The data retrieved from GDC Portal were from the miRNA profiling of patients with hematopoietic and reticuloendothelial cancer. This yielded a dataset of 587 miRNAs elevated in myeloid cancers. The sequences of these miRNAs were extracted from miRBase, a sample of data is given in
Table 1.
3.1. miRNA Sequence Length and Nucleotide Frequency Analysis
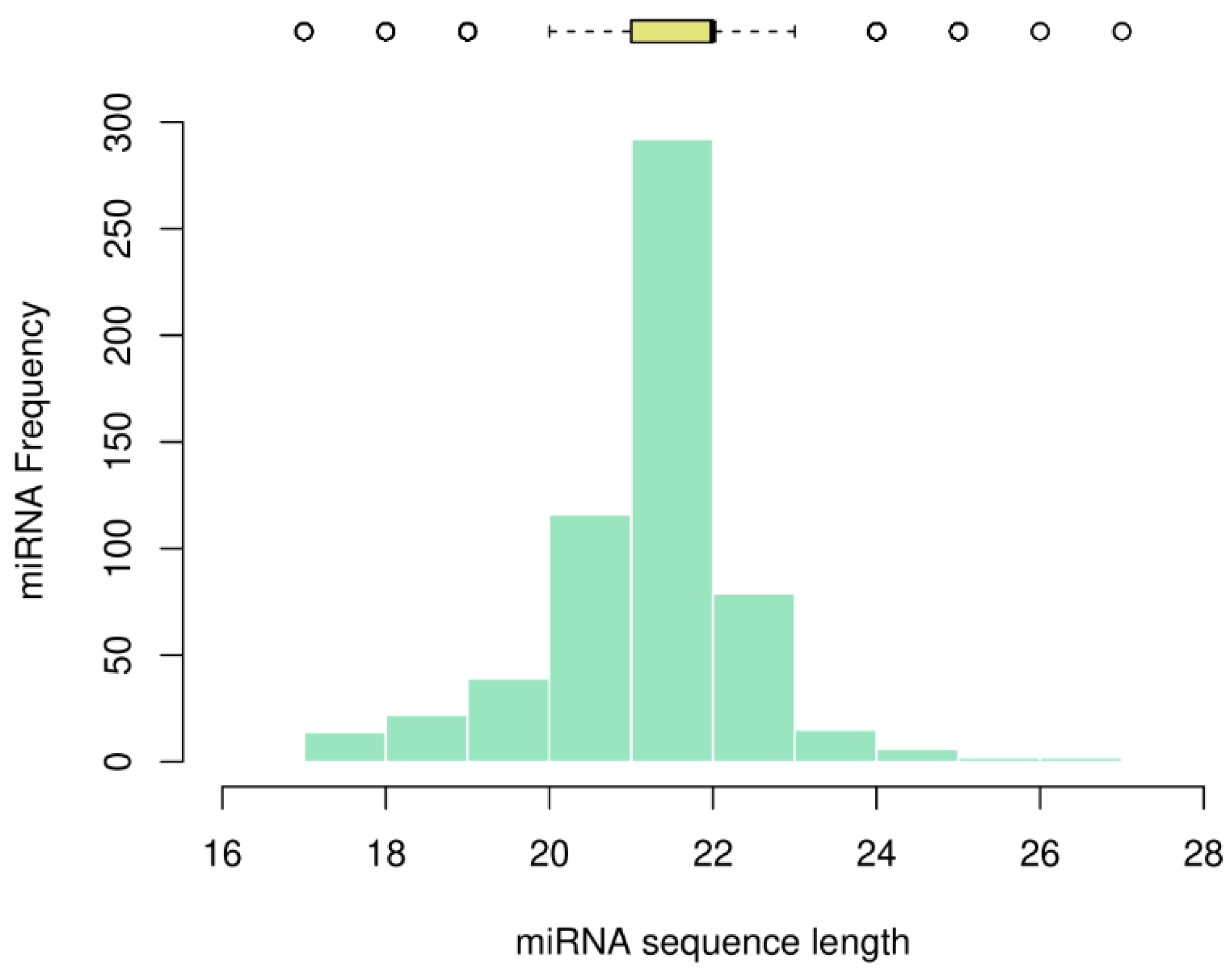
We analyzed the sequence length of the miRNAs and plotted the frequency as a histogram, together with the descriptive statistics as a boxplot on top (
Figure 2).
