mitchell, M.E. Investigation of somatic NKX2-5 mutations in congenital heart disease. J. Med. Genet. 2009, 46, 115–122. [Google Scholar] [CrossRef]
- Salazar, M.; Consoli, F.; Villegas, V.; Caicedo, V.; Maddaloni, V.; Daniele, P.; Caianiello, G.; Pachón, S.; Nuñez, F.; Limongelli, G.; et al. Search of somatic GATA4 and NKX2.5 gene mutations in sporadic septal heart defects. Eur. J. Med. Genet. 2011, 54, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, Y.; Chen, H.; Yin, M.; Yu, T.; Fu, Q. Investigation of somatic NKX2-5, GATA4 and HAND1 mutations in patients with tetralogy of Fallot. Pathology 2011, 43, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Butler, T.L.; Blue, G.M.; Cole, A.D.; Sholler, G.F.; Kirk, E.P.; Grossfeld, P.; Perryman, B.M.; Harvey, R.P.; Winlaw, D.S. Somatic mutations in NKX2–5, GATA4, and HAND1 are not a common cause of tetralogy of Fallot or hypoplastic left heart. Am. J. Med. Genet. Part A 2011, 155a, 2416–2421. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.T.; Xue, S.; Xu, Y.J.; Yang, Y.Q. Somatic mutations in the GATA6 gene underlie sporadic tetralogy of Fallot. Int. J. Mol. Med. 2013, 31, 51–58. [Google Scholar] [CrossRef]
- Huang, R.T.; Xue, S.; Xu, Y.J.; Zhou, M.; Yang, Y.Q. Somatic GATA5 mutations in sporadic tetralogy of Fallot. Int. J. Mol. Med. 2014, 33, 1227–1235. [Google Scholar] [CrossRef]
- Zheng, J.; Li, F.; Liu, J.; Xu, Z.; Zhang, H.; Fu, Q.; Wang, J.; Sun, K. Investigation of Somatic NKX2-5 Mutations in Chinese Children with Congenital Heart Disease. Int. J. Med. Sci. 2015, 12, 538–543. [Google Scholar] [CrossRef][Green Version]
- Durbin, M.D.; Cadar, A.G.; Williams, C.H.; Guo, Y.; Bichell, D.P.; Su, Y.R.; Hong, C.C. Hypoplastic Left Heart Syndrome Sequencing Reveals a Novel NOTCH1 Mutation in a Family with Single Ventricle Defects. Pediatr. Cardiol. 2017, 38, 1232–1240. [Google Scholar] [CrossRef]
- Yin, J.; Qian, J.; Dai, G.; Wang, C.; Qin, Y.; Xu, T.; Li, Z.; Zhang, H.; Yang, S. Search of Somatic Mutations of NKX2-5 and GATA4 Genes in Chinese Patients with Sporadic Congenital Heart Disease. Pediatr. Cardiol. 2019, 40, 17–22. [Google Scholar] [CrossRef]
- Fardoun, M.; Dehaini, H.; Kamar, A.; Bitar, F.; Majdalani, M.; El-Rassi, I.; Nemer, G.; Arabi, M. A Novel Somatic Variant in HEY2 Unveils an Alternative Splicing Isoform Linked to Ventricular Septal Defect. Pediatr. Cardiol. 2019, 40, 1084–1091. [Google Scholar] [CrossRef]
- Hsieh, A.; Morton, S.U.; Willcox, J.A.L.; Gorham, J.M.; Tai, A.C.; Qi, H.; DePalma, S.; McKean, D.; Griffin, E.; Manheimer, K.B.; et al. EM-mosaic detects mosaic point mutations that contribute to congenital heart disease. Genome. Med. 2020, 12, 42. [Google Scholar] [CrossRef]
- Spielman, R.S.; McGinnis, R.E.; Ewens, W.J. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am. J. Hum. Genet. 1993, 52, 506–516. [Google Scholar]
- Patterson, N.; Price, A.L.; Reich, D. Population structure and eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome. Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome. Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Rentzsch, P.; Schubach, M.; Shendure, J.; Kircher, M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome. Med. 2021, 13, 31. [Google Scholar] [CrossRef]
