Effect of Developmental Stages on Genes Involved in Middle and Downstream Pathway of Volatile Terpene Biosynthesis in Rose Petals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Phylogenetic Analysis of RcTPT Gene Family
2.2. Re-Identification and Sequence Analysis of the RcTPS Gene Family
2.3. Chromosomal Localization and Collinearity Analyses
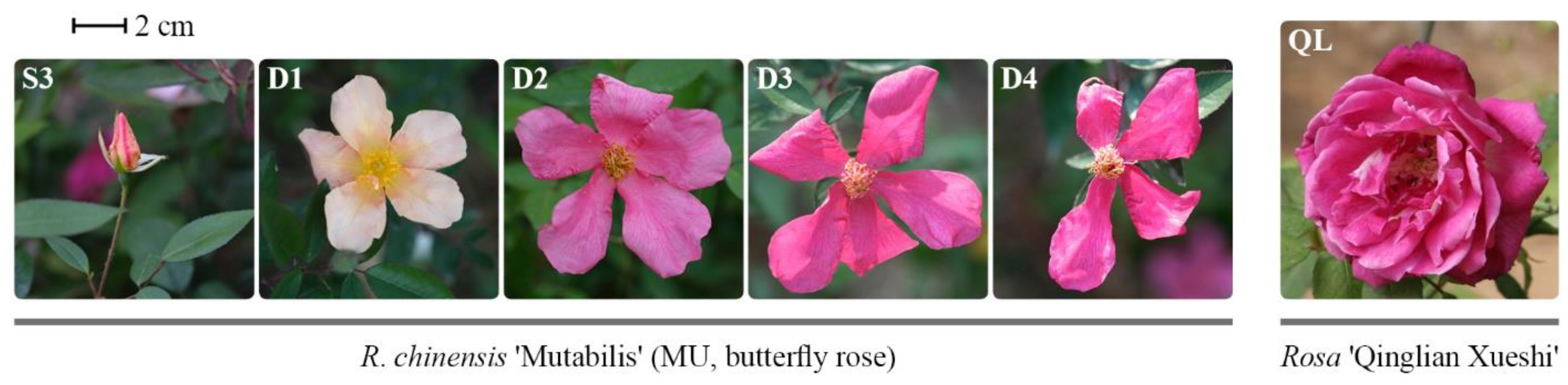
2.4. Plant Materials
2.5. RNA-Seq Analysis
2.6. Volatile Sampling and Gas Chromatography–Mass Spectrometry (GC-MS) Analysis
2.7. qRT-PCR Analysis
2.8. Data Analyses
3. Results
3.1. Identification of RcTPT Genes
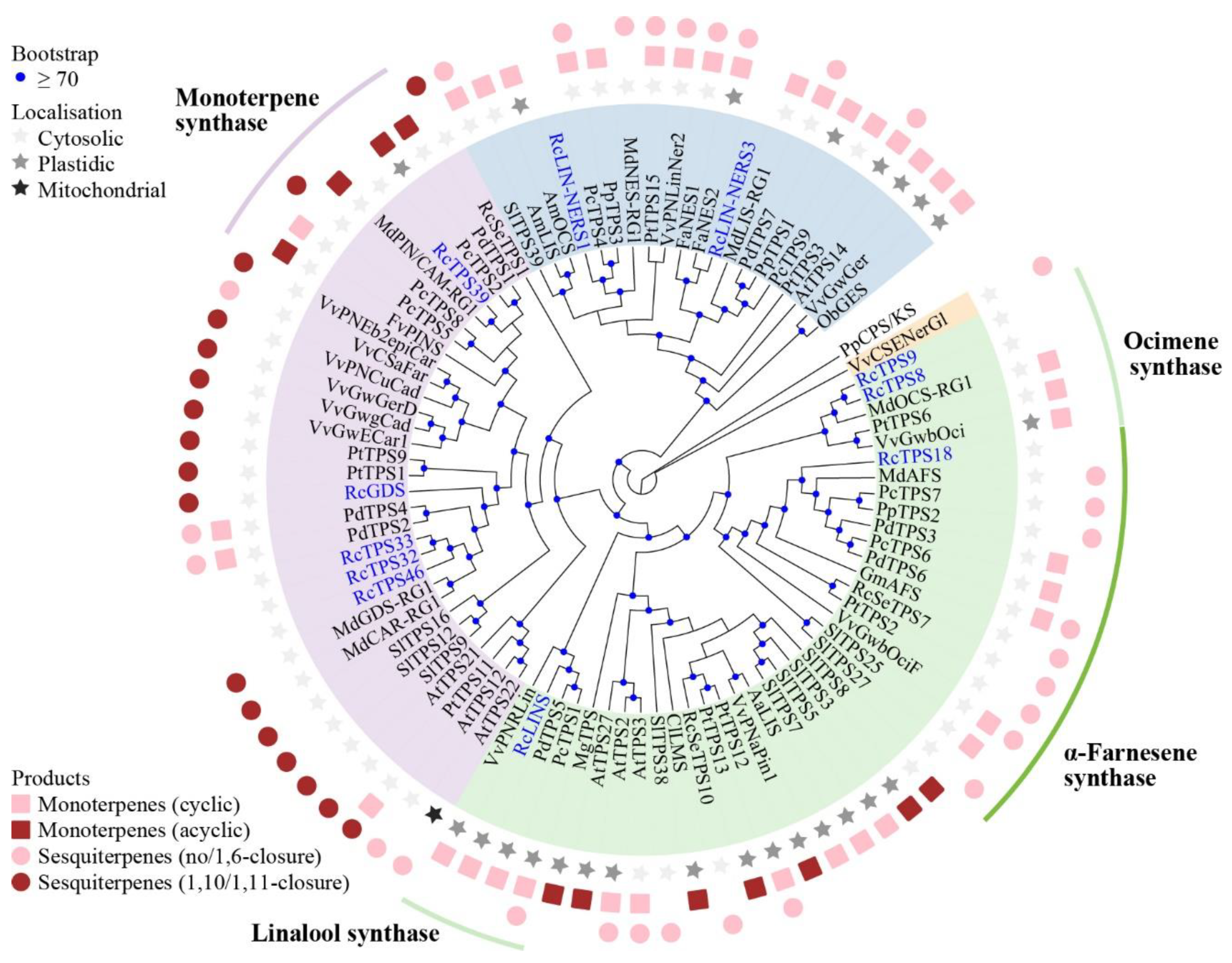
3.2. Re-Identification of RcTPS Genes
3.3. Chromosomal Localization and Gene Duplication
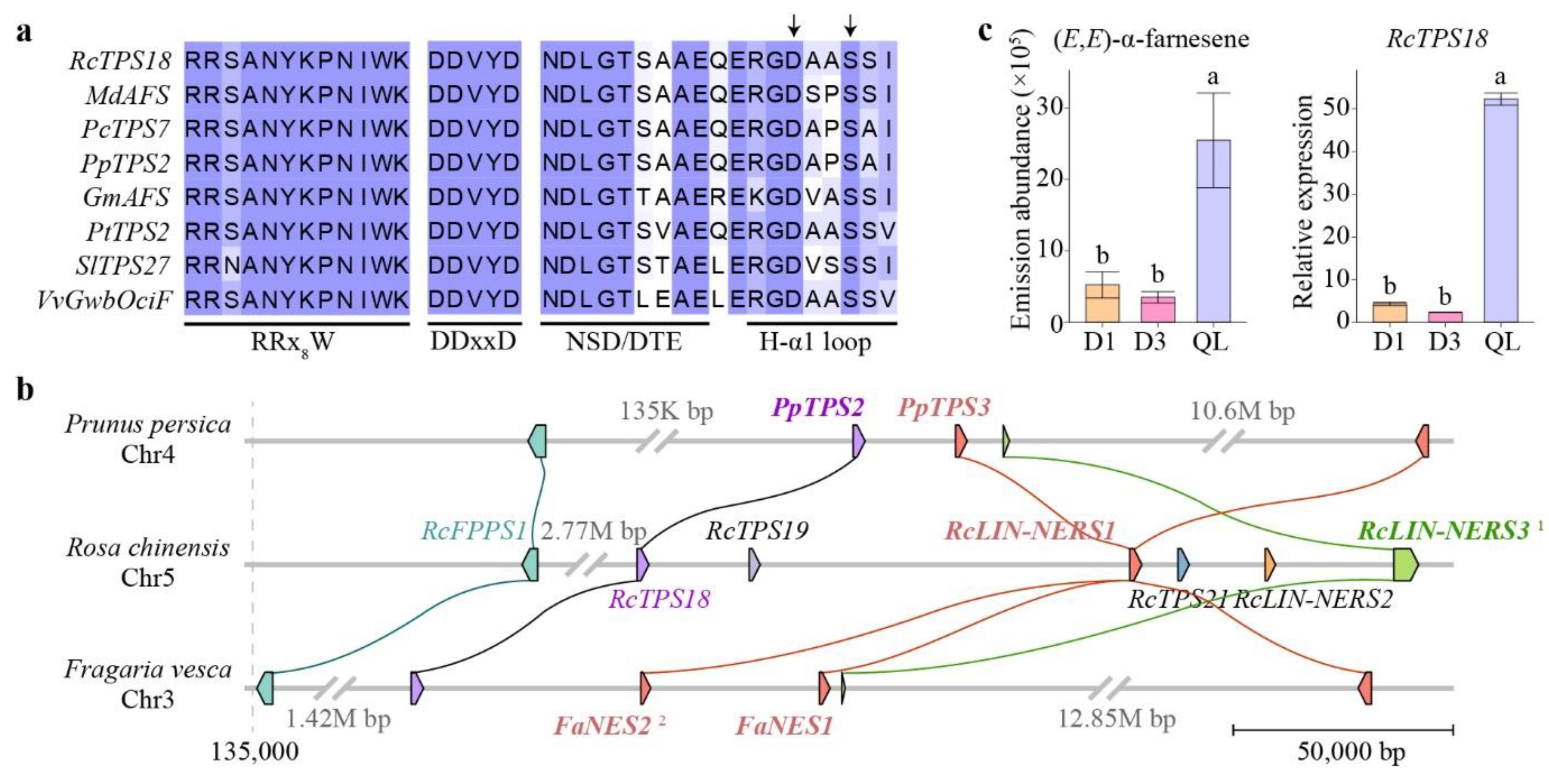
3.4. Collinearity Analysis of RcTPT and RcTPS Genes
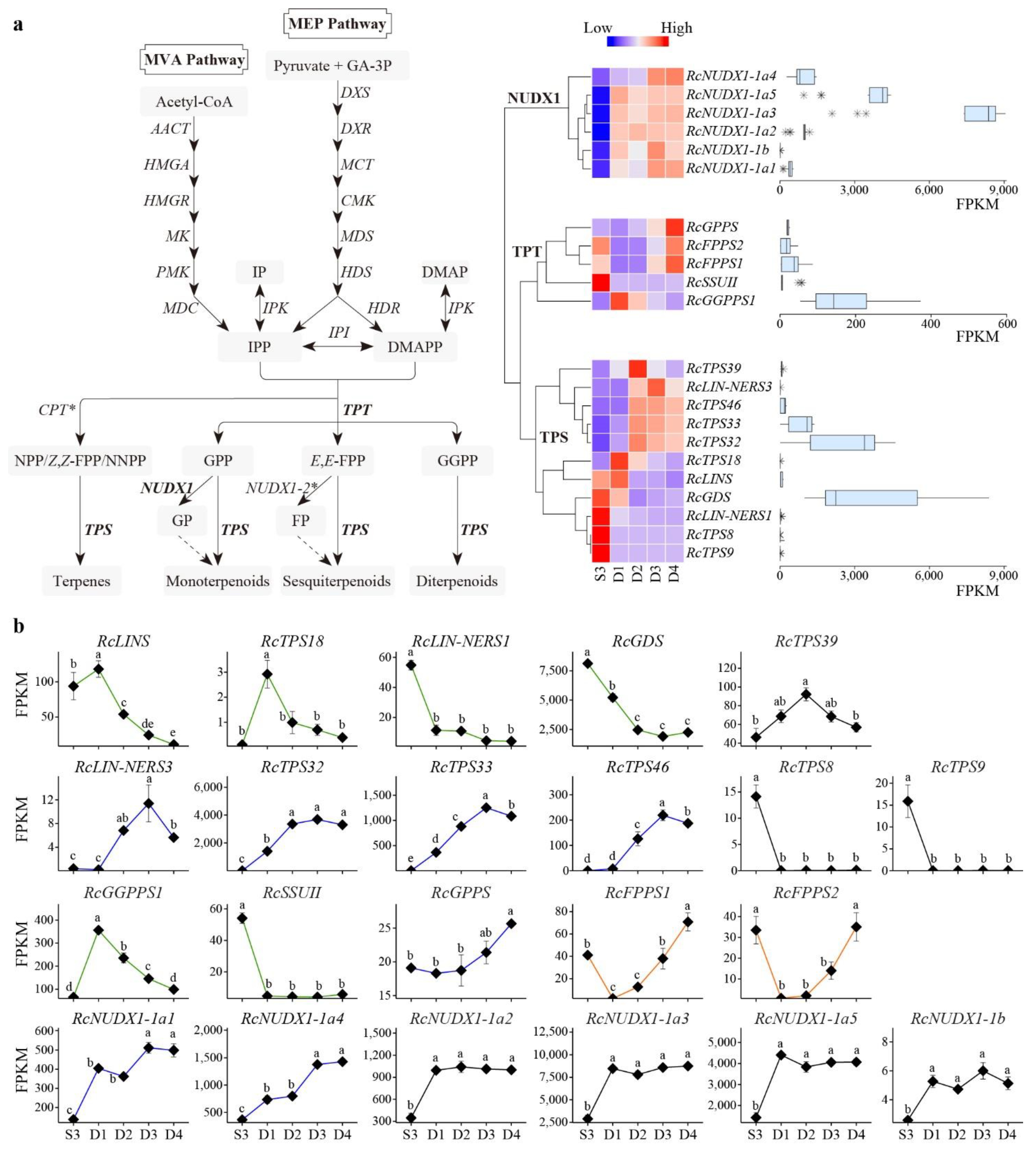
3.5. Effect of Developmental Stages on RcTPS Expression
3.6. Effect of Developmental Stages on RcTPT and RcNUDX1 Expression
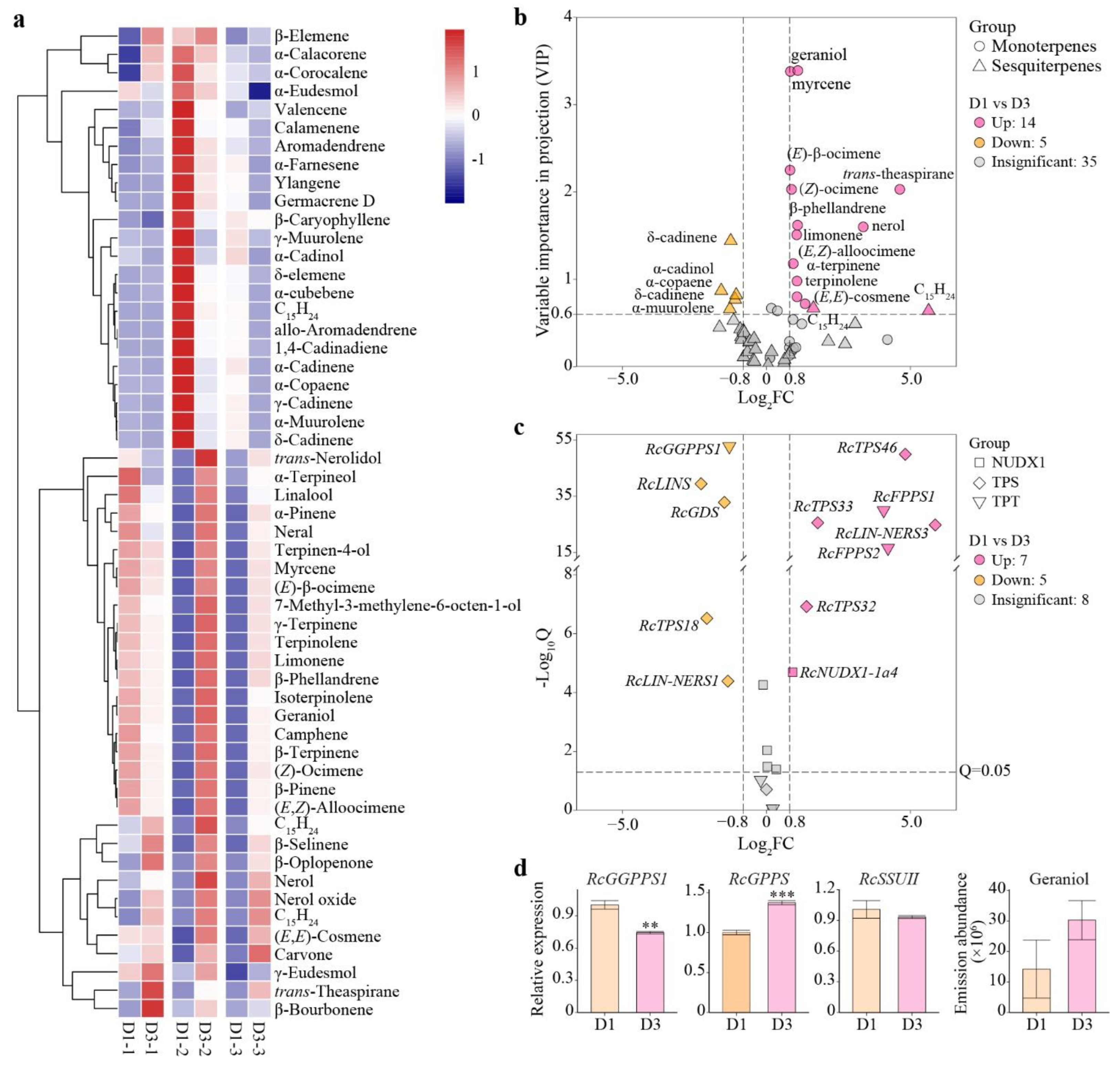
3.7. Analysis of Differential Volatile Terpenes and Differential Gene Expression
3.8. Functional Analysis of Petal-Expressed RcTPSs in Butterfly Rose
4. Discussion
4.1. Evolution and Function of the RcTPT Genes
4.2. Evolution of the TPS-b and TPS-g Genes in R. chinensis
4.3. Different TPS Expression Profiles during Flower Developmental Stages
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbas, F.; Ke, Y.; Yu, R.; Yue, Y.; Amanullah, S.; Jahangir, M.M.; Fan, Y. Volatile terpenoids: Multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta 2017, 246, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Fineschi, S.; Loreto, F.; Staudt, M.; Peñuelas, J. Diversification of volatile isoprenoid emissions from trees: Evolutionary and ecological perspectives. In Biology, Controls and Models of Tree Volatile Organic Compound Emissions; Niinemets, Ü., Monson, R.K., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 1–20. [Google Scholar] [CrossRef]
- Bergougnoux, V.; Caissard, J.-C.; Jullien, F.; Magnard, J.-L.; Scalliet, G.; Cock, J.M.; Hugueney, P.; Baudino, S. Both the adaxial and abaxial epidermal layers of the rose petal emit volatile scent compounds. Planta 2007, 226, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Muhlemann, J.K.; Klempien, A.; Dudareva, N. Floral volatiles: From biosynthesis to function. Plant Cell Environ. 2014, 37, 1936–1949. [Google Scholar] [CrossRef] [PubMed]
- Wiese, N.; Fischer, J.; Heidler, J.; Lewkowski, O.; Degenhardt, J.; Erler, S. The terpenes of leaves, pollen, and nectar of thyme (Thymus vulgaris) inhibit growth of bee disease-associated microbes. Sci. Rep. 2018, 8, 14634. [Google Scholar] [CrossRef] [Green Version]
