Verification of Candidate SNP Effects Reveals Two QTLs on BTA7 for Beef Marbling in Two Japanese Black Cattle Populations
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Genotyping Using the Fluidigm Biomark HD System
3.2. Effect of Genotyped SNPs on BMS in Two Populations
3.3. LD Block Analysis
3.4. Statistical Analysis of the Most Significant SNPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wheeler, T.L.; Cundiff, L.V.; Shackelford, S.D.; Koohmaraie, M. Characterization of biological types of cattle (Cycle VI): Carcass, yield, and longissimus palatability traits. J. Anim. Sci. 2004, 82, 1177–1189. [Google Scholar] [CrossRef] [PubMed]
- Oyama, K. Genetic variability of Wagyu cattle estimated by statistical approaches. Anim. Sci. J. 2011, 82, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, S.; Yamada, T.; Sukegawa, S.; Miyake, T.; Fujita, T.; Morita, M. Association of a single nucleotide polymorphism in akirin 2 gene with marbling in Japanese Black beef cattle. BMC Res. Notes 2009, 2, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, T.; Sasaki, S.; Sukegawa, S.; Miyake, T.; Fujita, T.; Kose, H. Novel SNP in 5′ flanking region of EDG1 associated with marbling in Japanese Black beef cattle. J. Anim. Sci. 2009, 80, 486–489. [Google Scholar] [CrossRef]
- Nakajima, A.; Kawaguchi, F.; Uemoto, Y.; Fukushima, M.; Yoshida, E.; Iwamoto, E.; Akiyama, T.; Kohama, N.; Kobayashi, E.; Honda, T.; et al. A genome-wide association study for fat-related traits computed by image analysis in Japanese Black cattle. Anim. Sci. J. 2018, 89, 743–751. [Google Scholar] [CrossRef]
- Hirano, T.; Watanabe, T.; Inoue, K.; Sugimoto, Y. Fine-mapping of a marbling trait to a 2.9-cM region on bovine chromosome 7 in Japanese Black cattle. Anim. Genet. 2007, 39, 79–83. [Google Scholar] [CrossRef]
