Identification and Characterization Roles of Phytoene Synthase (PSY) Genes in Watermelon Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Isolation and cDNA Synthesis
2.3. Identification and Cloning of ClPSY Genes
2.4. Expression Pattern Analysis of ClPSYs
2.5. ClPSY Functional Complementation in E. coli
2.6. Phylogenetic Tree Construction and Genome Collinearity Analysis among Cucurbit Crops
2.7. Public RNA-Seq Data Analysis
3. Results
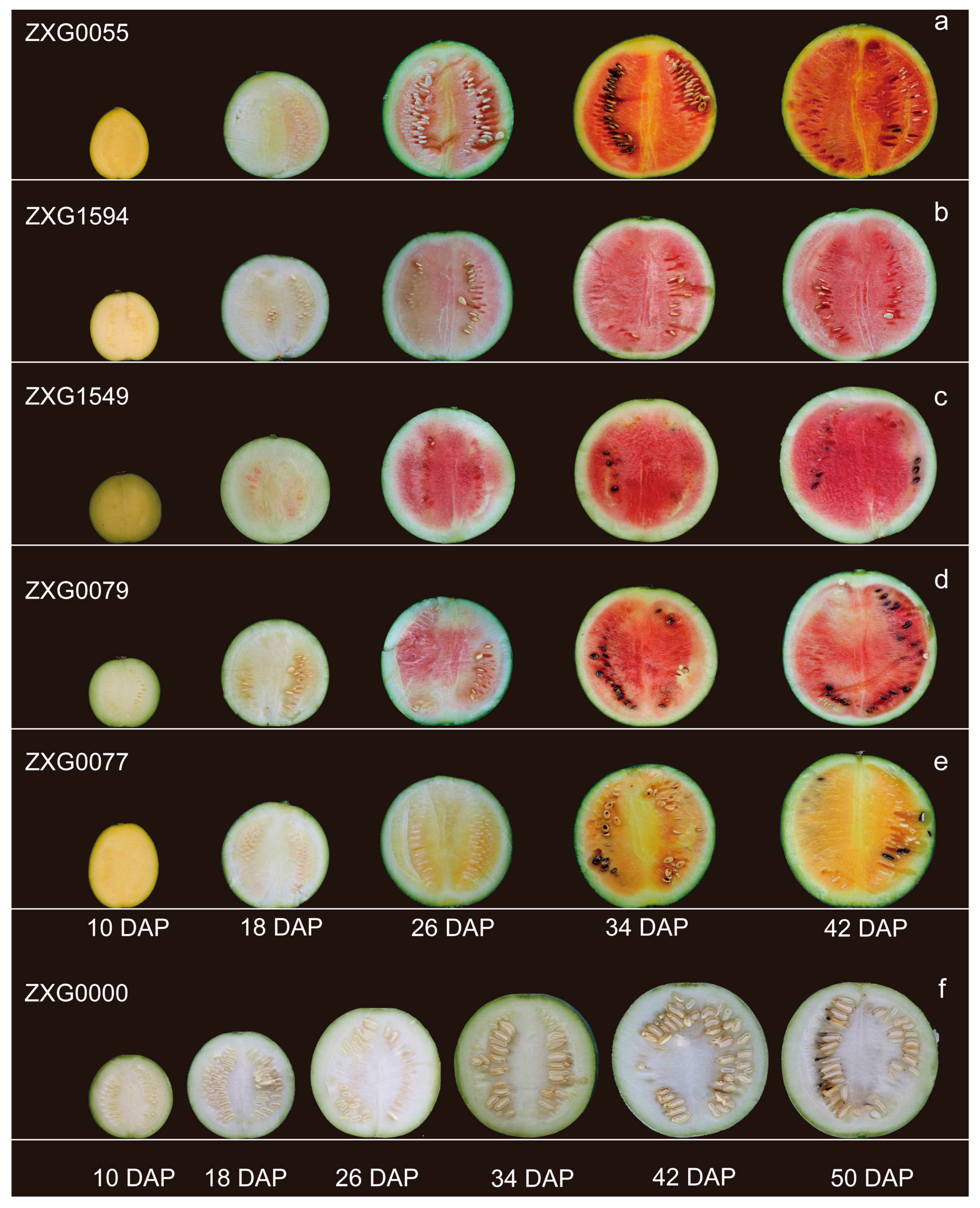
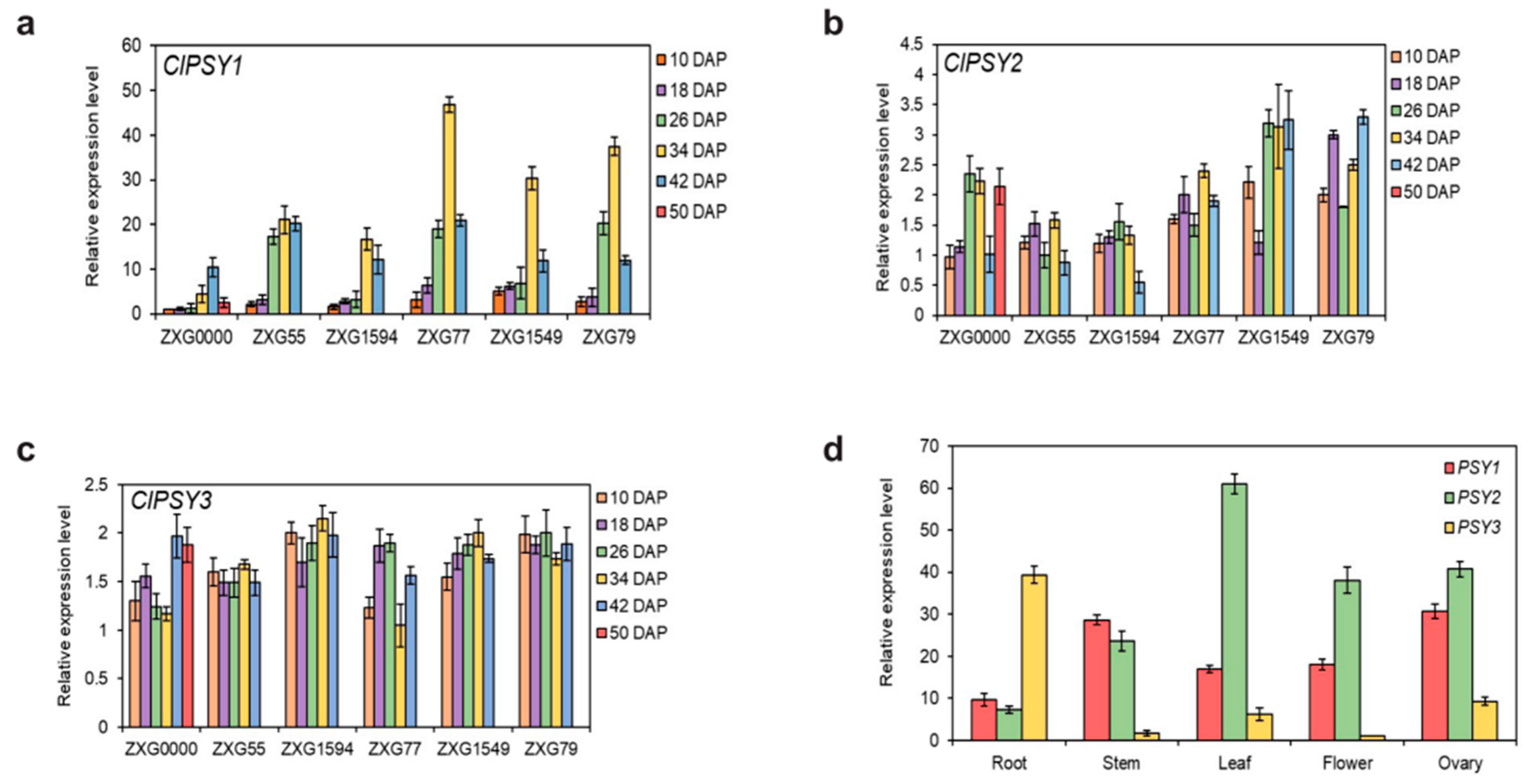
3.1. Sequence Analysis of ClPSY Genes and Genes Expression during Watermelon Flesh Development
3.2. Functional Analyses of ClPSY Proteins in Watermelon
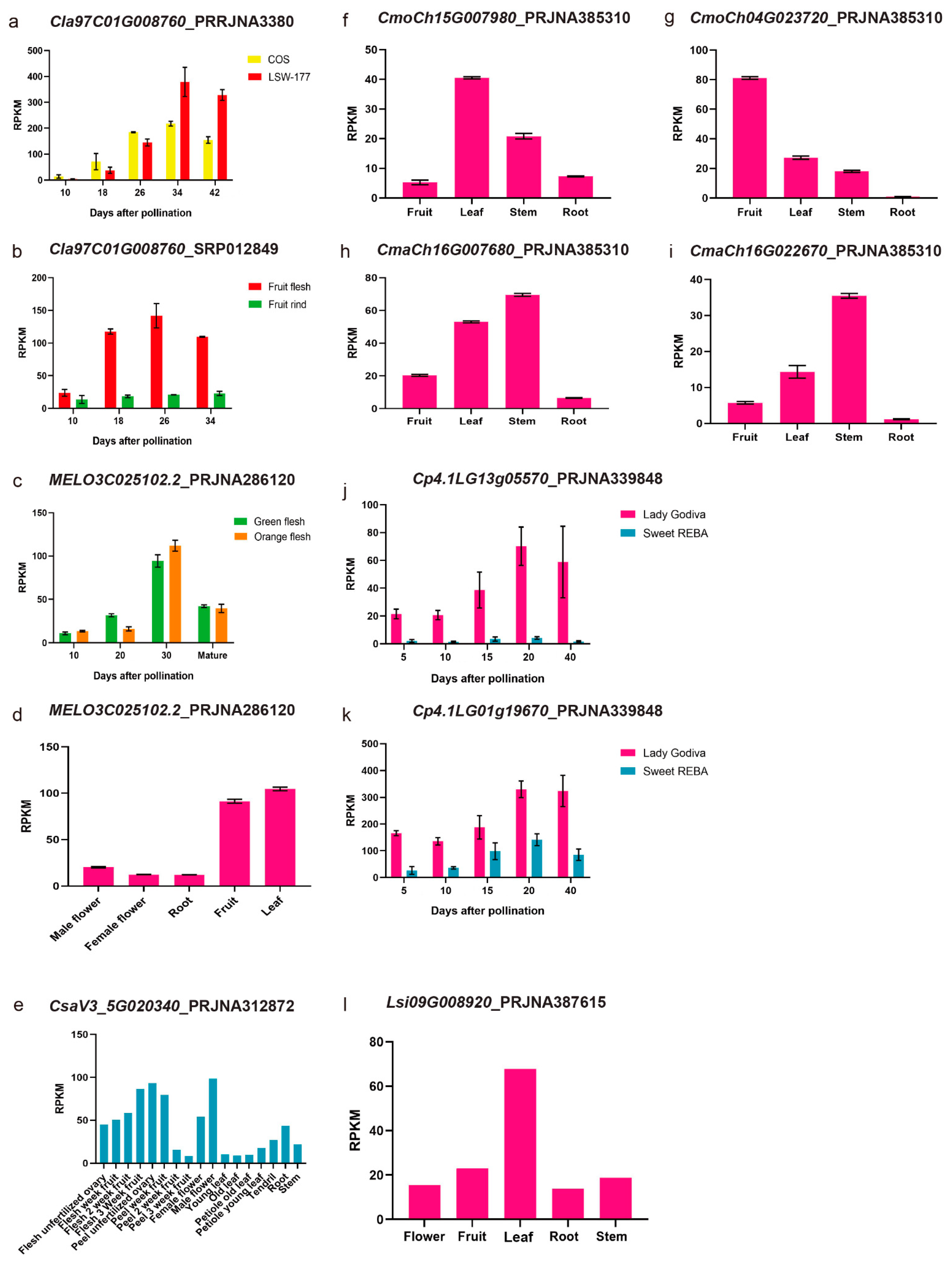
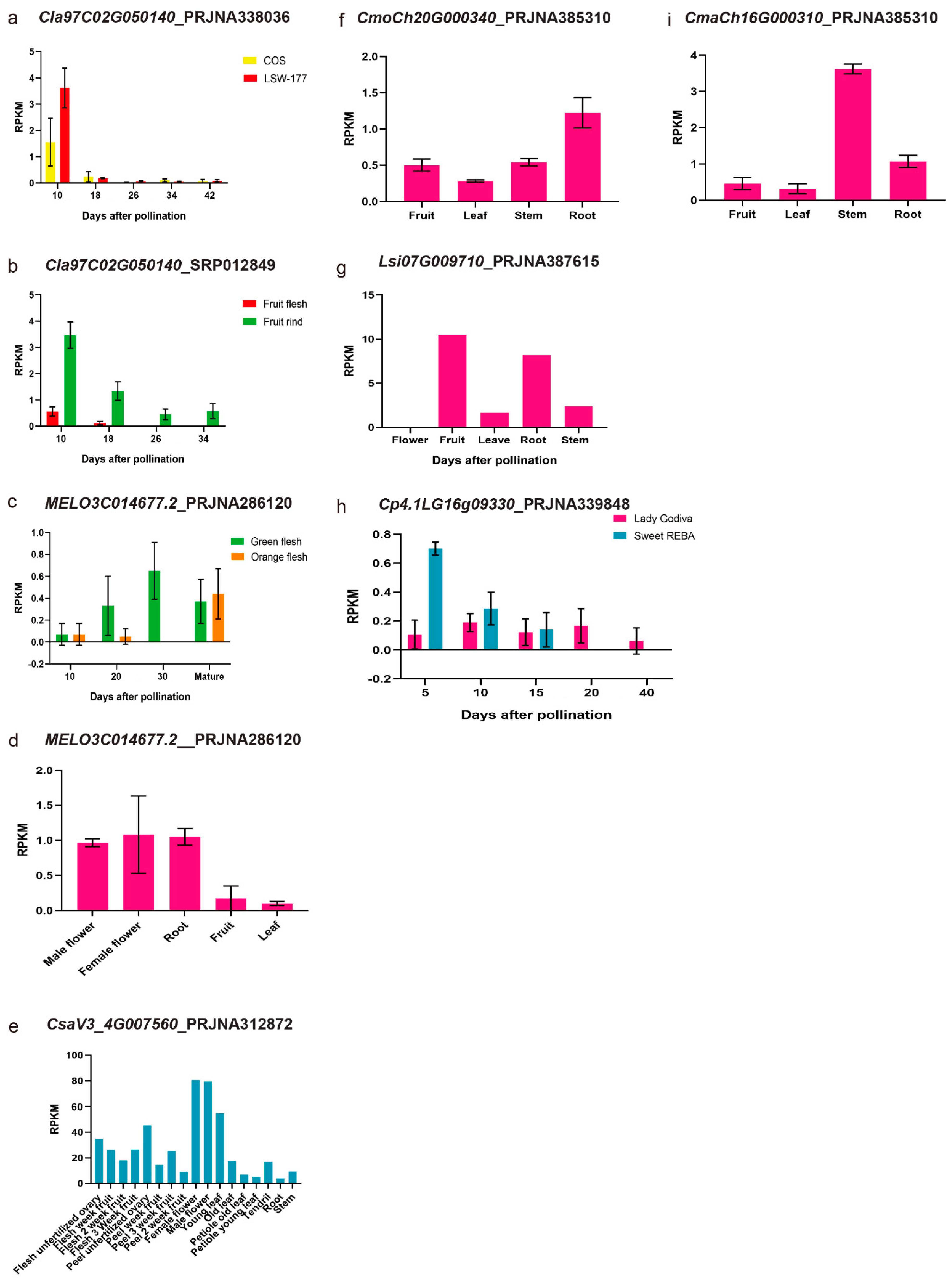
3.3. Synteny Analysis of PSY Members among Different Cucurbit Crops and Their Expression Patterns with the RNA-Seq Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid Metabolism and Regulation in Horticultural Crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Concepcion, M.; Avalos, J.; Bonet, M.L.; Boronat, A.; Gomez-Gomez, L.; Hornero-Mendez, D.; Limon, M.C.; Meléndez-Martínez, A.J.; Olmedilla-Alonso, B.; Palou, A.; et al. A Global Perspective on Carotenoids: Metabolism, Biotechnology, and Benefits for Nutrition and Health. Prog. Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cazzonelli, C.I.; Pogson, B.J. Source to Sink: Regulation of Carotenoid Biosynthesis in Plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Welsch, R.; Arango, J.; Bär, C.; Salazar, B.; Al-Babili, S.; Beltrán, J.; Chavarriaga, P.; Ceballos, H.; Tohme, J.; Beyera, P. Provitamin a Accumulation in Cassava (Manihot esculenta) Roots Driven by a Single Nucleotide Polymorphism in a Phytoene Synthase Gene. Plant Cell 2010, 22, 3348–3356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuliano, G. Provitamin A Biofortification of Crop Plants: A Gold Rush with Many Miners. Curr. Opin. Biotechnol. 2017, 44, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Kobayashi, K.; Nagata, N.; Masuda, T.; Wada, H. Digalactosyldiacylglycerol Is Essential for Organization of the Membrane Structure in Etioplasts. Plant Physiol. 2018, 177, 1487–1497. [Google Scholar] [CrossRef]