- Farré-Armengol, G.; Filella, I.; Llusià, J.; Peñuelas, J. β-Ocimene, a key floral and foliar volatile involved in multiple interactions between plants and other organisms. Molecules 2017, 22, 1148. [Google Scholar] [CrossRef] [Green Version]
- Raguso, R.A. More lessons from linalool: Insights gained from a ubiquitous floral volatile. Curr. Opin. Plant Biol. 2016, 32, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Knauer, A.C.; Schiestl, F.P. The effect of pollinators and herbivores on selection for floral signals: A case study in Brassica rapa. Evol. Ecol. 2017, 31, 285–304. [Google Scholar] [CrossRef]
- Huang, M.; Sanchez-Moreiras, A.M.; Abel, C.; Sohrabi, R.; Lee, S.; Gershenzon, J.; Tholl, D. The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 2012, 193, 997–1008. [Google Scholar] [CrossRef]
- Chen, X.; Köllner, T.G.; Jia, Q.; Norris, A.; Santhanam, B.; Rabe, P.; Dickschat, J.S.; Shaulsky, G.; Gershenzon, J.; Chen, F. Terpene synthase genes in eukaryotes beyond plants and fungi: Occurrence in social amoebae. Proc. Natl. Acad. Sci. USA 2016, 113, 12132–12137. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Chen, Q.; Fan, D.; Li, J.; Wang, G.; Zhang, P. Structural analyses of short-chain prenyltransferases identify an evolutionarily conserved GFPPS clade in Brassicaceae plants. Mol. Plant 2016, 9, 195–204. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Yu, R.; Fan, Y. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta 2014, 240, 745–762. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Pichersky, E. The complete functional characterization of the terpene synthase family in tomato. New Phytol. 2020, 226, 1341–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalit, M.; Guterman, I.; Volpin, H.; Bar, E.; Tamari, T.; Menda, N.; Adam, Z.; Zamir, D.; Vainstein, A.; Weiss, D.; et al. Volatile ester formation in roses. Identification of an acetyl-coenzyme A. geraniol/citronellol acetyltransferase in developing rose petals. Plant Physiol. 2003, 131, 1868–1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tholl, D.; Gershenzon, J. The flowering of a new scent pathway in rose. Science 2015, 349, 28–29. [Google Scholar] [CrossRef] [PubMed]
- Lewinsohn, E.; Vainstein, A.; Weiss, D. An integrated genomics approach to identifying floral scent genes in rose. In Biology of Floral Scent; Pichersky, E., Dudareva, N., Eds.; CRC Press: Boca Raton, FL, USA, 2006; pp. 91–102. [Google Scholar]
- Shalit, M.; Shafir, S.; Larkov, O.; Bar, E.; Kaslassi, D.; Adam, Z.; Zamir, D.; Vainstein, A.; Weiss, D.; Ravid, U.Z.I.; et al. Volatile compounds emitted by rose cultivars: Fragrance perception by man and honeybees. Isr. J. Plant Sci. 2004, 52, 245–255. [Google Scholar] [CrossRef]
- Joichi, A.; Yomogida, K.; Awano, K.-I.; Ueda, Y. Volatile components of tea-scented modern roses and ancient Chinese roses. Flavour Frag. J. 2005, 20, 152–157. [Google Scholar] [CrossRef]
