Overexpression of DfRaf from Fragrant Woodfern (Dryopteris fragrans) Enhances High-Temperature Tolerance in Tobacco (Nicotiana tabacum)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Rubisco Activity Measurements
2.3. RCA Activity Determination
2.4. PRK Activity Measurements
2.5. Assessment of Carbon Assimilation-Related Gene Expression by qRT-PCR
2.6. Subcellular Localization Assay
2.7. Generation of Transgenic Nicotiana tabacum Plants
2.8. Determination of Physiological Indices
2.9. Histochemical Staining of Hydrogen Peroxide (H2O2) and Superoxide (O2−)
2.10. Photosynthetic Pigments
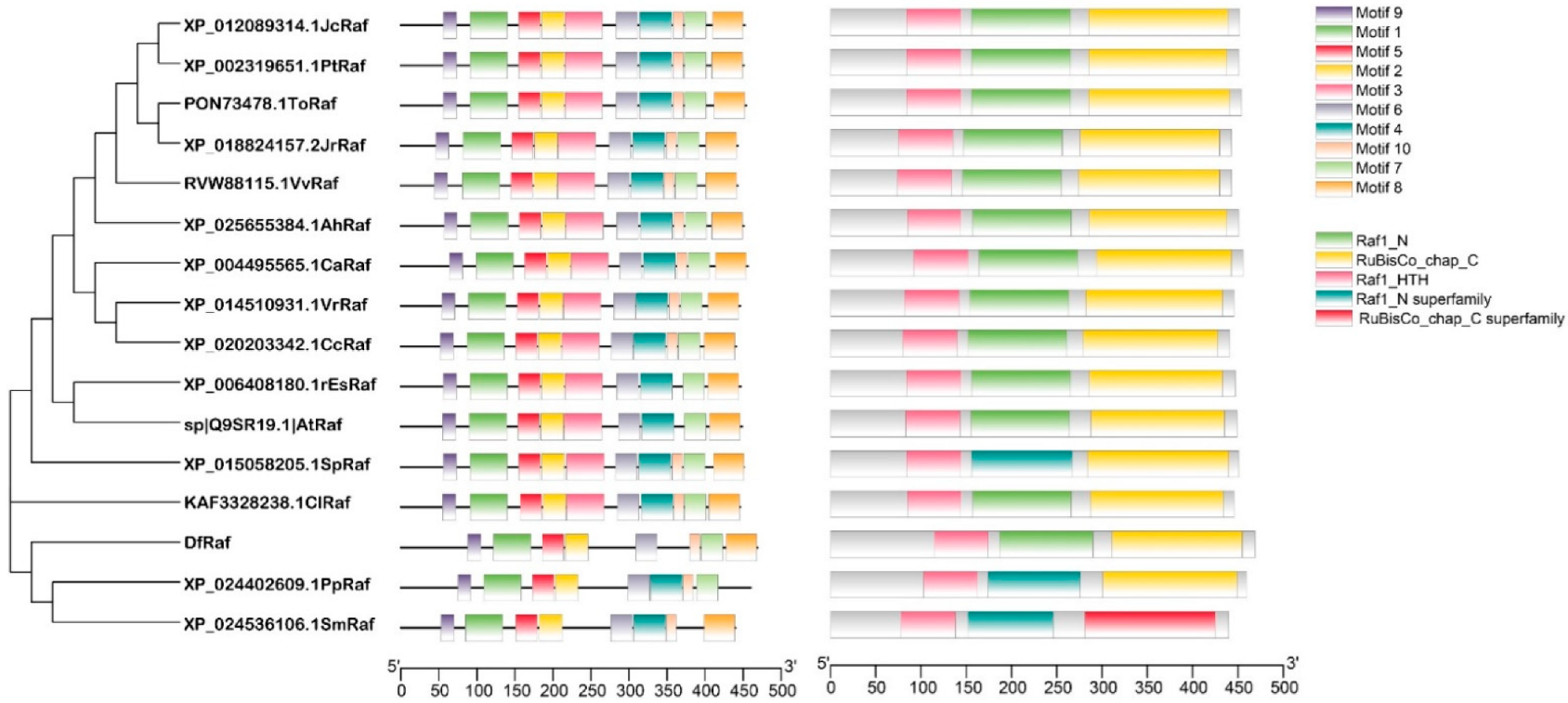
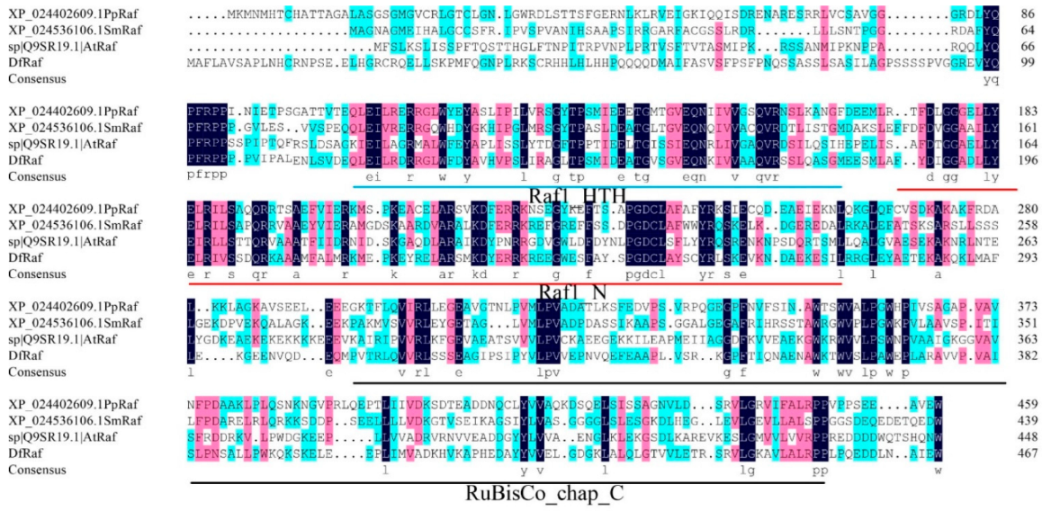
2.11. Phylogenetic, Multiple Sequence Alignment and Conservative Motif Analyses
2.12. Statistical Analysis
3. Results
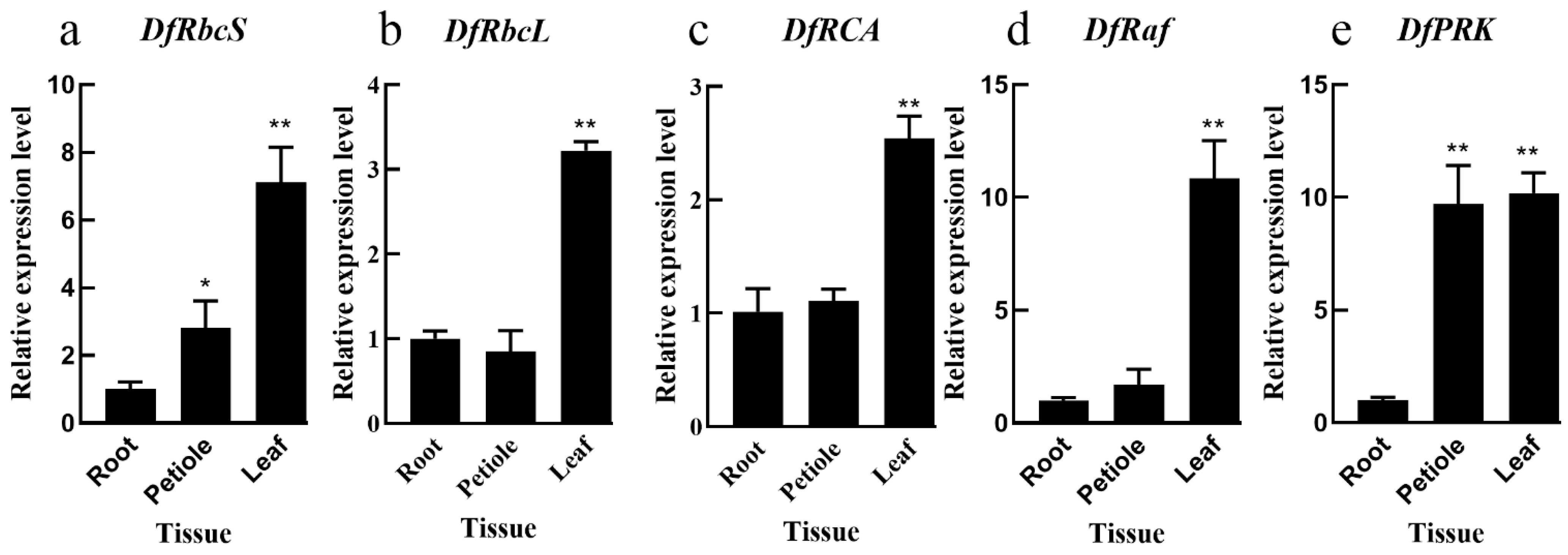
3.1. Carbon Assimilation Gene Expression Patterns in Different Fragrant Woodfern Tissues
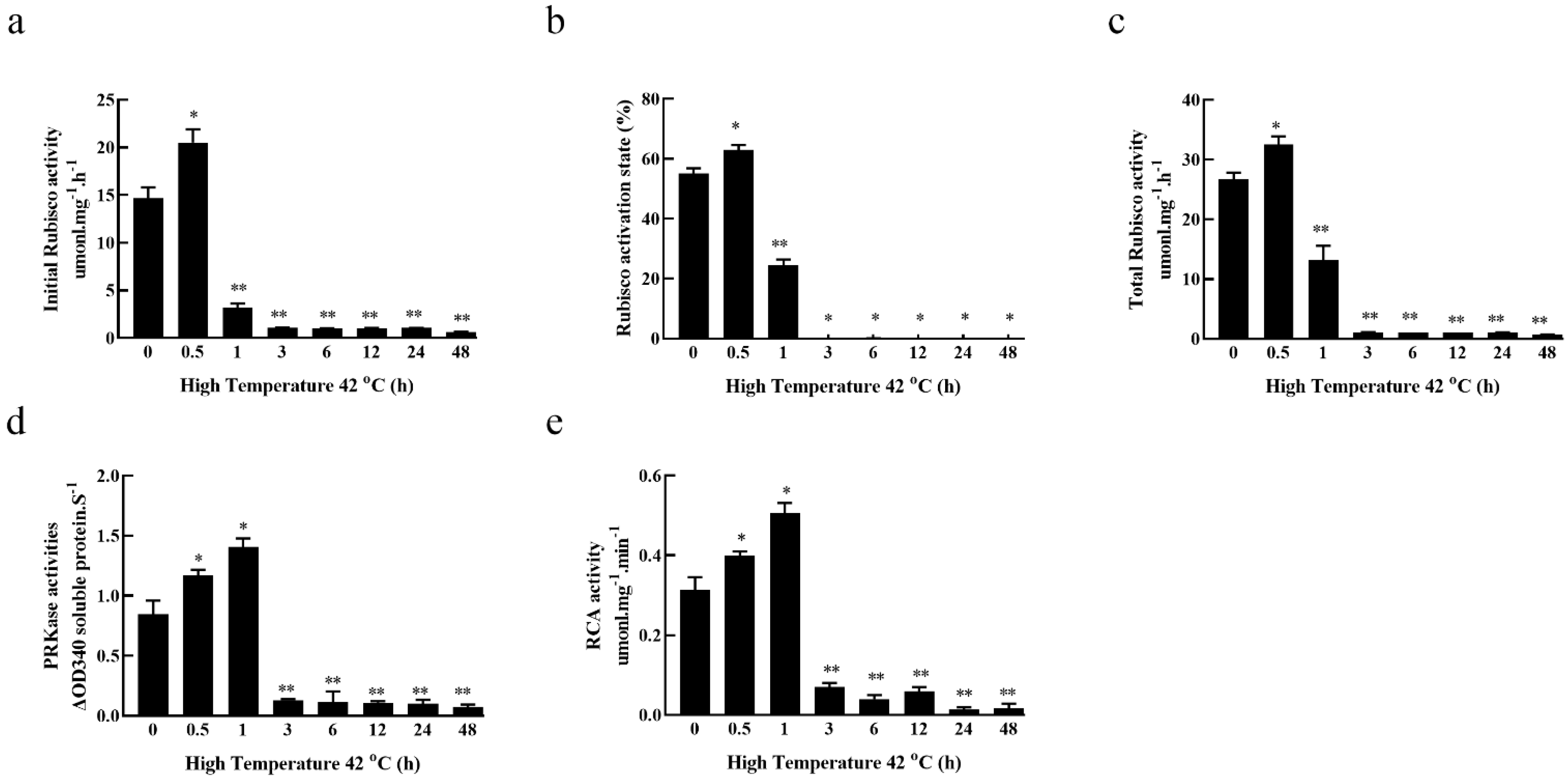
3.2. Effects of High-Temperature Stress on Carbon Assimilation Enzymes’ Activity in Fragrant Woodfern Leaves
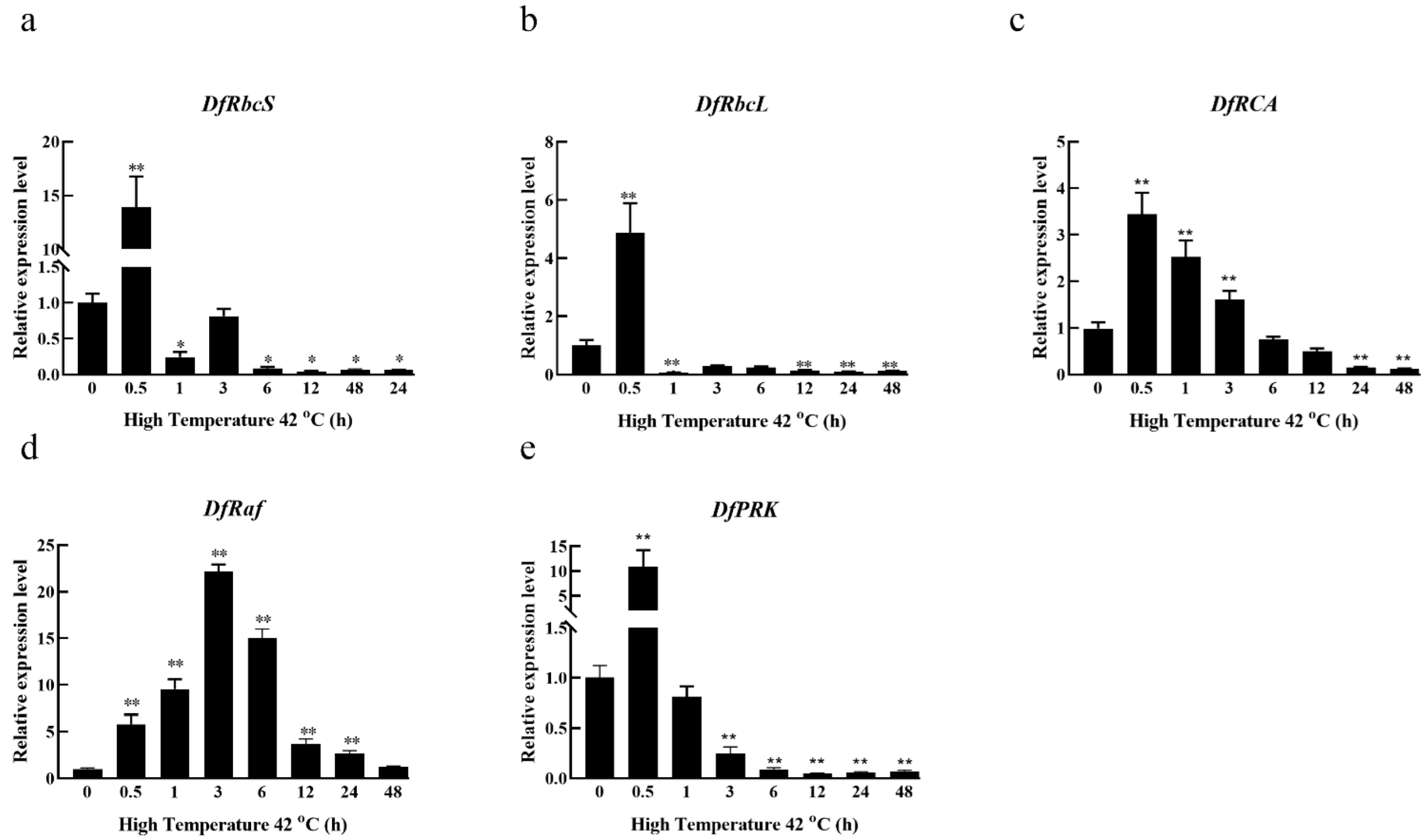
3.3. Effects of High-Temperature Stress on the Relative Expression of Carbon Assimilation Enzymes Genes in Fragrant Woodfern Leaves
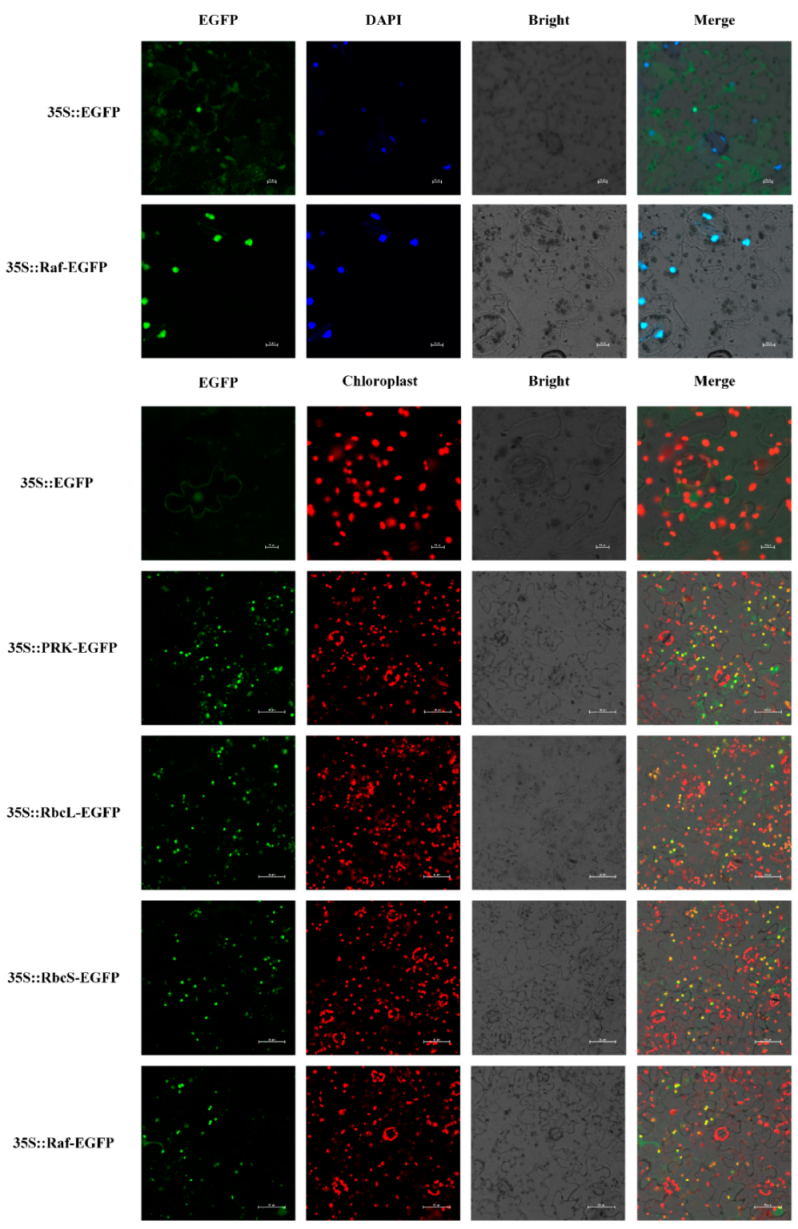
3.4. Subcellular Location of Carbon Assimilation Enzyme Genes
3.5. Phylogenetic, Multiple Sequence Alignment and Conservative Motif Analysis of Fragrant Woodfern Raf Proteins
3.6. Overexpression of DfRaf Increased the High-Temperature Tolerance of Transgenic Tobacco
3.7. Effects of High-Temperature Stress on Carbon Assimilation Enzyme Activity in Transgenic Tobacco
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sukhov, V.; Gaspirovich, V.; Mysyagin, S.; Vodeneev, V. High-Temperature Tolerance of Photosynthesis Can Be Linked to Local Electrical Responses in Leaves of Pea. Front. Physiol. 2017, 8, 763. [Google Scholar] [CrossRef] [PubMed]
- Hein, N.T.; Impa, S.M.; Wagner, D.; Bheemanahalli, R.; Kumar, R.; Tiwari, M.; Prasad, P.V.V.; Tilley, M.; Wu, X.; Neilsen, M.; et al. Grain micronutrient composition and yield components in field-grown wheat are negatively impacted by high night-time temperature. Cereal Chem. 2022, 99, 615–624. [Google Scholar] [CrossRef]
- Tiwari, M.; Kumar, R.; Min, D.; Jagadish, S.V.K. Genetic and molecular mechanisms underlying root architecture and function under heat stress-A hidden story. Plant Cell Environ. 2022, 45, 771–788. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Sharma, M.M.M.; Patra, A.; Vashisth, M.; Mehta, S.; Singh, B.; Tiwari, M.; Pandey, V. The role of key transcription factors for cold tolerance in plants. In Transcription Factors for Abiotic Stress Tolerance in Plants; Academic Press: Cambridge, MA, USA, 2020; pp. 123–152. [Google Scholar] [CrossRef]
