Genome-Wide Analysis of the GDSL Genes in Pecan (Carya illinoensis K. Koch): Phylogeny, Structure, Promoter Cis-Elements, Co-Expression Networks, and Response to Salt Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Sample Preparation under Salt Stress
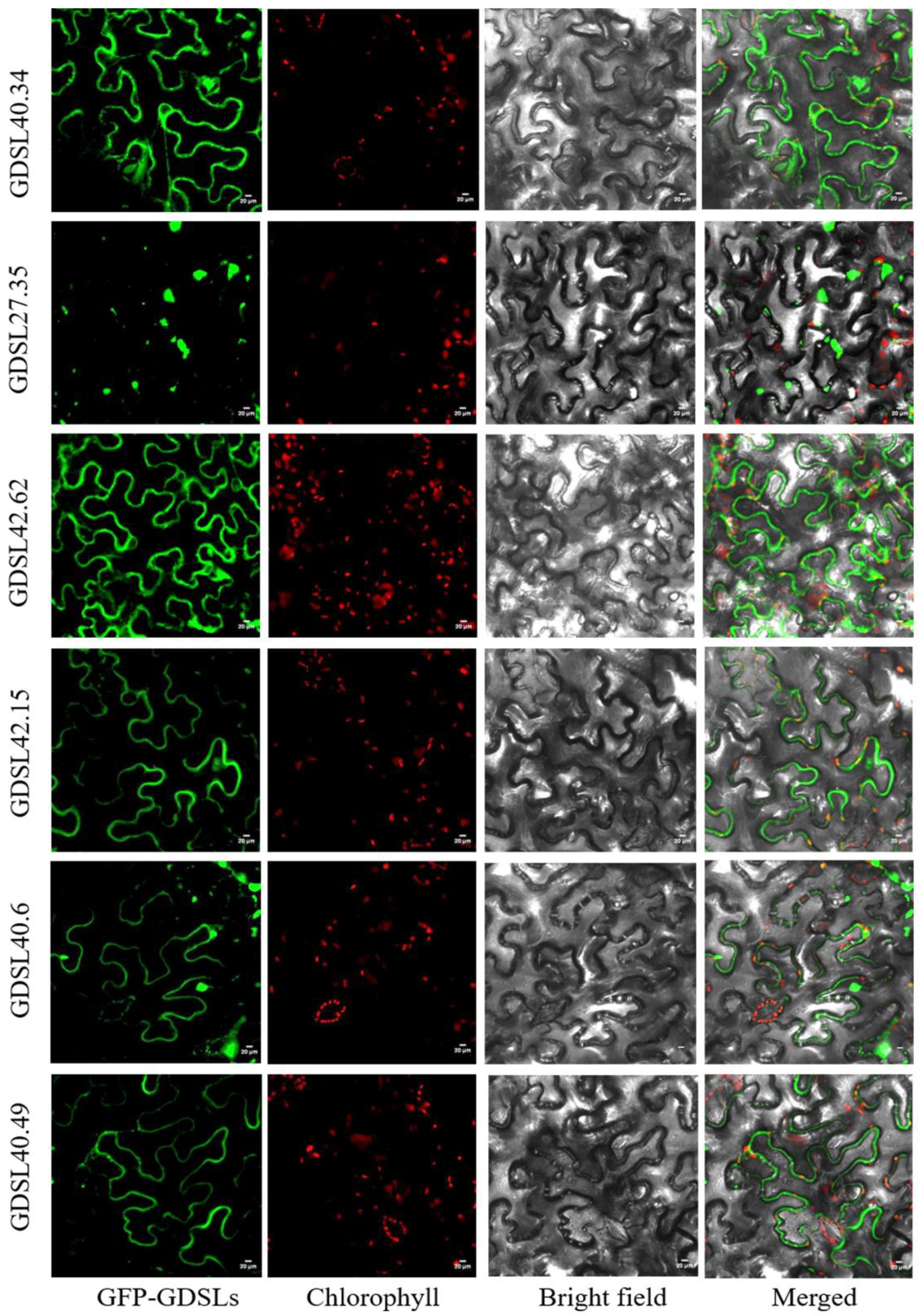
2.2. The GDSL-Type Esterase/Lipase Genes in the Pecan Genome and Subcellular Localization Analysis
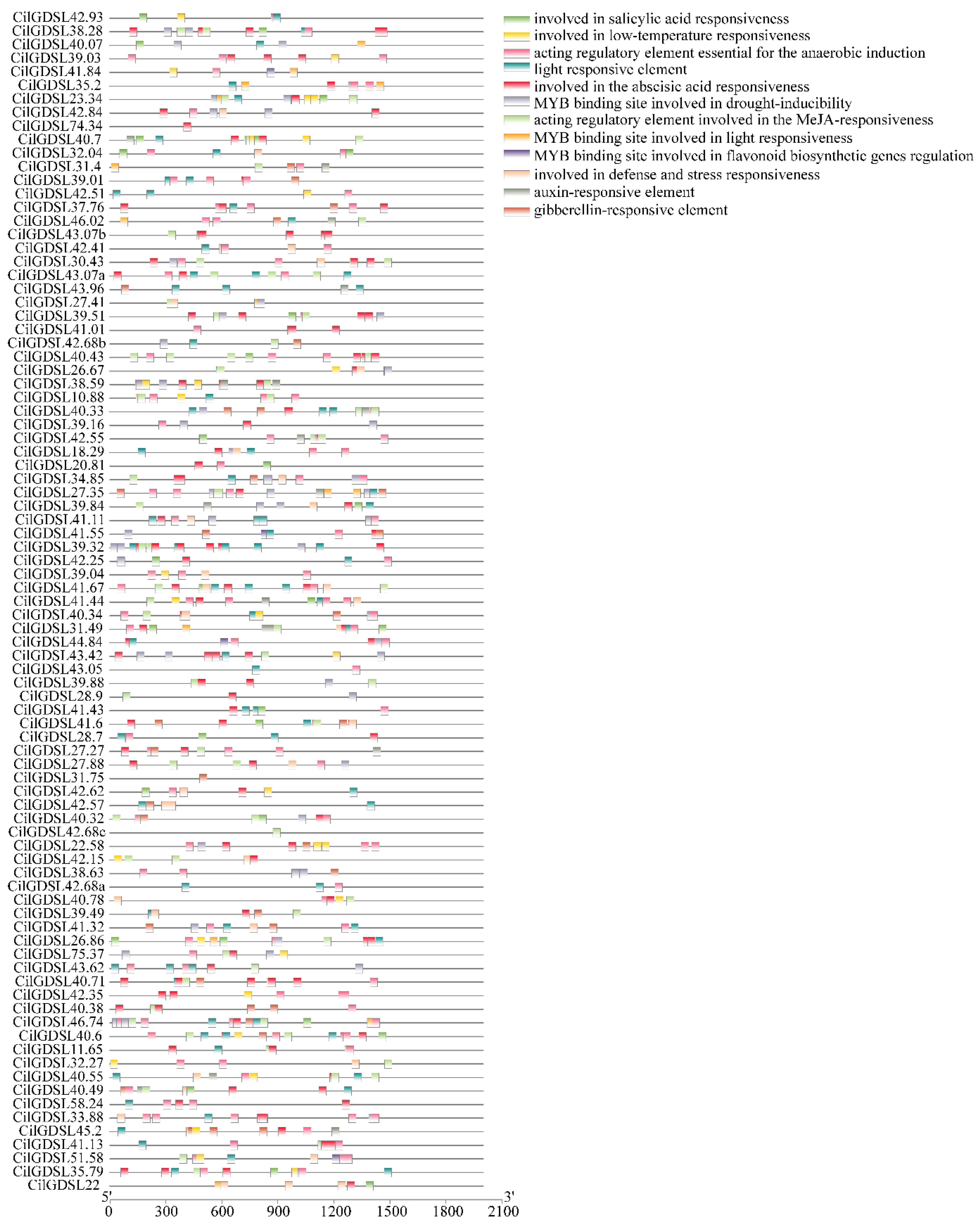
2.3. Phylogenetic Analysis, Gene Structure, and Cis-Acting Elements in GDSL Promoter Regions