- Helsper, J.P.F.G.; Davies, J.A.; Bouwmeester, H.J.; Krol, A.F.; van Kampen, M.H. Circadian rhythmicity in emission of volatile compounds by flowers of Rosa hybrida L. cv. Honesty. Planta 1998, 207, 88–95. [Google Scholar] [CrossRef]
- Guterman, I.; Shalit, M.; Menda, N.; Piestun, D.; Dafny-Yelin, M.; Shalev, G.; Bar, E.; Davydov, O.; Ovadis, M.; Emanuel, M.; et al. Rose scent: Genomics approach to discovering novel floral fragrance-related genes. Plant Cell 2002, 14, 2325–2338. [Google Scholar] [CrossRef] [Green Version]
- Magnard, J.-L.; Bony, A.R.; Bettini, F.; Campanaro, A.; Blerot, B.; Baudino, S.; Jullien, F. Linalool and linalool nerolidol synthases in roses, several genes for little scent. Plant Physiol. Bioch. 2018, 127, 74–87. [Google Scholar] [CrossRef]
- Magnard, J.-L.; Roccia, A.; Caissard, J.-C.; Vergne, P.; Sun, P.; Hecquet, R.; Dubois, A.; Hibrand-Saint Oyant, L.; Jullien, F.; Nicolè, F.; et al. Biosynthesis of monoterpene scent compounds in roses. Science 2015, 349, 81–83. [Google Scholar] [CrossRef] [Green Version]
- Sun, P.; Dégut, C.; Réty, S.; Caissard, J.-C.; Hibrand-Saint Oyant, L.; Bony, A.; Paramita, S.N.; Conart, C.; Magnard, J.-L.; Jeauffre, J.; et al. Functional diversification in the Nudix hydrolase gene family drives sesquiterpene biosynthesis in Rosa × wichurana. Plant J. 2020, 104, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Dani, K.G.S.; Fineschi, S.; Michelozzi, M.; Trivellini, A.; Pollastri, S.; Loreto, F. Diversification of petal monoterpene profiles during floral development and senescence in wild roses: Relationships among geraniol content, petal colour, and floral lifespan. Oecologia 2021, 197, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Jia, Q.; Chen, F. Catalytic Functions of the Isoprenyl Diphosphate Synthase Superfamily in Plants: A Growing Repertoire. Mol. Plant 2016, 9, 189–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, T.A.; Matsuba, Y.; Schauvinhold, I.; Yu, G.; Lees, H.A.; Klein, S.E.; Pichersky, E. The tomato cis–prenyltransferase gene family. Plant J. 2013, 73, 640–652. [Google Scholar] [CrossRef]
- Yan, Y.; Li, M.; Zhang, X.; Kong, W.; Bendahmane, M.; Bao, M.; Fu, X. Tissue-specific expression of the terpene synthase family genes in Rosa chinensis and effect of abiotic stress conditions. Genes 2022, 13, 547. [Google Scholar] [CrossRef]
- Yang, Z.; Xie, C.; Zhan, T.; Li, L.; Liu, S.; Huang, Y.; An, W.; Zheng, X.; Huang, S. Genome-wide identification and functional characterization of the trans-isopentenyl diphosphate synthases gene Family in Cinnamomum camphora. Front. Plant Sci. 2021, 12, 708697. [Google Scholar] [CrossRef]