- Ramachandra Reddy, A.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef]
- Mathur, S.; Jajoo, A.; Mehta, P.; Bharti, S. Analysis of elevated temperature-induced inhibition of photosystem II using chlorophyll a fluorescence induction kinetics in wheat leaves (Triticum aestivum). Plant Biol. 2011, 13, 1–6. [Google Scholar] [CrossRef]
- Mathur, S.; Mehta, P.; Jajoo, A. Effects of dual stress (high salt and high temperature) on the photochemical efficiency of wheat leaves (Triticum aestivum). Physiol. Mol. Biol. Plants 2013, 19, 179–188. [Google Scholar] [CrossRef]
- Haworth, M.; Marino, G.; Brunetti, C.; Killi, D.; de Carlo, A.; Centritto, M. The Impact of Heat Stress and Water Deficit on the Photosynthetic and Stomatal Physiology of Olive (Olea europaea L.)—A Case Study of the 2017 Heat Wave. Plants 2018, 7, 76. [Google Scholar] [CrossRef]
- Killi, D.; Bussotti, F.; Raschi, A.; Haworth, M. Adaptation to high temperature mitigates the impact of water deficit during combined heat and drought stress in C3 sunflower and C4 maize varieties with contrasting drought tolerance. Physiol. Plant 2017, 159, 130–147. [Google Scholar] [CrossRef]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 2012, 63, 695–709. [Google Scholar] [CrossRef]
- Feiz, L.; Williams-Carrier, R.; Wostrikoff, K.; Belcher, S.; Barkan, A.; Stern, D.B. Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell 2012, 24, 3435–3446. [Google Scholar] [CrossRef]
- Andersson, I.; Backlund, A. Structure and function of Rubisco. Plant Physiol. Biochem. 2008, 46, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Gujjar, R.S.; Banyen, P.; Chuekong, W.; Worakan, P.; Roytrakul, S.; Supaibulwatana, K. A Synthetic Cytokinin Improves Photosynthesis in Rice under Drought Stress by Modulating the Abundance of Proteins Related to Stomatal Conductance, Chlorophyll Contents, and Rubisco Activity. Plants 2020, 9, 1106. [Google Scholar] [CrossRef] [PubMed]
- Carmo-Silva, A.E.; Salvucci, M.E. The regulatory properties of Rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiol. 2013, 161, 1645–1655. [Google Scholar] [CrossRef]