Most miRNAs in myeloid cancer were found to be 21, 22, and 23 nucleotides in length, with percentages of 19.8%, 49.8%, and 13.5%, respectively. We calculated a mode and median of 22 nucleotides, with an average of 21.69 and a variance of 1.65. The shortest miRNAs consist of 17 nucleotides (namely, hsa-mir-1260, hsa-mir-1825, hsa-mir-1207, hsa-mir-453, hsa-mir-1268, hsa-mir-1306, hsa-mir-1321, and hsa-mir-1827) and the four longest miRNAs consist of 26 and 27 nucleotides (hsa-mir-1248, hsa-mir-1183, hsa-mir-1272, and hsa-mir-1244).
It is not yet known whether the nucleotide length of miRNAs plays an active role in cancer or other diseases. Perfect base-pairing leads to the degradation of mRNA (a mechanism mainly seen in plants) and imperfect base-pairing with the target mRNA leads to repression of translation [
31]. In this line of argumentation, it could be assumed that the longer the miRNA sequence, the higher the probability to complement the target mRNAs. However, this needs experimental proof. Based on the volume of literature published for each miRNA in miRBase, we noted a higher volume of research carried out on short miRNAs (consisting of 17 and 18 nucleotides) when compared to the ones with 26 and 27 nucleotides (
Table 2).
Next, the percentage of nucleotides in each position was quantified (
Figure 3). The analysis was done from the first (5′) to the last (3′) position of the miRNA’s nucleotides. Overall, Uracil was the most observed nucleotide, and Adenine was the least observed one with 27.8%, and 22.6%, respectively.
In the first nucleotide position, 183 miRNAs have an A and 186 miRNAs start with a U. There is a higher frequency of Adenines at the beginning of the sequences when compared to Uracil which is more frequent at the end of miRNA sequences. In particular, there is a high density of Uracils between nucleotide positions 22 and 25. Purine (A and G) and pyrimidine (C and U) nucleotide bases were analyzed for their frequency in the studied miRNA structures (
Supplementary Table S2). The highest and lowest purine-scoring miRNAs are listed in
Table 3. hsa-mir-765 (85.71% purines), hsa-mir-1468 (81.82%), and hsa-mir-1910 (80.00%) are some of the very high purine content miRNAs. The lowest purine content is present in hsa-mir-1281 (5.88%) and hsa-mir-483 (9.52%).
3.2. Motifs in microRNA Sequences Implicated in Myeloid Cancer
In addition to miRNA structure analysis, their common motifs were determined according to their length and frequencies. To find the most abundant motifs in miRNA sequences, we searched for nucleotide motifs containing 3, 4, 5, 6, and 7 nucleotides shared among all miRNA sequences. For this, the smallest motif encoding an amino acid, 3-nucleotide (3n), was searched. Then, the same analysis was done in the form of 4n, 5n, 6n, and 7n.
3.2.1. 3n miRNA Motifs
In the first round of motif search, we analyzed the sequences for 3n motifs, which are the smallest significant motifs. The most observed and the shortest motifs in the miRNA sequences were CUG, UGC, UGG, UGU, CAG, UUG, CCU, CUU, GUG, AGG, UCU, GCU, CGU, CGC, GCG, UGC, ACG, and CGA. These were found in 91.65% of miRNAs. Only 8.35% did not have any of these 3n motifs in their sequences (for example, hsa-mir-122 and hsa-mir-1181) (
Table 4).
3.2.2. 4n miRNA Motifs
We divided the 4n motifs identified into two groups, the ones occurring in more than 75 miRNAs (the most detected) and the ones occurring in less than 10 miRNAs (the least detected) (
Table 5).
CUGC, ACUG, and UGCA were found as the most detected motifs in 87 and 85 different miRNAs, respectively. On the other hand, CGAA, CGAG, CGUA, and UCGA were the least detected 4n motifs in 10 different miRNA sequences. Moreover, 112 sequences of 587 microRNAs (19%) do not have any of the top 4n motifs. In addition, 28 of 112 sequences have the least common motifs, and 84 of the miRNAs do not have any of the listed motifs (
Supplementary Table S3).