- Sperschneider, J.; Catanzariti, A.-M.; DeBoer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, 44598. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Huang, F.; Wang, X.; Zhang, M.; Zheng, R.; Wang, J.; Yu, D. Genome-wide analysis of terpene synthases in soybean: Functional characterization of GmTPS3. Gene 2014, 544, 83–92. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Lee, T.; Cheng, C.-H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Zhang, Y.; Song, S.; Su, W.; Chen, R.; Sun, G.; Hao, Y.; Liu, H. Comparative RNA-Seq analysis on the regulation of cucumber sex differentiation under different ratios of blue and red light. Bot. Stud. 2018, 59, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The genome sequence archive family: Toward explosive data growth and diverse data types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef] [PubMed]
- CNCB-NGDC Members and Partners. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2021, 50, D27–D38. [Google Scholar] [CrossRef]
- Wei, G.; Tian, P.; Zhang, F.; Qin, H.; Miao, H.; Chen, Q.; Hu, Z.; Cao, L.; Wang, M.; Gu, X.; et al. Integrative analyses of nontargeted volatile profiling and transcriptome data provide molecular Insight into VOC diversity in cucumber plants (Cucumis sativus). Plant Physiol. 2016, 172, 603–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, C.; Diao, W.; Zhu, H.; Umer, M.J.; Zhao, S.; He, N.; Lu, X.; Yuan, P.; Anees, M.; Yang, D.; et al. Metabolome and transcriptome integration reveals insights into flavor formation of ‘Crimson’ watermelon flesh during fruit development. Front. Plant Sci. 2021, 12, 629361. [Google Scholar] [CrossRef]
- Cheng, C.; Yu, Q.; Wang, Y.; Wang, H.; Dong, Y.; Ji, Y.; Zhou, X.; Li, Y.; Jiang, C.-Z.; Gan, S.-S.; et al. Ethylene-regulated asymmetric growth of the petal base promotes flower opening in rose (Rosa hybrida). Plant Cell 2021, 33, 1229–1251. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Kong, Y.; Wang, H.; Lang, L.; Dou, X.; Bai, J. Metabolome-based discrimination analysis of five Lilium bulbs associated with differences in secondary metabolites. Molecules 2021, 26, 1340. [Google Scholar] [CrossRef] [PubMed]
- Irmisch, S.; Jiang, Y.; Chen, F.; Gershenzon, J.; Köllner, T.G. Terpene synthases and their contribution to herbivore-induced volatile emission in western balsam poplar (Populus trichocarpa). BMC Plant Biol. 2014, 14, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aharoni, A.; Giri, A.P.; Verstappen, F.W.A.; Bertea, C.M.; Sevenier, R.; Sun, Z.; Jongsma, M.A.; Schwab, W.; Bouwmeester, H.J. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 2004, 16, 3110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Pichersky, E. More is better: The diversity of terpene metabolism in plants. Curr. Opin. Plant Biol. 2020, 55, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-F.; Wen, C.-H.; Lee, Y.-R.; Chu, F.-H. Cloning and characterization of terpene synthase genes from Taiwan cherry. Tree Genet. Genomes 2019, 15, 51. [Google Scholar] [CrossRef]