- Saatchi, M.; Schnabel, R.D.; Taylor, J.F.; Garrick, D.J. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMS Genom. 2014, 15, 442. [Google Scholar] [CrossRef] [Green Version]
- Peters, S.O.; Kizilkaya, K.; Garrick, D.J. Bayesian genome-wide association analysis of growth and yearling ultrasound measures of carcass traits in Brangus heifers. J. Anim. Sci. 2012, 90, 3398–3409. [Google Scholar] [CrossRef]
- Sasazaki, S.; Kawaguchi, F.; Nakajima, A.; Akiyama, T.; Kohama, N.; Yoshida, E.; Kobayashi, E.; Honda, T.; Oyama, K.; Mannen, H. Detection of candidate polymorphisms around the QTL for fat area ratio to rib eye area on BTA7 using whole-genome resequencing in Japanese Black cattle. Anim. Sci. J. 2020, 90, e13335. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [Green Version]
- Springer, T.A. Adhesion receptors of the immune system. Nature 1990, 2, 425–434. [Google Scholar] [CrossRef]
- Rothlein, R.; Dustin, M.L.; Marlin, S.D.; Springer, T.A. A human intercellular adhesion molecule (ICAM1) distinct from LFA-1. J. Immunol. 1986, 15, 1270–1274. [Google Scholar]
- Butini, L.; De Fougerolles, A.R.; Vaccarezza, M.; Graziosi, C.; Cohen, D.I.; Montroni, M.; Springer, T.A.; Pantaleo, G.; Fauci, A.S. Intercellular adhesion molecules (ICAM)-1 ICAM-2 and ICAM-3 function as counter-receptors for lymphocyte function-associated molecule 1 in human immunodeficiency virus-mediated syncytia formation. Eur. J. Immunol. 1994, 9, 2191–2195. [Google Scholar] [CrossRef]
- Yang, L.; Froio, R.M.; Sciuto, T.E.; Dvorak, A.M.; Alon, R.; Luscinskas, F.W. ICAM1 regulates neutrophil adhesion and transcellular migration of TNF-alpha-activated vascular endothelium under flow. Blood 2005, 106, 584–592. [Google Scholar] [CrossRef] [Green Version]
- Langer, H.F.; Chavakis, T. Leukocyte-endothelial interactions in inflammation. J. Cell Mol. Med. 2009, 13, 1211–1220. [Google Scholar] [CrossRef]