- Perdomo, J.A.; Degen, G.E.; Worrall, D.; Carmo-Silva, E. Rubisco activation by wheat Rubisco activase isoform 2β is insensitive to inhibition by ADP. Biochem. J. 2019, 476, 2595–2606. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.F.; Way, D.A.; Kubien, D.S. Rubisco, Rubisco activase, and global climate change. J. Exp. Bot. 2008, 59, 1581–1595. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.F.; Kubien, D.S. The temperature response of C(3) and C(4) photosynthesis. Plant Cell Environ. 2007, 30, 1086–1106. [Google Scholar] [CrossRef] [PubMed]
- Hauser, T.; Bhat, J.Y.; Miličić, G.; Wendler, P.; Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Structure and mechanism of the Rubisco-assembly chaperone Raf1. Nat. Struct. Mol. Biol. 2015, 22, 720–728. [Google Scholar] [CrossRef]
- Kolesinski, P.; Belusiak, I.; Czarnocki-Cieciura, M.; Szczepaniak, A. Rubisco Accumulation Factor 1 from Thermosynechococcus elongatus participates in the final stages of ribulose-1,5-bisphosphate carboxylase/oxygenase assembly in Escherichia coli cells and in vitro. FEBS J. 2014, 281, 3920–3932. [Google Scholar] [CrossRef]
- Rouzeau, C.; Dagkesamanskaya, A.; Langer, K.; Bibette, J.; Baudry, J.; Pompon, D.; Anton-Leberre, V. Adaptive response of yeast cells to triggered toxicity of phosphoribulokinase. Res. Microbiol. 2018, 169, 335–342. [Google Scholar] [CrossRef]
- Wilson, R.H.; Hayer-Hartl, M.; Bracher, A. Crystal structure of phosphoribulokinase from Synechococcus sp. strain PCC 6301. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2019, 75, 278–289. [Google Scholar] [CrossRef]
- Harrison, E.P.; Olcer, H.; Lloyd, J.C.; Long, S.P.; Raines, C.A. Small decreases in SBPase cause a linear decline in the apparent RuBP regeneration rate, but do not affect Rubisco carboxylation capacity. J. Exp. Bot. 2001, 52, 1779–1784. [Google Scholar] [CrossRef] [PubMed]
- Lawson, T.; Bryant, B.; Lefebvre, S.; Lloyd, J.C.; Raines, C.A. Decreased SBPase activity alters growth and development in transgenic tobacco plants. Plant Cell Environ. 2006, 29, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Banks, F.M.; Driscoll, S.P.; Parry, M.A.; Lawlor, D.W.; Knight, J.S.; Gray, J.C.; Paul, M.J. Decrease in phosphoribulokinase activity by antisense RNA in transgenic tobacco. Relationship between photosynthesis, growth, and allocation at different nitrogen levels. Plant Physiol. 1999, 119, 1125–1136. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lefebvre, S.; Lawson, T.; Zakhleniuk, O.V.; Lloyd, J.C.; Raines, C.A.; Fryer, M. Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol. 2005, 138, 451–460. [Google Scholar] [CrossRef]
- Rosenthal, D.M.; Locke, A.M.; Khozaei, M.; Raines, C.A.; Long, S.P.; Ort, D.R. Over-expressing the C(3) photosynthesis cycle enzyme Sedoheptulose-1-7 Bisphosphatase improves photosynthetic carbon gain and yield under fully open air CO(2) fumigation (FACE). BMC Plant Biol. 2011, 11, 123. [Google Scholar] [CrossRef]