- Nawade, B.; Yahyaa, M.; Reuveny, H.; Shaltiel-Harpaz, L.; Eisenbach, O.; Faigenboim, A.; Bar-Yaakov, I.; Holland, D.; Ibdah, M. Profiling of volatile terpenes from almond (Prunus dulcis) young fruits and characterization of seven terpene synthase genes. Plant Sci. 2019, 287, 110187. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuizen, N.J.; Green, S.A.; Chen, X.; Bailleul, E.J.D.; Matich, A.J.; Wang, M.Y.; Atkinson, R.G. Functional genomics reveals that a compact terpene synthase gene family can account for terpene volatile production in apple. Plant Physiol. 2013, 161, 787–804. [Google Scholar] [CrossRef] [Green Version]
- Luck, K.; Chen, X.; Norris, A.M.; Chen, F.; Gershenzon, J.; Köllner, T.G. The reconstruction and biochemical characterization of ancestral genes furnish insights into the evolution of terpene synthase function in the Poaceae. Plant Mol. Biol. 2020, 104, 203–215. [Google Scholar] [CrossRef]
- Pechous, S.W.; Whitaker, B.D. Cloning and functional expression of an (E,E)-α-farnesene synthase cDNA from peel tissue of apple fruit. Planta 2004, 219, 84–94. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Cao, X.; Liu, X.; Xin, R.; Wang, J.; Gao, J.; Wu, B.; Gao, L.; Xu, C.; Zhang, B.; et al. UV-B irradiation differentially regulates terpene synthases and terpene content of peach. Plant Cell Environ. 2017, 40, 2261–2275. [Google Scholar] [CrossRef]
- Lin, J.; Wang, D.; Chen, X.; Köllner, T.G.; Mazarei, M.; Guo, H.; Pantalone, V.R.; Arelli, P.; Stewart, C.N., Jr.; Wang, N.; et al. An (E,E)-α-farnesene synthase gene of soybean has a role in defence against nematodes and is involved in synthesizing insect-induced volatiles. Plant Biotechnol. J. 2017, 15, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Danner, H.; Boeckler, G.A.; Irmisch, S.; Yuan, J.S.; Chen, F.; Gershenzon, J.; Unsicker, S.B.; Köllner, T.G. Four terpene synthases produce major compounds of the gypsy moth feeding-induced volatile blend of Populus trichocarpa. Phytochemistry 2011, 72, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 2010, 10, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, X.; Kirby, J.; Keasling, J.D. Functional characterization of four sesquiterpene synthases from Ricinus communis (Castor bean). Phytochemistry 2012, 78, 20–28. [Google Scholar] [CrossRef]
- Green, S.; Squire, C.J.; Nieuwenhuizen, N.J.; Baker, E.N.; Laing, W. Defining the potassium binding region in an apple terpene synthase. J. Biol. Chem. 2009, 284, 8661–8669. [Google Scholar] [CrossRef] [Green Version]
- You, M.K.; Lee, Y.J.; Yu, J.S.; Ha, S.-H. The predicted functional compartmentation of rice terpenoid metabolism by trans-prenyltransferase structural analysis, expression and localization. Int. J. Mol. Sci. 2020, 21, 8927. [Google Scholar] [CrossRef]