- Obrero, A.; González-Verdejo, C.I.; Román, B.; Gómez, P.; Die, J.V.; Ampomah-Dwamena, C. Identification, Cloning, and Expression Analysis of Three Phytoene Synthase Genes from Cucurbita Pepo. Biol. Plant 2015, 59, 201–210. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Driedonks, N.; Lewis, D.; Shumskaya, M.; Chen, X.; Wurtzel, E.T.; Espley, R.V.; Allan, A.C. The Phytoene Synthase Gene Family of Apple (Malus x Domestica) and Its Role in Controlling Fruit Carotenoid Content. BMC Plant Biol. 2015, 15, 185. [Google Scholar] [CrossRef] [Green Version]
- Fantini, E.; Falcone, G.; Frusciante, S.; Giliberto, L.; Giuliano, G. Dissection of Tomato Lycopene Biosynthesis through Virus-Induced Gene Silencing. Plant Physiol. 2013, 163, 986–998. [Google Scholar] [CrossRef]
- Peng, G.; Wang, C.; Song, S.; Fu, X.; Azam, M.; Grierson, D.; Xu, C. The Role of 1-Deoxy-d-Xylulose-5-Phosphate Synthase and Phytoene Synthase Gene Family in Citrus Carotenoid Accumulation. Plant Physiol. Biochem. 2013, 71, 67–76. [Google Scholar] [CrossRef]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in Planta Gene Targeting in Tomato Using Geminiviral Replicons and the CRISPR/Cas9 System. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, P.; Li, N.; Liu, H.; Gu, H.; Zhao, W.E. Changes in Carotenoid Profiles and in the Expression Pattern of the Genes in Carotenoid Metabolisms during Fruit Development and Ripening in Four Watermelon Cultivars. Food Chem. 2015, 174, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, S.; Gao, P.; Luan, F.; Davis, A.R. Developmental Changes in Gene Expression Drive Accumulation of Lycopene and β-Carotene in Watermelon. J. Am. Soc. Hortic. Sci. 2016, 141, 434–443. [Google Scholar] [CrossRef] [Green Version]