- Ding, F.; Wang, M.; Zhang, S.; Ai, X. Changes in SBPase activity influence photosynthetic capacity, growth, and tolerance to chilling stress in transgenic tomato plants. Sci. Rep. 2016, 6, 32741. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, L.; Duan, D.H.; Zhang, Y.H.; Huang, S.X.; Chang, Y. Cytotoxicity-Guided Isolation of Two New Phenolic Derivatives from Dryopteris fragrans (L.) Schott. Molecules 2018, 23, 1652. [Google Scholar] [CrossRef]
- Gao, R.; Wang, W.; Huang, Q.; Fan, R.; Wang, X.; Feng, P.; Zhao, G.; Bian, S.; Ren, H.; Chang, Y. Complete chloroplast genome sequence of Dryopteris fragrans (L.) Schott and the repeat structures against the thermal environment. Sci. Rep. 2018, 8, 16635. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, D.; Song, C.; Wang, H.; Tang, X.; Chang, Y. Transcriptomic Analysis and Specific Expression of Transcription Factor Genes in the Root and Sporophyll of Dryopteris fragrans (L.) Schott. Int. J. Mol. Sci. 2020, 21, 7296. [Google Scholar] [CrossRef]
- Guan, Y.; Zhao, X.; Song, N.; Cui, Y.; Chang, Y. Albicanol antagonizes Cd-induced apoptosis through a NO/iNOS-regulated mitochondrial pathway in chicken liver cells. Food Funct. 2021, 12, 1757–1768. [Google Scholar] [CrossRef]
- Hamachi, A.; Nisihara, M.; Saito, S.; Rim, H.; Takahashi, H.; Islam, M.; Uemura, T.; Ohnishi, T.; Ozawa, R.; Maffei, M.E.; et al. Overexpression of geraniol synthase induces heat stress susceptibility in Nicotiana tabacum. Planta 2019, 249, 235–249. [Google Scholar] [CrossRef]
- Jiang, Y.P.; Cheng, F.; Zhou, Y.H.; Xia, X.J.; Mao, W.H.; Shi, K.; Chen, Z.; Yu, J.Q. Cellular glutathione redox homeostasis plays an important role in the brassinosteroid-induced increase in CO2 assimilation in Cucumis sativus. New Phytol. 2012, 194, 932–943. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, T. Isolation and purification of ribulose-5-phosphate kinase from Nicotiana glutinosa. In Methods in Chloroplast Molecular Biology; Elsevier: Amsterdam, The Netherlands, 1982. [Google Scholar]
- Lin, G.Y.; Xun, T.; Dongrui, Z.; Dexin, X.; Jie, L.; Shouyin, L.; Chunhua, S.; Ying, C. Bioinformatics and Expression Pattern Analysis of DfTCP in Dryopteris fragrans. Acta Agriculturae Boreali-Sinica 2020, 35, 43–50. [Google Scholar]
- Tang, X.; Guan, Y.; Chen, L.; Dexin, X.; Song, C.; Liu, L.; Chang, Y. Cloning and Expression of DfDREB Gene from Dryopteris fragrans. Acta Botanica Boreali-Occidentalia Sinica 2020, 7, 1105–1113. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Wallroth, M.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar] [CrossRef]