- Chen, F.; Su, L.; Hu, S.; Xue, J.-Y.; Liu, H.; Liu, G.; Jiang, Y.; Du, J.; Qiao, Y.; Fan, Y.; et al. A chromosome-level genome assembly of rugged rose (Rosa rugosa) provides insights into its evolution, ecology, and floral characteristics. Hortic. Res. 2021, 8, 141. [Google Scholar] [CrossRef]
- Kulkarni, R.; Pandit, S.; Chidley, H.; Nagel, R.; Schmidt, A.; Gershenzon, J.; Pujari, K.; Giri, A.; Gupta, V. Characterization of three novel isoprenyl diphosphate synthases from the terpenoid rich mango fruit. Plant Physiol. Bioch. 2013, 71, 121–131. [Google Scholar] [CrossRef]
- Wang, G.; Dixon Richard, A. Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. USA 2009, 106, 9914–9919. [Google Scholar] [CrossRef] [Green Version]
- Tholl, D.; Kish, C.M.; Orlova, I.; Sherman, D.; Gershenzon, J.; Pichersky, E.; Dudareva, N. Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 2004, 16, 977–992. [Google Scholar] [CrossRef] [Green Version]
- Rai, A.; Smita, S.S.; Singh, A.K.; Shanker, K.; Nagegowda, D.A. Heteromeric and homomeric geranyl diphosphate synthases from Catharanthus roseus and their role in monoterpene indole alkaloid biosynthesis. Mol. Plant 2013, 6, 1531–1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.O.; Perez-Fons, L.; Robertson, F.P.; Bramley, P.M.; Fraser, P.D. Functional characterization of long-chain prenyl diphosphate synthases from tomato. Biochem. J. 2013, 449, 729–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.M.; Chiang, A.; Lund, S.T.; Bohlmann, J. Biosynthesis of wine aroma: Transcript profiles of hydroxymethylbutenyl diphosphate reductase, geranyl diphosphate synthase, and linalool/nerolidol synthase parallel monoterpenol glycoside accumulation in Gewürztraminer grapes. Planta 2012, 236, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, F.-L.; Chang, T.-H.; Ko, T.-P.; Wang, A.H.J. Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase. Plant Physiol. 2011, 155, 1079–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhandapani, S.; Tjhang, J.G.; Jang, I.-C. Production of multiple terpenes of different chain lengths by subcellular targeting of multi-substrate terpene synthase in plants. Metab. Eng. 2020, 61, 397–405. [Google Scholar] [CrossRef]
- Ding, G.; Zhang, S.; Ma, B.; Liang, J.; Li, H.; Luo, Y.; He, N. Origin and functional differentiation of (E)-β-ocimene synthases reflect the expansion of monoterpenes in angiosperms. J. Exp. Bot. 2020, 71, 6571–6586. [Google Scholar] [CrossRef]
- Lynch, J.H.; Pichersky, E.; Dudareva, N. Floral scent metabolic pathways and their regulation. In Biology of Plant Volatiles, 2nd ed.; Pichersky, E., Dudareva, N., Eds.; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar] [CrossRef]