- Van Wijk, K.J.; Kessler, F. Plastoglobuli: Plastid Microcompartments with Integrated Functions in Metabolism, Plastid Developmental Transitions, and Environmental Adaptation. Annu. Rev. Plant Biol. 2017, 68, 253–289. [Google Scholar] [CrossRef] [PubMed]
- Shumskaya, M.; Bradbury, L.M.T.; Monaco, R.R.; Wurtzela, E.T. Plastid Localization of the Key Carotenoid Enzyme Phytoene Synthase Is Altered by Isozyme, Allelic Variation, and Activity. Plant Cell 2012, 24, 3725–3741. [Google Scholar] [CrossRef] [Green Version]
- Lätari, K.; Wüst, F.; Hübner, M.; Schaub, P.; Beisel, K.G.; Matsubara, S.; Beyer, P.; Welsch, R. Tissue-Specific Apocarotenoid Glycosylation Contributes to Carotenoid Homeostasis in Arabidopsis Leaves. Plant Physiol. 2015, 168, 1550–1562. [Google Scholar] [CrossRef]
- Martel, C.; Vrebalov, J.; Tafelmeyer, P.; Giovannoni, J.J. The Tomato MADS-Box Transcription Factor RIPENING INHIBITOR Interacts with Promoters Involved in Numerous Ripening Processes in a COLORLESS NONRIPENING-Dependent Manner. Plant Physiol. 2011, 157, 1568–1579. [Google Scholar] [CrossRef] [Green Version]
- Xiong, C.; Luo, D.; Lin, A.; Zhang, C.; Shan, L.; He, P.; Li, B.; Zhang, Q.; Hua, B.; Yuan, Z.; et al. A Tomato B-Box Protein SlBBX20 Modulates Carotenoid Biosynthesis by Directly Activating PHYTOENE SYNTHASE 1, and Is Targeted for 26S Proteasome-Mediated Degradation. New Phytol. 2019, 221, 279–294. [Google Scholar] [CrossRef] [Green Version]
- Ampomah-Dwamena, C.; Thrimawithana, A.H.; Dejnoprat, S.; Lewis, D.; Espley, R.V.; Allan, A.C. A Kiwifruit (Actinidia deliciosa) R2R3-MYB Transcription Factor Modulates Chlorophyll and Carotenoid Accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The Citrus Transcription Factor CsMADS6 Modulates Carotenoid Metabolism by Directly Regulating Carotenogenic Genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef] [Green Version]