- Huang, A.Y.; Zhang, Z.; Ye, A.Y.; Dou, Y.; Yan, L.; Yang, X.; Zhang, Y.; Wei, L. MosaicHunter: Accurate detection of postzygotic single-nucleotide mosaicism through next-generation sequencing of unpaired, trio, and paired samples. Nucleic Acids Res. 2017, 45, e76. [Google Scholar] [CrossRef]
- Laird, N.M.; Horvath, S.; Xu, X. Implementing a unified approach to family-based tests of association. Genet. Epidemiol. 2000, 19 (Suppl. S1), S36–S42. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Cardoso-Moreira, M.; Halbert, J.; Valloton, D.; Velten, B.; Chen, C.; Shao, Y.; Liechti, A.; Ascenção, K.; Rummel, C.; Ovchinnikova, S.; et al. Gene expression across mammalian organ development. Nature 2019, 571, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Asiri, A.; Aloyouni, E.; Umair, M.; Alyafee, Y.; Al Tuwaijri, A.; Alhamoudi, K.M.; Almuzzaini, B.; Al Baz, A.; Alwadaani, D.; Nashabat, M.; et al. Mutated RAP1GDS1 causes a new syndrome of dysmorphic feature, intellectual disability & speech delay. Ann. Clin. Transl. Neurol. 2020, 7, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Imanaka-Yoshida, K.; Wang, L.; Zhan, S.; Schneider, M.D.; DeMayo, F.J.; Schwartz, R.J. Inhibition of Rho family GTPases by Rho GDP dissociation inhibitor disrupts cardiac morphogenesis and inhibits cardiomyocyte proliferation. Development 2002, 129, 1705–1714. [Google Scholar] [CrossRef] [PubMed]
- Kaarbø, M.; Crane, D.I.; Murrell, W.G. RhoA is highly up-regulated in the process of early heart development of the chick and important for normal embryogenesis. Dev. Dyn. 2003, 227, 35–47. [Google Scholar] [CrossRef]
- Phillips, H.M.; Murdoch, J.N.; Chaudhry, B.; Copp, A.J.; Henderson, D.J. Vangl2 acts via RhoA signaling to regulate polarized cell movements during development of the proximal outflow tract. Circ. Res. 2005, 96, 292–299. [Google Scholar] [CrossRef]
- Kruszka, P.; Tanpaiboon, P.; Neas, K.; Crosby, K.; Berger, S.I.; Martinez, A.F.; Addissie, Y.A.; Pongprot, Y.; Sittiwangkul, R.; Silvilairat, S.; et al. Loss of function in ROBO1 is associated with tetralogy of Fallot and septal defects. J. Med. Genet. 2017, 54, 825–829. [Google Scholar] [CrossRef]
- Vaqueiro, A.C.; de Oliveira, C.P.; Cordoba, M.S.; Versiani, B.R.; de Carvalho, C.X.; Alves Rodrigues, P.G.; de Oliveira, S.F.; Mazzeu, J.F.; Pic-Taylor, A. Expanding the spectrum of TBL1XR1 deletion: Report of a patient with brain and cardiac malformations. Eur. J. Med. Genet. 2018, 61, 29–33. [Google Scholar] [CrossRef]
- Adams, R.H.; Wilkinson, G.A.; Weiss, C.; Diella, F.; Gale, N.W.; Deutsch, U.; Risau, W.; Klein, R. Roles of ephrinB ligands and EphB receptors in cardiovascular development: Demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes Dev. 1999, 13, 295–306. [Google Scholar] [CrossRef]
- Gerety, S.S.; Wang, H.U.; Chen, Z.F.; Anderson, D.J. Symmetrical mutant phenotypes of the receptor EphB4 and its specific transmembrane ligand ephrin-B2 in cardiovascular development. Mol. Cell 1999, 4, 403–414. [Google Scholar] [CrossRef]
- Cohen-Barak, O.; Yi, Z.; Hagiwara, N.; Monzen, K.; Komuro, I.; Brilliant, M.H. Sox6 regulation of cardiac myocyte development. Nucleic Acids Res. 2003, 31, 5941–5948. [Google Scholar] [CrossRef] [PubMed]