- Shi, S.; Duan, G.; Li, D.; Wu, J.; Liu, X.; Hong, B.; Yi, M.; Zhang, Z. Two-dimensional analysis provides molecular insight into flower scent of Lilium ‘Siberia’. Sci. Rep. 2018, 8, 5352. [Google Scholar] [CrossRef] [Green Version]
- Abbas, F.; Ke, Y.; Zhou, Y.; Ashraf, U.; Li, X.; Yu, Y.; Yue, Y.; Ahmad, K.W.; Yu, R.; Fan, Y. Molecular cloning, characterization and expression analysis of LoTPS2 and LoTPS4 involved in floral scent formation in oriental hybrid Lilium variety ‘Siberia’. Phytochemistry 2020, 173, 112294. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.; Yu, R.; Fan, Y. Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium ‘Siberia’. Planta 2019, 249, 71–93. [Google Scholar] [CrossRef]
- Zeng, X.; Liu, C.; Zheng, R.; Cai, X.; Luo, J.; Zou, J.; Wang, C. Emission and accumulation of monoterpene and the key terpene synthase (TPS) associated with monoterpene biosynthesis in Osmanthus fragrans Lour. Front. Plant Sci. 2016, 6, 1232. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Yu, R.; Fan, Y. Transcriptome profiling provides new insights into the formation of floral scent in Hedychium coronarium. BMC Genom. 2015, 16, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
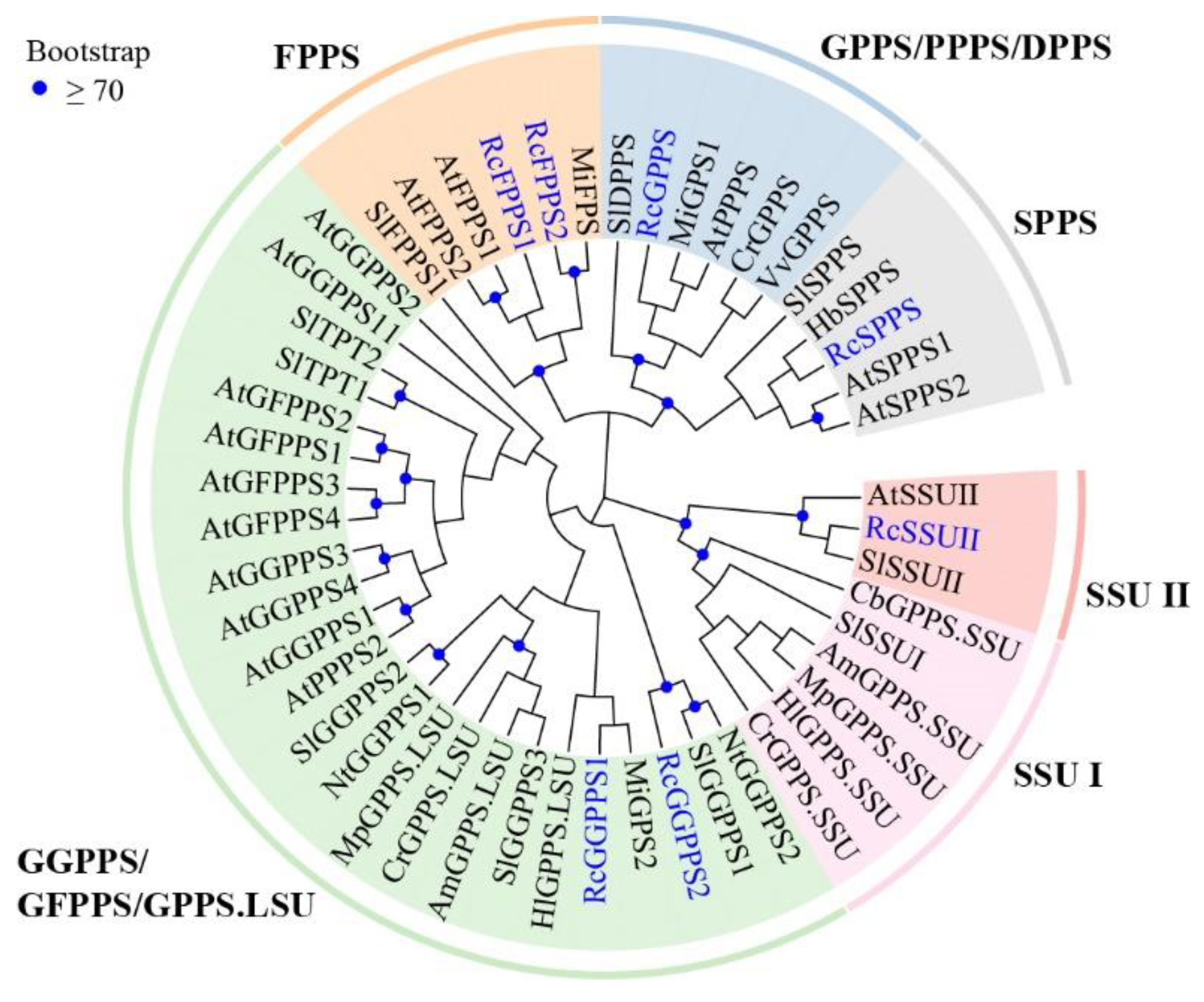
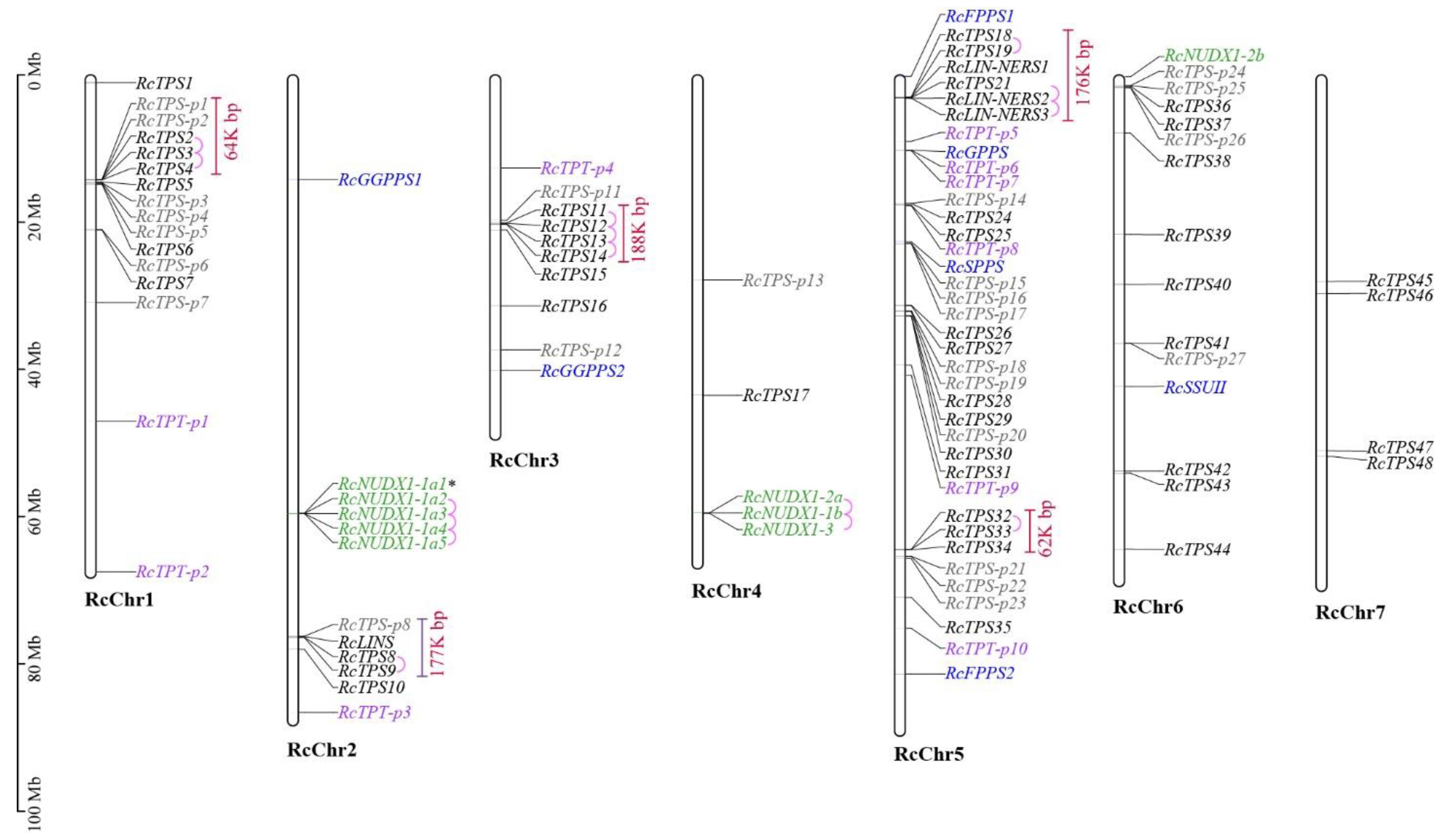
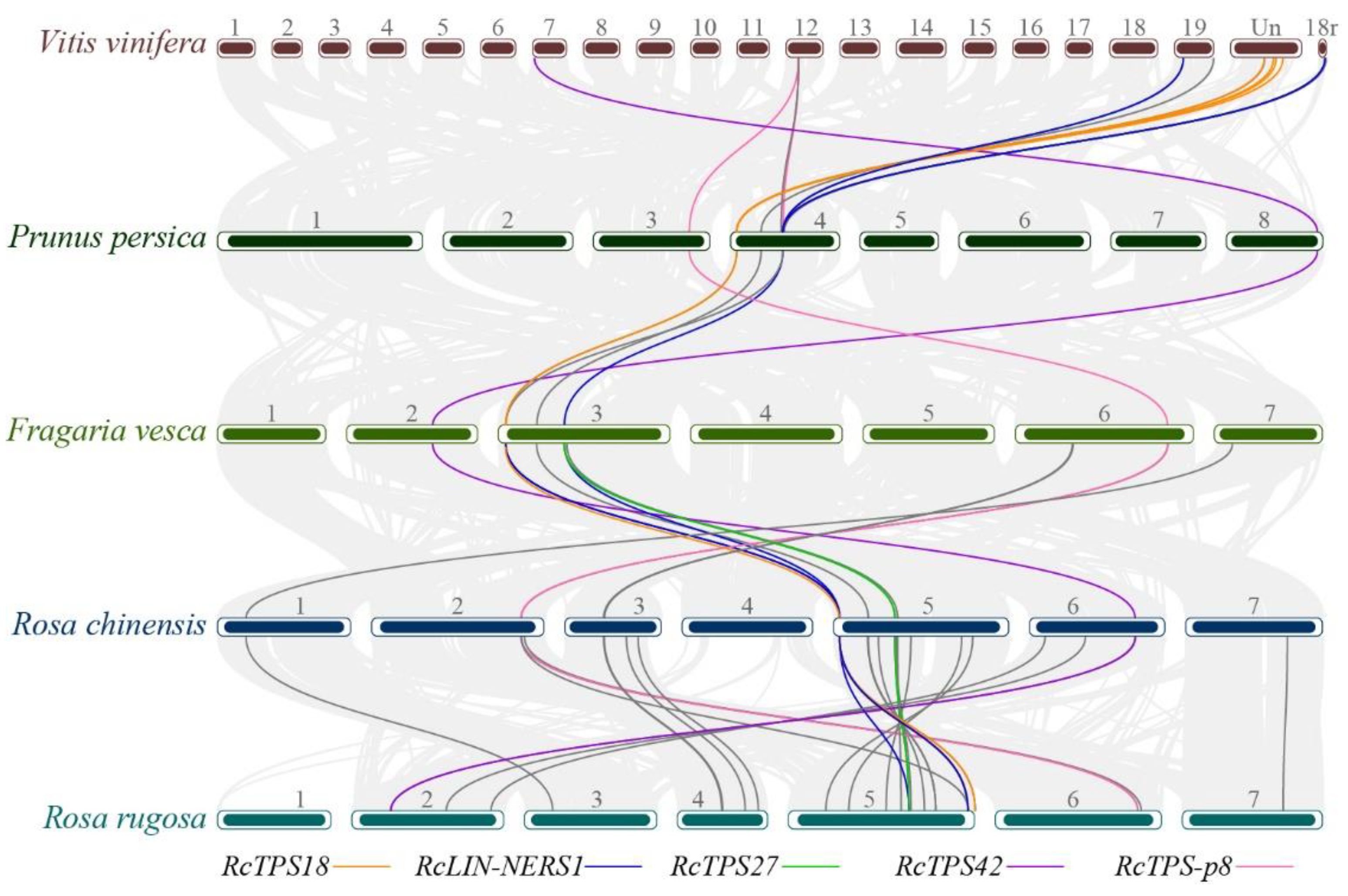
| Group | Name | Id | Chr | Amino Acid | Exon Number | Localization | Conserved Motif | |
|---|---|---|---|---|---|---|---|---|
| TargetP | LOCALIZER | |||||||
| SC 1 | RcGGPPS1 | RchiOBHmChr2g0102671 | 2 | 363 | 1 | C 3 | C | DDX(2–4)D, DDXXD, CXXXC |
| RcGGPPS2 | RchiOBHmChr3g0493061 | 3 | 360 | 1 | C | C | DDX(2–4)D, DDXXD, CXXXC | |
| RcSSUII | RchiOBHmChr6g0279181 | 6 | 329 | 2 | C | C | DDX(2–4)D, DDXXE, CXXXC, CXXXC | |
| RcGPPS | RchiOBHmChr5g0014811 | 5 | 426 | 12 | M 4 | M | DDX(2–4)D, DDXXD | |
| RcFPPS1 | RchiOBHmChr5g0000321 | 5 | 342 | 12 | / | / | DDX(2–4)D, DDXXD | |
| RcFPPS2 | RchiOBHmChr5g0075621 | 5 | 342 | 11 | / | / | DDX(2–4)D, DDXXD | |
| LC 2 | RcSPPS | RchiOBHmChr5g0028851 | 5 | 421 | 6 | / | C | DDX(2–4)D, DDXXD |
| Name | Id | Chr | Sub-Family | Amino Acid | Name | Id | Chr | Sub-Family | Amino Acid |
|---|---|---|---|---|---|---|---|---|---|
| RcTPS1 | RchiOBHmChr1g0313881 | 1 | a | 562 | RcTPS24 | RchiOBHmChr5g0023471 | 5 | e/f | 799 |
| RcTPS2 | RchiOBHmChr1g0326051 | 1 | a | 581 | RcTPS25 | RchiOBHmChr5g0023641 | 5 | e/f | 724 |
| RcTPS3 | RchiOBHmChr1g0326061 | 1 | a | 580 | RcTPS26 | RchiOBHmChr5g0036921 | 5 | a | 561 |
| RcTPS4 | RchiOBHmChr1g0326071 | 1 | a | 546 | RcTPS27 | RchiOBHmChr5g0037011 | 5 | a | 555 |
| RcTPS5 | RchiOBHmChr1g0326251 | 1 | a | 556 | RcTPS28 | RchiOBHmChr5g0037601 | 5 | a | 549 |
| RcTPS6 | RchiOBHmChr1g0326391 | 1 | a | 556 | RcTPS29 | RchiOBHmChr5g0038021 | 5 | a | 565 |
| RcTPS7 | RchiOBHmChr1g0331211 | 1 | b | 583 | RcGDS | RchiOBHmChr5g0038101 | 5 | a | 565 |