- Enfissi, E.M.A.; Nogueira, M.; Bramley, P.M.; Fraser, P.D. The Regulation of Carotenoid Formation in Tomato Fruit. Plant J. 2017, 89, 774–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arango, J.; Jourdan, M.; Geoffriau, E.; Beyer, P.; Welsch, R. Carotene Hydroxylase Activity Determines the Levels of Both α-Carotene and Total Carotenoids in Orange Carrots. Plant Cell 2014, 26, 2223–2233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Fish, T.; Liu, J.; Thannhauser, T.W.; Li, L. Arabidopsis OR Proteins Are the Major Posttranscriptional Regulators of Phytoene Synthase in Controlling Carotenoid Biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 3558–3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Kim, H.S.; Jung, Y.J.; Kim, S.H.; Ji, C.Y.; Wang, Z.; Jeong, J.C.; Lee, H.S.; Lee, S.Y.; Kwak, S.S. Orange Protein Has a Role in Phytoene Synthase Stabilization in Sweetpotato. Sci. Rep. 2016, 6, 33563. [Google Scholar] [CrossRef] [PubMed]
- Bou-Torrent, J.; Toledo-Ortiz, G.; Ortiz-Alcaide, M.; Cifuentes-Esquivel, N.; Halliday, K.J.; Martinez-García, J.F.; Rodriguez-Concepcion, M. Regulation of Carotenoid Biosynthesis by Shade Relies on Specific Subsets of Antagonistic Transcription Factors and Cofactors. Plant Physiol. 2015, 169, 1584–1594. [Google Scholar] [CrossRef]
- Tzuri, G.; Zhou, X.; Chayut, N.; Yuan, H.; Portnoy, V.; Meir, A.; Sa’Ar, U.; Baumkoler, F.; Mazourek, M.; Lewinsohn, E.; et al. A “golden” SNP in CmOr Governs the Fruit Flesh Color of Melon (Cucumis melo). Plant J. 2015, 82, 267–279. [Google Scholar] [CrossRef]
- Chayut, N.; Yuan, H.; Ohali, S.; Meir, A.; Sa’ar, U.; Tzuri, G.; Zheng, Y.; Mazourek, M.; Gepstein, S.; Zhou, X.; et al. Distinct Mechanisms of the ORANGE Protein in Controlling Carotenoid Flux. Plant Physiol. 2017, 173, 376–389. [Google Scholar] [CrossRef]