- Tiwari, M.; Yadav, M.; Singh, B.; Pandey, V.; Nawaz, K.; Bhatia, S. Evolutionary and functional analysis of two-component system in chickpea reveals CaRR13, a TypeB RR, as positive regulator of symbiosis. Plant Biotechnol. J. 2021, 19, 2415–2427. [Google Scholar] [CrossRef]
- Tiwari, M.; Pandey, V.; Singh, B.; Yadav, M.; Bhatia, S. Evolutionary and expression dynamics of LRR-RLKs and functional establishment of KLAVIER homolog in shoot mediated regulation of AON in chickpea symbiosis. Genomics 2021, 113, 4313–4326. [Google Scholar] [CrossRef]
- Mishra, D.; Suri, G.S.; Kaur, G.; Tiwari, M. Comprehensive analysis of structural, functional, and evolutionary dynamics of Leucine Rich Repeats-RLKs in Thinopyrum elongatum. Int. J. Biol. Macromol. 2021, 183, 513–527. [Google Scholar] [CrossRef]
- Pareek, A.; Rathi, D.; Mishra, D.; Chakraborty, S.; Chakraborty, N. Physiological plasticity to high temperature stress in chickpea: Adaptive responses and variable tolerance. Plant Sci. 2019, 289, 110258. [Google Scholar] [CrossRef]
- Tian, Y.; Peng, K.; Bao, Y.; Zhang, D.; Meng, J.; Wang, D.; Wang, X.; Cang, J. Glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase genes of winter wheat enhance the cold tolerance of transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 161, 86–97. [Google Scholar] [CrossRef]
- Shekhar, S.; Mishra, D.; Buragohain, A.K.; Chakraborty, S.; Chakraborty, N. Comparative analysis of phytochemicals and nutrient availability in two contrasting cultivars of sweet potato (Ipomoea batatas L.). Food Chem. 2015, 173, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.; Shekhar, S.; Chakraborty, S.; Chakraborty, N. Wheat 2-Cys peroxiredoxin plays a dual role in chlorophyll biosynthesis and adaptation to high temperature. Plant J. 2021, 105, 1374–1389. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Zhang, Y.L.; Lu, J.; Shao, H.B. Roles of plant soluble sugars and their responses to plant cold stress. Afr. J. Biotechnol. 2009, 8, 2004–2010. [Google Scholar]
- Sun, N.; Song, T.; Ma, Z.; Dong, L.; Zhan, L.; Xing, Y.; Liu, J.; Song, J.; Wang, S.; Cai, H. Overexpression of MsSiR enhances alkali tolerance in alfalfa (Medicago sativa L.) by increasing the glutathione content. Plant Physiol. Biochem. 