| RcLINS | RchiOBHmChr2g0160421(RcTPS-p9)+ RchiOBHmChr2g0160441(RcTPS-p10) | 2 | b | 601 | RcTPS31 | RchiOBHmChr5g0044191 | 5 | a | 565 |
| RcTPS32 | RchiOBHmChr5g0059501 | 5 | a | 557 | |||||
| RcTPS8 | RchiOBHmChr2g0160561 | 2 | b | 570 | RcTPS33 | RchiOBHmChr5g0059511 | 5 | a | 557 |
| RcTPS9 | RchiOBHmChr2g0160591 | 2 | b | 501 | RcTPS34 | RchiOBHmChr5g0059541 | 5 | a | 544 |
| RcTPS10 | RchiOBHmChr2g0162311 | 2 | e/f | 852 | RcTPS35 | RchiOBHmChr5g0065101 | 5 | a | 557 |
| RcTPS11 | RchiOBHmChr3g0474411 | 3 | a | 560 | RcTPS36 | RchiOBHmChr6g0245751 | 6 | a | 557 |
| RcTPS12 | RchiOBHmChr3g0474441 | 3 | a | 560 | RcTPS37 | RchiOBHmChr6g0246001 | 6 | a | 559 |
| RcTPS13 | RchiOBHmChr3g0474501 | 3 | a | 560 | RcTPS38 | RchiOBHmChr6g0252721 | 6 | b | 569 |
| RcTPS14 | RchiOBHmChr3g0474541 | 3 | a | 557 | RcTPS39 | RchiOBHmChr6g0265741 | 6 | a | 567 |
| RcTPS15 | RchiOBHmChr3g0475221 | 3 | a | 560 | RcTPS40 | RchiOBHmChr6g0270581 | 6 | a | 564 |
| RcTPS16 | RchiOBHmChr3g0484891 | 3 | b | 566 | RcTPS41 | RchiOBHmChr6g0274871 | 6 | a | 553 |
| RcTPS17 | RchiOBHmChr4g0418071 | 4 | a | 564 | RcTPS42 | RchiOBHmChr6g0290871 | 6 | c | 857 |
| RcTPS18 | RchiOBHmChr5g0004591 | 5 | b | 580 | RcTPS43 | RchiOBHmChr6g0290941 | 6 | c | 825 |
| RcTPS19 | RchiOBHmChr5g0004631 | 5 | b | 583 | RcTPS44 | RchiOBHmChr6g0305391 | 6 | a | 570 |
| RcLIN-NERS1 | RchiOBHmChr5g0004711 | 5 | g | 544 | RcTPS45 | RchiOBHmChr7g0210371 | 7 | a | 565 |
| RcTPS21 | RchiOBHmChr5g0004731 | 5 | g | 509 | RcTPS46 | RchiOBHmChr7g0212441 | 7 | a | 558 |
| RcLIN-NERS2 | RchiOBHmChr5g0004761 | 5 | g | 580 | RcTPS47 | RchiOBHmChr7g0227831 | 7 | a | 571 |
| RcLIN-NERS3 | RchiOBHmChr5g0004801 | 5 | g | 580 | RcTPS48 | RchiOBHmChr7g0228501 | 7 | a | 539 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, Y.; Wang, H.; Lang, L.; Dou, X.; Bai, J. Effect of Developmental Stages on Genes Involved in Middle and Downstream Pathway of Volatile Terpene Biosynthesis in Rose Petals. Genes 2022, 13, 1177. https://doi.org/10.3390/genes13071177
Kong Y, Wang H, Lang L, Dou X, Bai J. Effect of Developmental Stages on Genes Involved in Middle and Downstream Pathway of Volatile Terpene Biosynthesis in Rose Petals. Genes. 2022; 13(7):1177. https://doi.org/10.3390/genes13071177
Chicago/Turabian StyleKong, Ying, Huan Wang, Lixin Lang, Xiaoying Dou, and Jinrong Bai. 2022. "Effect of Developmental Stages on Genes Involved in Middle and Downstream Pathway of Volatile Terpene Biosynthesis in Rose Petals" Genes 13, no. 7: 1177. https://doi.org/10.3390/genes13071177
APA StyleKong, Y., Wang, H., Lang, L., Dou, X., & Bai, J. (2022). Effect of Developmental Stages on Genes Involved in Middle and Downstream Pathway of Volatile Terpene Biosynthesis in Rose Petals. Genes, 13(7), 1177. https://doi.org/10.3390/genes13071177