- Wiemer-Kruel, A.; Mayer, H.; Ewert, P.; Martinoff, S.; Eckstein, H.H.; Kriebel, T.; Bissler, J.; Franz, D.; Bast, T. Congenital Lymphatic Malformation and Aortic Aneurysm in a Patient with TSC2 Mutation. Neuropediatrics 2020, 51, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Homsy, J.; Zaidi, S.; Shen, Y.; Ware, J.S.; Samocha, K.E.; Karczewski, K.J.; DePalma, S.R.; McKean, D.; Wakimoto, H.; Gorham, J.; et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 2015, 350, 1262–1266. [Google Scholar] [CrossRef] [PubMed]
- Sifrim, A.; Hitz, M.P.; Wilsdon, A.; Breckpot, J.; Turki, S.H.; Thienpont, B.; McRae, J.; Fitzgerald, T.W.; Singh, T.; Swaminathan, G.J.; et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat. Genet. 2016, 48, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.P.; Omran, H.; Leigh, M.W.; Dell, S.; Morgan, L.; Molina, P.L.; Robinson, B.V.; Minnix, S.L.; Olbrich, H.; Severin, T.; et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation 2007, 115, 2814–2821. [Google Scholar] [CrossRef] [PubMed]
- French, V.M.; van de Laar, I.M.; Wessels, M.W.; Rohe, C.; Roos-Hesselink, J.W.; Wang, G.; Frohn-Mulder, I.M.; Severijnen, L.A.; de Graaf, B.M.; Schot, R.; et al. NPHP4 variants are associated with pleiotropic heart malformations. Circ. Res. 2012, 110, 1564–1574. [Google Scholar] [CrossRef]
- Burnicka-Turek, O.; Steimle, J.D.; Huang, W.; Felker, L.; Kamp, A.; Kweon, J.; Peterson, M.; Reeves, R.H.; Maslen, C.L.; Gruber, P.J.; et al. Cilia gene mutations cause atrioventricular septal defects by multiple mechanisms. Hum. Mol. Genet. 2016, 25, 3011–3028. [Google Scholar] [CrossRef]
- Gabriel, G.C.; Young, C.B.; Lo, C.W. Role of cilia in the pathogenesis of congenital heart disease. Semin. Cell Dev. Biol. 2021, 110, 2–10. [Google Scholar] [CrossRef]
- Vecoli, C.; Pulignani, S.; Foffa, I.; Andreassi, M.G. Congenital heart disease: The crossroads of genetics, epigenetics and environment. Curr. Genom. 2014, 15, 390–399. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meerschaut, I.; Steyaert, W.; Bové, T.; François, K.; Martens, T.; De Groote, K.; De Wilde, H.; Muiño Mosquera, L.; Panzer, J.; Vandekerckhove, K.; et al. Exploring the Mutational Landscape of Isolated Congenital Heart Defects: An Exome Sequencing Study Using Cardiac DNA. Genes 2022, 13, 1214. https://doi.org/10.3390/genes13071214
Meerschaut I, Steyaert W, Bové T, François K, Martens T, De Groote K, De Wilde H, Muiño Mosquera L, Panzer J, Vandekerckhove K, et al. Exploring the Mutational Landscape of Isolated Congenital Heart Defects: An Exome Sequencing Study Using Cardiac DNA. Genes. 2022; 13(7):1214. https://doi.org/10.3390/genes13071214
Chicago/Turabian StyleMeerschaut, Ilse, Wouter Steyaert, Thierry Bové, Katrien François, Thomas Martens, Katya De Groote, Hans De Wilde, Laura Muiño Mosquera, Joseph Panzer, Kristof Vandekerckhove, and et al. 2022. "Exploring the Mutational Landscape of Isolated Congenital Heart Defects: An Exome Sequencing Study Using Cardiac DNA" Genes 13, no. 7: 1214. https://doi.org/10.3390/genes13071214
APA StyleMeerschaut, I., Steyaert, W., Bové, T., François, K., Martens, T., De Groote, K., De Wilde, H., Muiño Mosquera, L., Panzer, J., Vandekerckhove, K., Moons, L., Vermassen, P., Symoens, S., Coucke, P. J., De Wolf, D., & Callewaert, B. (2022). Exploring the Mutational Landscape of Isolated Congenital Heart Defects: An Exome Sequencing Study Using Cardiac DNA. Genes, 13(7), 1214. https://doi.org/10.3390/genes13071214