2020, 154, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.; Shekhar, S.; Agrawal, L.; Chakraborty, S.; Chakraborty, N. Cultivar-specific high temperature stress responses in bread wheat (Triticum aestivum L.) associated with physicochemical traits and defense pathways. Food Chem. 2017, 221, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Rane, J.; Singh, A.K.; Tiwari, M.; Prasad, P.V.V.; Jagadish, S.V.K. Effective Use of Water in Crop Plants in Dryland Agriculture: Implications of Reactive Oxygen Species and Antioxidative System. Front. Plant Sci. 2022, 12, 778270. [Google Scholar] [CrossRef]
- Ye, Z.P. A new model for relationship between irradiance and the rate of photosynthesis in Oryza sativa. Photosynthetica 2007, 45, 637–640. [Google Scholar] [CrossRef]
- Peng, K.; Tian, Y.; Jing, C.; Yu, J.; Tan, Y. Overexpression of TaFBA-A10 from Winter Wheat Enhances Freezing Tolerance in Arabidopsis thaliana. J. Plant Growth Regul. 2022, 41, 314–326. [Google Scholar] [CrossRef]
- Vitlin Gruber, A.; Nisemblat, S.; Azem, A.; Weiss, C. The complexity of chloroplast chaperonins. Trends Plant Sci. 2013, 18, 688–694. [Google Scholar] [CrossRef]
- Mccourt, R.M.; Karol, K.G.; Kaplan, S.; Hoshaw, R.W. Using rbcL sequences to test hypotheses of chloroplast and thallus evolution in conjugating green algae (Zygnematales, Charophyceae). J. Phycol. 1995, 31, 989–995. [Google Scholar] [CrossRef]
- Nozaki, H.; Ito, M.; Uchida, H.; Watanabe, M.M.; Kuroiwa, T. Phylogenetic analysis of Eudorina species (Valvocaceae, Chlorophyta) based on rbcL gene sequences. J. Phycol. 2010, 33, 859–863. [Google Scholar] [CrossRef]
- Bouma, W.L.M.; Ritchie, P.; Perrie, L.R. Phylogeny and generic taxonomy of the New Zealand Pteridaceae ferns from chloroplast rbcL DNA sequences. Aust. Syst. Bot. 2010, 23, 143–151. [Google Scholar] [CrossRef]
- Jeroni, G.; Miquel, R.-C.; Hipólito, M.; Jaume, F. Rubisco activity in Mediterranean species is regulated by the chloroplastic CO2 concentration under water stress. J. Exp. Bot. 2011, 62, 653–665. [Google Scholar] [CrossRef]
- Zhang, N.; Kallis, R.P.; Ewy, R.G.; Portis, A.R. Light modulation of Rubisco in Arabidopsis requires a capacity for redox regulation of the larger Rubisco activase isoform. Proc. Natl. Acad. Sci. USA 2002, 99, 3330–3334. [Google Scholar] [CrossRef]
- Portis, A.R. Rubisco activase—Rubisco’s catalytic chaperone. Photosynth. Res. 2003, 75, 11–27. [Google Scholar] [CrossRef]
- Yin, Z.; Meng, F.; Song, H.; Wang, X.; Yu, X.D. Expression Quantitative Trait Loci Analysis of Two Genes Encoding Rubisco Activase in Soybean. Plant Physiol. 2010, 152, 1625–1637. [Google Scholar] [CrossRef]
- Law, R.D.; Crafts-Brandner, S.J. High temperature stress increases the expression of wheat leaf ribulose-1,5-bisphosphate carboxylase/oxygenase activase protein. Arch. Biochem. Biophys. 2001, 386, 261–267. [Google Scholar] [CrossRef]