3.2.3. Longer Motifs
The purpose of finding longer motifs such as 5n, 6n, and 7n new motifs was to find potentially conserved or master sites in miRNAs. A total of 271 different 5n motifs were detected (
Supplementary Table S3). AGUGC was the most frequent 5n motif found in 36 miRNAs (6%). A total of 38 different 5n motifs were unique (
Table 6). The other mostly detected long motifs are made of 6n and 7n sequences. The most frequently observed 6n motifs were AAGUGC and GCUUCC (detected in 22 different miRNAs, 4%), while UUUAGAG was the most detected 7n motif in our dataset (found in 19 miRNAs, 3%). Finally, the longest motifs were detected, 8n (AAGUGCUU), 9n (AAGUGCUUC), 10n (AAAGUGCUUC), and 20n (AAAGUGCUUCCCUUUAGAGU). hsa-mir-106a, hsa-mir-302a, b, c, d, e, hsa-mir-526b have the 8n, 9n, and 10n motifs, but hsa-mir-520a, b, c, d, e, g, and h include all the long motifs in their structures (
Supplementary Table S3).
3.2.4. Consensus miRNA Sequences Having Many Motifs
Consensus motifs were analyzed in the miRNA sequences, elucidating the consecutive alignment of our motifs in different miRNA sequences to different degrees. In this way, detailed results were obtained about where the identified motifs are located in the miRNA, and how they appear in high-consensus sequences (
Table 7).
The results of this analysis show that a miRNA can be associated with one or more mRNA targets, using the common motifs it has in the sequence. Apart from the importance of motifs and consensus sequences in the miRNA binding on their target, the secondary importance of our results may arise in these sequences being a target of RNA binding proteins (RBPs), which recognize specific sequence motifs and are key factors to regulate the miRNA function. Although the transcription factors and epigenetic modifications control the synthesis of miRNAs, their regulation after synthesis is highly controlled with RBPs [
32]. Overall studies regarding the RBP binding and regulation of miRNAs are insufficient. Among more than 500 identified human RBPs, only a few have been characterized in terms of functioning in oncogene and tumor suppressor mRNAs [
33]. There are many secrets to be revealed behind the miRNA processing by RBPs in healthy and disease states for research to be carried out in the future. The complexity of regulation is further increased with the clues on the cooperative work of miRNAs and RBPs in controlling common mRNA targets [
34]. Taking all these into account, a detailed study of the structure of these short RNA molecules, which can perform so many functions, is crucial, and the results presented in our study, can serve as a starting point and raw material for these studies, especially in cancer models.
3.2.5. Target Genes of 7n Motifs
We next analyzed the target genes of the miRNAs that share common motifs, if they give hints on the functional aspects of the motifs we identified in myeloid cancer. For this study, 7n motifs were selected as they are longer and can be more specific in their targets [
1]. Using the miRNA-target prediction tool MirTar database, we identified the targets of our miRNAs, which are experimentally validated in different studies. The list of overlapping genes is listed in
Table 8, and all the detected targets are given in
Supplementary Table S4.
Our 7n motif GUGCUUC is present in 15 different miRNA sequences and all of them target the same six genes (EIF2S1, SPRED1, HIP1, YOD1, ELK4, ABHD15). We see that this is the case for many motifs in different degrees. This presumes that the motifs which are identified are an important factor for target recognition and small changes in the sequences can impact the specificity of binding/regulation.
3.2.6. Conserved 5n and 6n Motifs
We further wanted to test our motif-finding script in analyzing the conserved miRNA motifs in different species. For this, 5n and 6n motifs were searched in the available miRNA sequences from different species; 645 miRNAs for
Pongo pygmaeus, 1978 for
Mus musculus, 437 for
Caenorhabditis elegans, 690 for
Equus caballus, 469 for
Drosophila melanogaster, 600 for
Picea abies, 695 for
Oreochromis niloticus, 1138 for
Monodelphis domestica, 1232 for
Gallus gallus, 548 for
Ciona intestinalis, and 2654 for humans (
Supplementary Table S5). Among the top 15 identified motifs, we combined the ones that were common to all species and derived their percentages for specific species studied (
Table 9).
MiRNAs are key regulators of many cellular processes and may be one of the main players in post-transcriptional regulation. Because they also influence vital biological processes, they tend to be conserved between species. However, there have been contradictory reports on the SNP density of these regions when compared to control [
35]. Here, we show that the conservation may happen with certain motifs inside the miRNAs and the higher SNP density may be present in other parts of the miRNA, which add to the list of target genes of miRNA without disturbing the main target interactions.