- Welsch, R.; Zhou, X.; Yuan, H.; Álvarez, D.; Sun, T.; Schlossarek, D.; Yang, Y.; Shen, G.; Zhang, H.; Rodriguez-Concepcion, M.; et al. Clp Protease and OR Directly Control the Proteostasis of Phytoene Synthase, the Crucial Enzyme for Carotenoid Biosynthesis in Arabidopsis. Mol. Plant 2018, 11, 149–162. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.; Liu, S.; Gao, P.; Liu, H.; Wang, X.; Luan, F.; Zhang, Q.; Dai, Z. Expression of ClPAP and ClPSY1 in Watermelon Correlates with Chromoplast Differentiation, Carotenoid Accumulation, and Flesh Color Formation. Sci. Hortic. (Amst.) 2020, 270, 109437. [Google Scholar] [CrossRef]
- Kong, Q.; Yuan, J.; Gao, L.; Zhao, S.; Jiang, W.; Huang, Y.; Bie, Z. Identification of Suitable Reference Genes for Gene Expression Normalization in QRT-PCR Analysis in Watermelon. PLoS ONE 2014, 9, e90612. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; Gonźalez, V.M.; Heńaff, E.; Camara, F.; Cozzuto, L.; Lowy, E.; et al. The Genome of Melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Li, H.; Huang, W.; Xu, Y.; Zhou, Q.; Wang, S.; Ruan, J.; Huang, S.; Zhang, Z. A Chromosome-Scale Genome Assembly of Cucumber (Cucumis sativus L.). Gigascience 2019, 8, giz072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, J.; Liu, X.; Shen, D.; Miao, H.; Xie, B.; Li, X.; Zeng, P.; Wang, S.; Shang, Y.; Gu, X.; et al. A Genomic Variation Map Provides Insights into the Genetic Basis of Cucumber Domestication and Diversity. Nat. Genet. 2013, 45, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z.; et al. Karyotype Stability and Unbiased Fractionation in the Paleo-Allotetraploid Cucurbita Genomes. Mol. Plant 2017, 10, 1293–1306. [Google Scholar] [CrossRef] [Green Version]
- Montero-Pau, J.; Blanca, J.; Bombarely, A.; Ziarsolo, P.; Esteras, C.; Martí-Gómez, C.; Ferriol, M.; Gómez, P.; Jamilena, M.; Mueller, L.; et al. De Novo Assembly of the Zucchini Genome Reveals a Whole-Genome Duplication Associated with the Origin of the Cucurbita Genus. Plant Biotechnol. J. 2018, 16, 1161–1171. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Shamimuzzaman, M.; Sun, H.; Salse, J.; Sui, X.; Wilder, A.; Wu, Z.; Levi, A.; Xu, Y.; Ling, K.S.; et al. The Bottle Gourd Genome Provides Insights into Cucurbitaceae Evolution and Facilitates Mapping of a Papaya Ring-Spot Virus Resistance Locus. Plant J. 2017, 92, 963–975. [Google Scholar] [CrossRef] [Green Version]