- Salesse-Smith, C.E.; Sharwood, R.E.; Busch, F.A.; Kromdijk, J.; Bardal, V.; Stern, D.B. Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat. Plants 2018, 4, 802–810. [Google Scholar] [CrossRef]
- Whitney, S.M.; Birch, R.; Kelso, C.; Beck, J.L.; Kapralov, M.V. Improving recombinant Rubisco biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proc. Natl. Acad. Sci. USA 2015, 112, 3564–3569. [Google Scholar] [CrossRef]
- Spreitzer, R.J. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 2003, 414, 141–149. [Google Scholar] [CrossRef]
- Fristedt, R.; Hu, C.; Wheatley, N.; Roy, L.M.; Wachter, R.M.; Savage, L.; Harbinson, J.; Kramer, D.M.; Merchant, S.S.; Yeates, T.; et al. RAF2 is a RuBisCO assembly factor in Arabidopsis thaliana. Plant J. 2018, 94, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mcclung, C.C.T.R. Identification of an Arabidopsis thaliana ribulose-1,5-bisphosphate carboxylase/oxygenase activase (RCA) minimal promoter regulated by light and the circadian clock. Plant Physiol. 1996, 112, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Watillon, B.; Kettmann, R.; Boxus, P.; Burny, A. Developmental and circadian pattern of rubisco activase mRNA accumulation in apple plants. Plant Mol. Biol. 1993, 23, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef]
- Jordan, D.B.; Ogren, W.L. Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 1983, 227, 425–433. [Google Scholar] [CrossRef]
- Vitlin Gruber, A.; Feiz, L. Rubisco Assembly in the Chloroplast. Front. Mol. Biosci. 2018, 5, 24. [Google Scholar] [CrossRef]




| Gene ID | GenBank | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Purpose |
|---|---|---|---|---|
| DfRbcL | MW524049 | GGAGACTAAAGCAGGTGTTG | GTCAACCCATCTGTCCATAC | qRT-PCR |
| DfRbcS | MW524050 | ACTGGTAAGGACAGGTGGAA | CCATGAAAGCAGGAGCAA | qRT-PCR |
| DfRCA | MW524051 | CGTCCACTGCCACTACTCCT | TCGTAAGCAAGGCCACCC | qRT-PCR |
| DfPRK | MW524052 | CTGCCCTCCCTTCTTCGT | GCAGCACCTCCGAAGACA | qRT-PCR |
| DfRaf | MW524053 | TGGGCTGTGGTTTGACTA | AGGCGAGCATACTTTCTTC | qRT-PCR |
| Df18sRNA | GCTTTCGCAGTAGTTCGTCTTTC | TGGTCCTATTATGTTGGTCTTCGG | qRT-PCR | |
| DfRbcL | ATGTCACCACAAACGGAGACT | TTACAACGTATCAATTGTCTCG | Gene cloning | |
| DfRbcS | ATGGCTACTACTGTAGCTGCT | CTCCCGCACTCATCATGATTG | Gene cloning | |
| DfRCA | ATGGCGTCCACTGCCACTACTC | TCACTTGGGGTCTGCAAAATCA | Gene cloning | |
| DfPRK | ATGAAACGCATTCACATATTC | TCAAGCCTTAGTGGATTGCAAA | Gene cloning | |
| DfRaf | ATGGCGTTTTTGGCGGTCTCA | TTAGTCCCATTCTATAGCATTC | Gene cloning | |