- Rosen, E.D.; Walkey, C.J.; Puigserver, P.; Spiegelman, B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000, 14, 1293–1307. [Google Scholar] [CrossRef]
- Mandrup, S.; Lane, M.D. Regulating adipogenesis. J. Biol. Chem. 1997, 28, 5367–5370. [Google Scholar] [CrossRef] [Green Version]
- Engelman, J.A.; Lisanti, M.P.; Scherer, P.E. Specific inhibitors of p38 mitogen-activated protein kinase block 3T3-L1 adipogenesis. J. Biol. Chem. 1998, 273, 32111–32120. [Google Scholar] [CrossRef] [Green Version]
- Bost, F.; Aouadi, M.; Caron, L.; Binétruy, B. The role of MAPKs in adipocyte differentiation and obesity. Biochimie 2005, 87, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.F.; Zhu, H.; Li, X.M.; Yang, F.; Chen, J.D.; Tang, B.; Sun, H.G.; Chu, Y.N.; Zheng, R.X.; Liu, Y.L.; et al. Intercellular adhesion molecule-1 inhibits osteogenic differentiation of mesenchymal stem cells and impairs bio-scaffold-mediated bone regeneration in vivo. Tissue Eng. Part A 2014, 20, 2768–2782. [Google Scholar] [CrossRef] [Green Version]
- Merrick, D.; Sakers, A.; Irgebay, Z.; Okada, C.; Calvert, C.; Morley, M.; Percec, I.; Seale, P. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 2019, 364, 6438. [Google Scholar] [CrossRef]
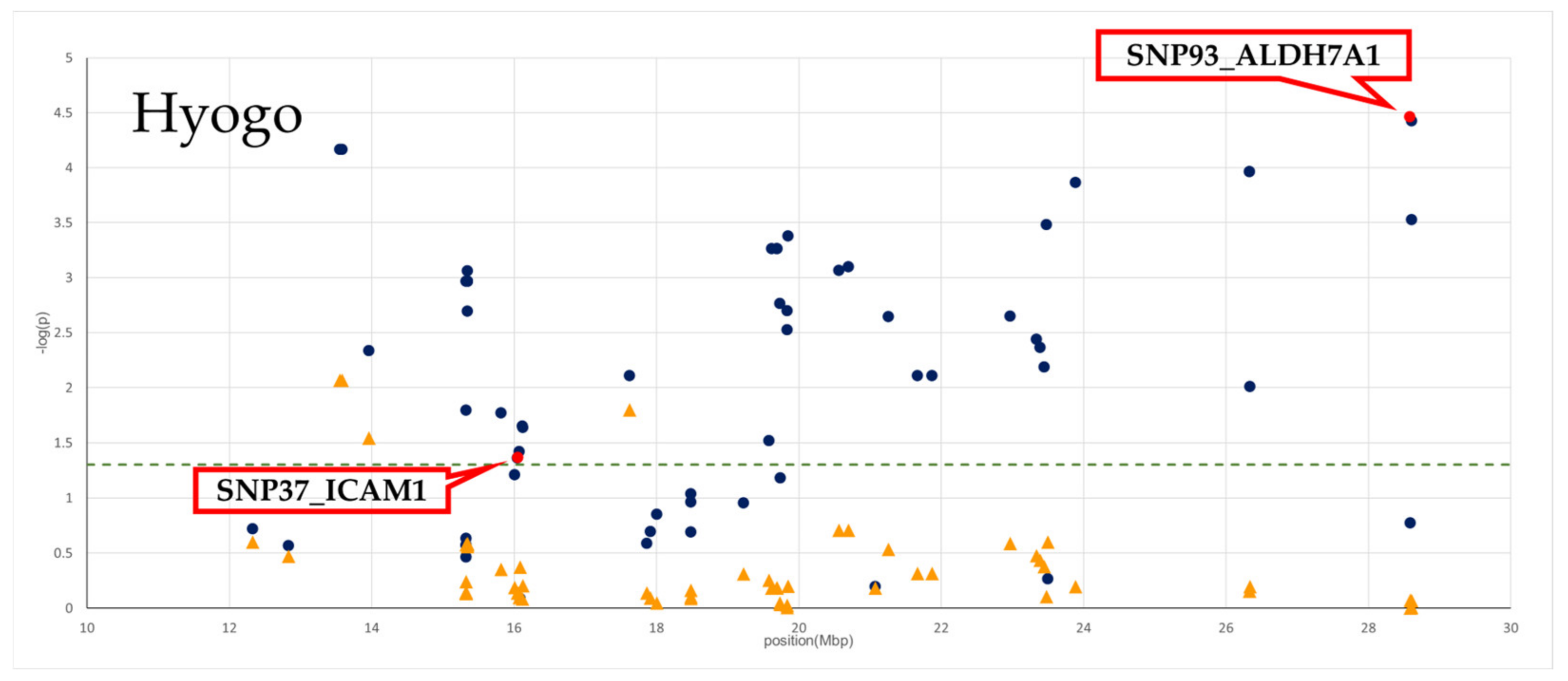

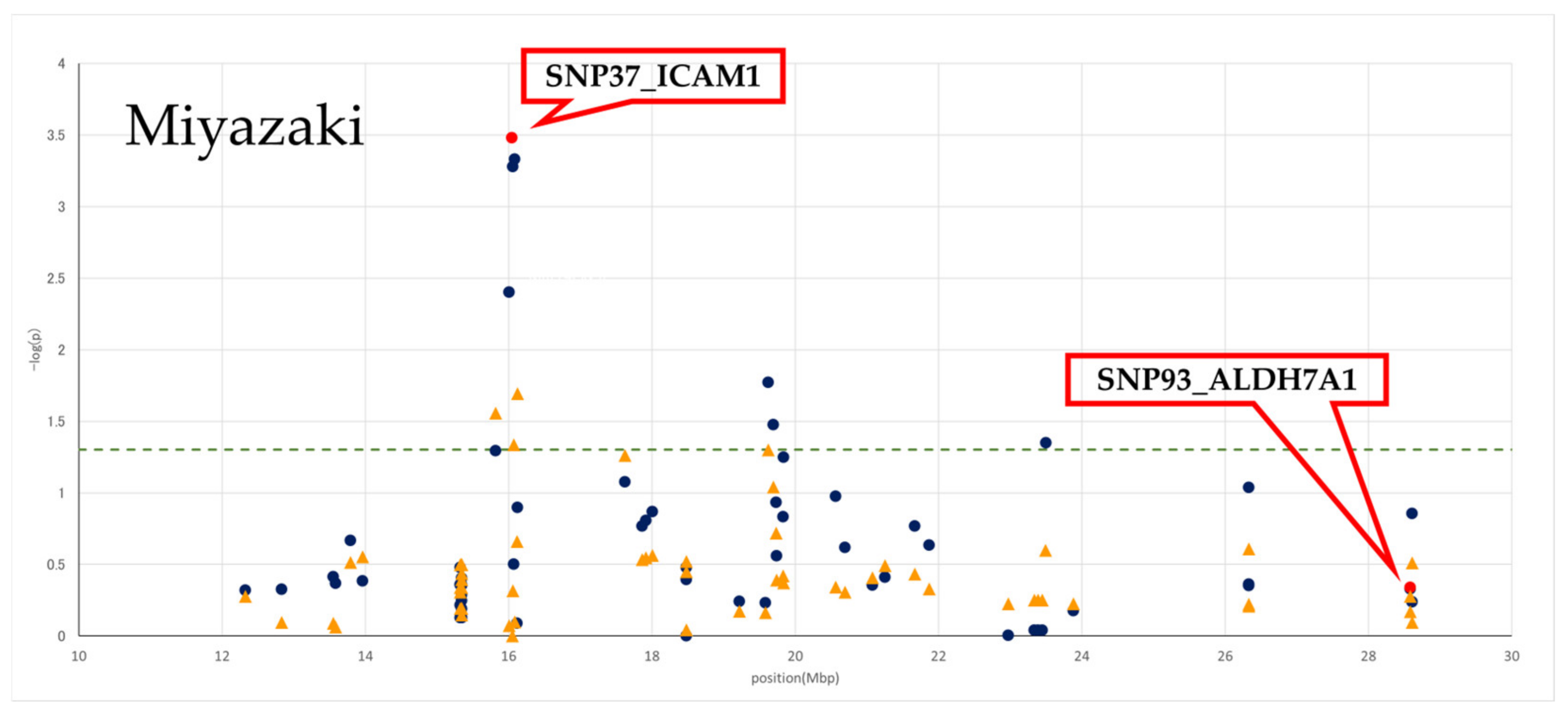

| Population | N | Genotype Frequency (the Number of Animals) | Allele Frequency | BMS (Least Square Mean) ± SE | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | A | C | AA | AC | CC | |||
| Hyogo | 441 | 0.03 | 0.42 | 0.55 | 0.24 | 0.76 | 6.86 a | 5.92 a | 5.25 b | 3.75 × 10−5 |
| (15) | (184) | (242) | ±0.45 | ±0.17 | ±0.15 | |||||
| Miyazaki | 529 | 0.01 | 0.24 | 0.74 | 0.13 | 0.87 | 6.50 | 6.30 | 6.07 | 0.459 |
| (6) | (129) | (394) | ±0.80 | ±0.22 | ±0.16 | |||||