- Barrera-Redondo, J.; Ibarra-Laclette, E.; Vázquez-Lobo, A.; Gutiérrez-Guerrero, Y.T.; Sánchez de la Vega, G.; Piñero, D.; Montes-Hernández, S.; Lira-Saade, R.; Eguiarte, L.E. The Genome of Cucurbita Argyrosperma (Silver-Seed Gourd) Reveals Faster Rates of Protein-Coding Gene and Long Noncoding RNA Turnover and Neofunctionalization within Cucurbita. Mol. Plant 2019, 12, 506–520. [Google Scholar] [CrossRef] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the Functional Annotation of Proteins. Nucleic Acids Res. 2011, 39, 225–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebastian, P.; Schaefer, H.; Telford, I.R.H.; Renner, S.S. Cucumber (Cucumis sativus) and Melon (C. melo) Have Numerous Wild Relatives in Asia and Australia, and the Sister Species of Melon Is from Australia. Proc. Natl. Acad. Sci. USA 2010, 107, 14269–14273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Koo, D.H.; Li, D.; Zhang, T.; Jiang, J.; Luan, F.; Renner, S.S.; Hénaff, E.; Sanseverino, W.; Garcia-Mas, J.; et al. Next-Generation Sequencing, FISH Mapping and Synteny-Based Modeling Reveal Mechanisms of Decreasing Dysploidy in Cucumis. Plant J. 2014, 77, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Pan, Y.; Wen, C.; Li, Y.; Liu, X.; Zhang, X.; Behera, T.K.; Xing, G.; Weng, Y. Integrated Analysis in Bi-Parental and Natural Populations Reveals CsCLAVATA3 (CsCLV3) Underlying Carpel Number Variations in Cucumber. Theor. Appl. Genet. 2016, 129, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, Y.; McGregor, C.; Liu, S.; Luan, F.; Gao, M.; Weng, Y. Genetic Architecture of Fruit Size and Shape Variation in Cucurbits: A Comparative Perspective. Theor. Appl. Genet. 2020, 133, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gao, M.; Liang, X.; Xu, M.; Liu, X.; Zhang, Y.; Liu, X.; Liu, J.; Gao, Y.; Qu, S.; et al. Quantitative Trait Loci for Seed Size Variation in Cucurbits—A Review. Front. Plant Sci. 2020, 11, 304. [Google Scholar] [CrossRef] [PubMed]