| DfRbcL-EGFP | CTCGGTACCCGGGGATCC ATGTCACCACAAACGGAGACT | GGTGTCGACTCTAGAGGATCC TTACAACGTATCAATTGTCTCG | Gene cloning | |
| DfRbcS-EGFP | CTCGGTACCCGGGGATCC ATGGCTACTACTGTAGCTGCT | GGTGTCGACTCTAGAGGATCC CTCCCGCACTCATCATGATTG | Gene cloning | |
| DfRCA-EGFP | CTCGGTACCCGGGGATCC ATGGCGTCCACTGCCACTACTC | GGTGTCGACTCTAGAGGATCC TCACTTGGGGTCTGCAAAATCA | Gene cloning | |
| DfPRK-EGFP | CTCGGTACCCGGGGATCC ATGAAACGCATTCACATATTC | GGTGTCGACTCTAGAGGATCC TCAAGCCTTAGTGGATTGCAAA | Gene cloning | |
| DfRaf-EGFP | CTCGGTACCCGGGGATCC ATGGCGTTTTTGGCGGTCTCA | GGTGTCGACTCTAGAGGATCC TTAGTCCCATTCTATAGCATTC | Gene cloning | |
| pCAMBIA2301-DfRaf | GGGCATCGATACGGGATCCAT ATGGCGTTTTTGGCGGTCTCA | TCGAGCTCGATGGATCCCGTA TTAGTCCCATTCTATAGCATTC | Gene cloning |
| No. | Sequence (5′ to 3′) | Number | Pfam |
|---|---|---|---|
| Motif1 | ILANRLGLWYEYAPLIPSLIREGFTPPSIEEATGISGVEQNRLVVAAQVR | 50 | Raf1_HTH |
| Motif2 | NRFDPKGAQDLARAMKDFPRRRGDKGWESFD | 31 | Raf1_N |
| Motif3 | YTLPGDCLSFMYYRQSREHKNPSEQRTAALEQALEVAETEKAKNRILEEL | 50 | Raf1_N |
| Motif4 | KEILEAPWECRSEGEFGVVVAEKGWKRWVVLPGWEPVVGLGKG | 43 | RuBisCo_chap_C |
| Motif5 | AFDTGGAELLYEIRLLSASQRAAAARFIV | 29 | - |
| Motif6 | VRVPVVRLKIGEVAEASSVVVLPVCKAEE | 29 | RuBisCo_chap_C |
| Motif7 | NRWYKEEPILVVADRGRKEVEADDGFYLV | 29 | RuBisCo_chap_C |
| Motif8 | GLKVERGSALKERGVKESLGTVVLVVRPPREETDDQLSDED | 41 | RuBisCo_chap_C |
| Motif9 | PSPPPPQQQLYQPFRPPP | 18 | - |
| Motif10 | VVVSFPDARVLPWK | 14 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, C.; Fan, Q.; Tang, Y.; Sun, Y.; Wang, L.; Wei, M.; Chang, Y. Overexpression of DfRaf from Fragrant Woodfern (Dryopteris fragrans) Enhances High-Temperature Tolerance in Tobacco (Nicotiana tabacum). Genes 2022, 13, 1212. https://doi.org/10.3390/genes13071212
Song C, Fan Q, Tang Y, Sun Y, Wang L, Wei M, Chang Y. Overexpression of DfRaf from Fragrant Woodfern (Dryopteris fragrans) Enhances High-Temperature Tolerance in Tobacco (Nicotiana tabacum). Genes. 2022; 13(7):1212. https://doi.org/10.3390/genes13071212
Chicago/Turabian StyleSong, Chunhua, Qi Fan, Yuqing Tang, Yanan Sun, Li Wang, Mingchu Wei, and Ying Chang. 2022. "Overexpression of DfRaf from Fragrant Woodfern (Dryopteris fragrans) Enhances High-Temperature Tolerance in Tobacco (Nicotiana tabacum)" Genes 13, no. 7: 1212. https://doi.org/10.3390/genes13071212
APA StyleSong, C., Fan, Q., Tang, Y., Sun, Y., Wang, L., Wei, M., & Chang, Y. (2022). Overexpression of DfRaf from Fragrant Woodfern (Dryopteris fragrans) Enhances High-Temperature Tolerance in Tobacco (Nicotiana tabacum). Genes, 13(7), 1212. https://doi.org/10.3390/genes13071212