| Population | N | Genotype Frequency (the Number of Animals) | Allele Frequency | BMS (Least Square Mean) ± SE | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | A | G | AA | AG | GG | |||
| Hyogo | 441 | 0.46 | 0.47 | 0.07 | 0.69 | 0.31 | 5.35 | 5.63 | 6.17 | 0.043 |
| (202) | (208) | (31) | ±0.17 | ±0.16 | ±0.33 | |||||
| Miyazaki | 529 | 0.34 | 0.56 | 0.10 | 0.62 | 0.38 | 5.64 b | 6.37 a | 6.05 ab | 3.33 × 10−4 |
| (179) | (297) | (53) | ±0.20 | ±0.17 | ±0.30 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasazaki, S.; Yamamoto, R.; Toyomoto, S.; Kondo, H.; Akiyama, T.; Kohama, N.; Yoshida, E.; Kawaguchi, F.; Oyama, K.; Mannen, H. Verification of Candidate SNP Effects Reveals Two QTLs on BTA7 for Beef Marbling in Two Japanese Black Cattle Populations. Genes 2022, 13, 1190. https://doi.org/10.3390/genes13071190
Sasazaki S, Yamamoto R, Toyomoto S, Kondo H, Akiyama T, Kohama N, Yoshida E, Kawaguchi F, Oyama K, Mannen H. Verification of Candidate SNP Effects Reveals Two QTLs on BTA7 for Beef Marbling in Two Japanese Black Cattle Populations. Genes. 2022; 13(7):1190. https://doi.org/10.3390/genes13071190
Chicago/Turabian StyleSasazaki, Shinji, Raito Yamamoto, Shintaro Toyomoto, Hina Kondo, Takayuki Akiyama, Namiko Kohama, Emi Yoshida, Fuki Kawaguchi, Kenji Oyama, and Hideyuki Mannen. 2022. "Verification of Candidate SNP Effects Reveals Two QTLs on BTA7 for Beef Marbling in Two Japanese Black Cattle Populations" Genes 13, no. 7: 1190. https://doi.org/10.3390/genes13071190
APA StyleSasazaki, S., Yamamoto, R., Toyomoto, S., Kondo, H., Akiyama, T., Kohama, N., Yoshida, E., Kawaguchi, F., Oyama, K., & Mannen, H. (2022). Verification of Candidate SNP Effects Reveals Two QTLs on BTA7 for Beef Marbling in Two Japanese Black Cattle Populations. Genes, 13(7), 1190. https://doi.org/10.3390/genes13071190