| Gene | Gene ID | Chromosome | Positions |
|---|---|---|---|
| ClPSY1 | Cla97C01G008760 | 1 | Chr01: 9448426 ~ 9451311 (+) |
| ClPSY2 | Cla97C02G050140 | 7 | Chr07: 25039090 ~ 25042269 (+) |
| ClPSY3 | Cla97C07G137500 | 2 | Chr02: 37419674 ~ 37421665 (−) |
| ClYLS8 | Cla97C02G038590 | 2 | Chr02: 22823405 ~ 22824590 (−) |
| Primer Name | Amplicon Size (bp) | Primer Sequence (5′-3′) |
|---|---|---|
| ClPSY1 | 1266 | F: GGAATTCATGTCTTTTGCTCCTTCGTTGG R: CCTCGAGAATTCATGAAGGGCCAAG |
| ClPSY2 | 1194 | F: GGAATTCATGTCTGGTGTGAATGCCAACTCTC R: CCTCGAGCTATCTTGTTACCAAATCTTT |
| ClPSY3 | 1068 | F: CGGGATCCATGAGCAAAAGTGGGGTAATT R: CCTCGAGTCAATGGAAGACTAGACTGGGTGCC |
| qPSY1 | 179 | F: TAAGTTTCCAGTTGATATTCAGCCG R: GTGCTTGCTTGGGAGTCAGG |
| qPSY2 | 132 | F: CCTGTCATGGGATTGGCACC R: CTCTTCCCCTCCTAGCATCTTCTCC |
| qPSY3 | 118 | F: GCACCTGATTCTTCACTTCCTACTC R: ATCCTACCCCTTATAGCATCCTCTC |
| ClYLS8 | 74 | F: AGAACGGCTTGTGGTCATTC R: GAGGCCAACACTTCATCCAT |
| Species | Gene ID | Position |
|---|---|---|
| Cucumis melo | MELO3C025102.2 | Chr09: 14553133 ~ 14556862 (+) |
| MELO3C014677.2 | Chr05: 487434 ~ 489932 (+) | |
| MELO3C016185.2 | Chr07: 20315779 ~ 20319205 (+) | |
| Cucumis sativus | CsaV3_5G020340 | Chr05: 15246891 ~ 15249568 (+) |
| CsaV3_4G023380 | Chr04: 13627834 ~ 13631557 (+) | |
| CsaV3_4G007560 | Chr04: 5185258 ~ 5189102 (−) | |
| Cucumis sativus var. hardwickii | CSPI05G13040 | Chr05: 12570152 ~ 12574113 (+) |
| CSPI04G07340 | Chr04: 5188671 ~ 5192373 (−) | |
| CSPI04G13030 | Chr04: 11240846 ~ 11244106 (+) | |
| Lagenaria siceraria | Lsi07G008190 | Chr07: 9123302 ~ 9126554 (−) |
| Lsi09G008920 | Chr09: 10525942 ~ 10528939 (+) | |
| Lsi07G008170 | Chr07: 9114307 ~ 9117400 (−) | |
| Lsi10G009710 | Chr10: 14062470 ~ 14067420 (+) | |
| Cucurbita maxima var. Rimu | CmaCh16G004720 | Chr16: 2390509 ~ 2394475 (+) |
| CmaCh15G007680 | Chr15: 3777993 ~ 3782451 (−) | |
| CmaCh04G022670 | Chr04: 15833307 ~ 15835991 (+) | |
| CmaCh04G006250 | Chr04: 3203989 ~ 3206323 (−) | |
| CmaCh20G000310 | Chr20: 140674 ~ 142255 (+) | |
| Cucurbita moschata var. Rifu | CmoCh04G023720 | Chr04: 17691476 ~ 17694558 (+) |
| CmoCh16G005120 | Chr16: 2472841 ~ 2477006 (+) | |
| CmoCh15G007980 | Chr15: 3929562 ~ 3934079 (−) | |
| CmoCh04G006730 | Chr04: 3346882 ~ 3349011 (−) | |
| CmoCh20G000340 | Chr20: 224511 ~ 225982 (+) | |
| Cucurbita pepo subsp. pepo | Cp4.1LG14g03180 | Chr14: 2471728 ~ 2475409 (+) |
| Cp4.1LG13g05570 | Chr13: 5416613 ~ 5421307 (+) | |
| Cp4.1LG01g19670 | Chr01: 16838967 ~ 16841930 (+) | |
| Cp4.1LG01g00990 | Chr01: 3271567 ~ 3274741 (−) | |
| Cp4.1LG16g09330 | Chr16: 8547559 ~ 8548969 (−) | |
| Cucurbita argyrosperma | Carg18294 | Scaffold_120: 491768 ~ 494735 (−) |
| Carg06947 | Scaffold_065: 466719 ~ 468129 (−) | |
| Carg13119 | Scaffold_101: 425459 ~ 427786 (+) | |
| Carg01435 | Scaffold_004: 945560 ~ 949123 (+) | |
| Carg11395 | Scaffold_025: 739796 ~ 743637 (+) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, X.; Gao, P.; Luan, F.; Liu, S. Identification and Characterization Roles of Phytoene Synthase (PSY) Genes in Watermelon Development. Genes 2022, 13, 1189. https://doi.org/10.3390/genes13071189
Fang X, Gao P, Luan F, Liu S. Identification and Characterization Roles of Phytoene Synthase (PSY) Genes in Watermelon Development. Genes. 2022; 13(7):1189. https://doi.org/10.3390/genes13071189
Chicago/Turabian StyleFang, Xufeng, Peng Gao, Feishi Luan, and Shi Liu. 2022. "Identification and Characterization Roles of Phytoene Synthase (PSY) Genes in Watermelon Development" Genes 13, no. 7: 1189. https://doi.org/10.3390/genes13071189
APA StyleFang, X., Gao, P., Luan, F., & Liu, S. (2022). Identification and Characterization Roles of Phytoene Synthase (PSY) Genes in Watermelon Development. Genes, 13(7), 1189. https://doi.org/10.3390/genes13071189