2.4. Analysis of GDSL Expression Profiles Using qRT-PCR and Isobaric Tags for Relative and Absolute Quantification (iTRAQ)
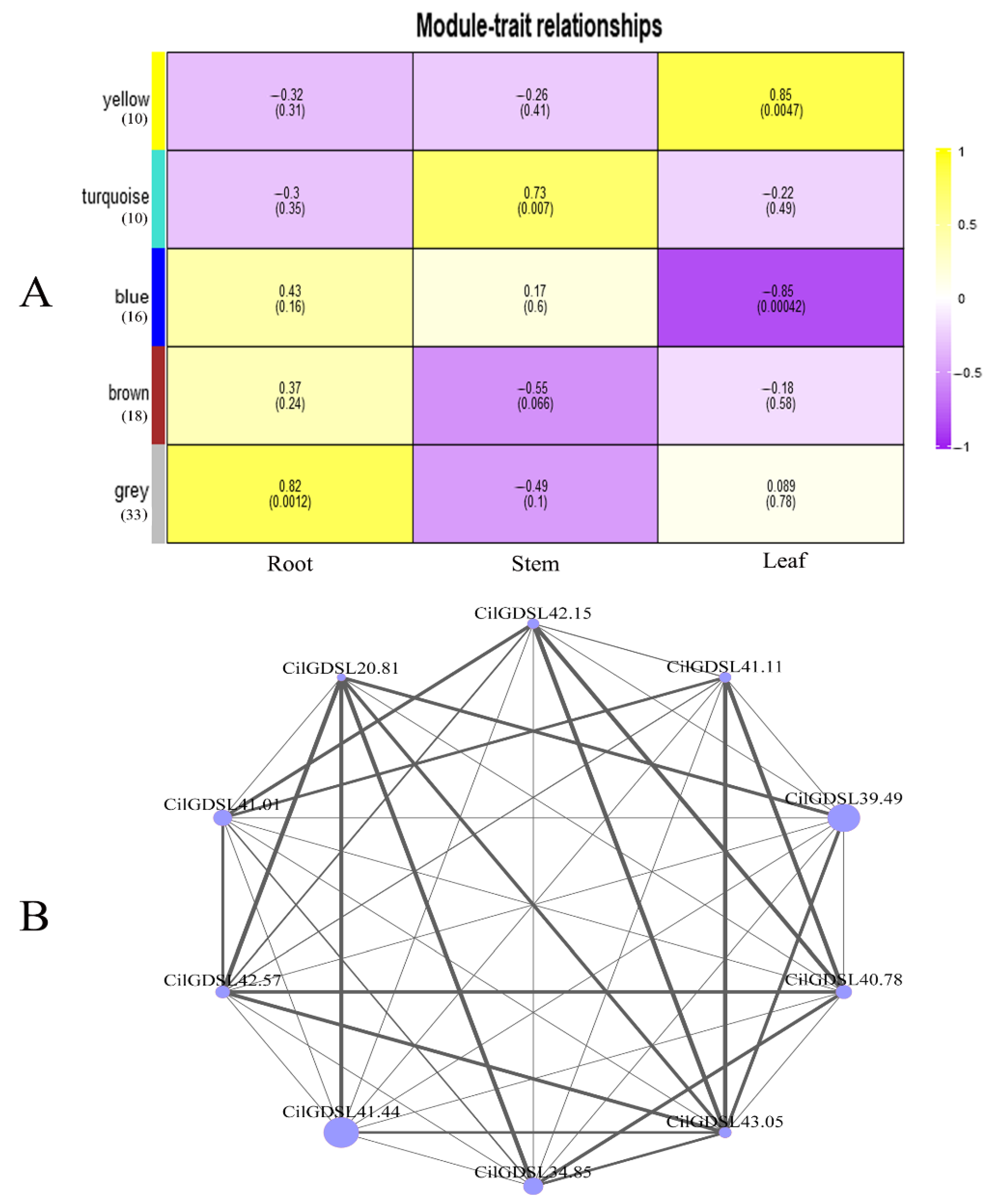
2.5. The Weighted Gene Co-Expression Network Analyses (WGCNA)
3. Results
3.1. Characterization of the GDSL-Type Lipase Genes in Pecan
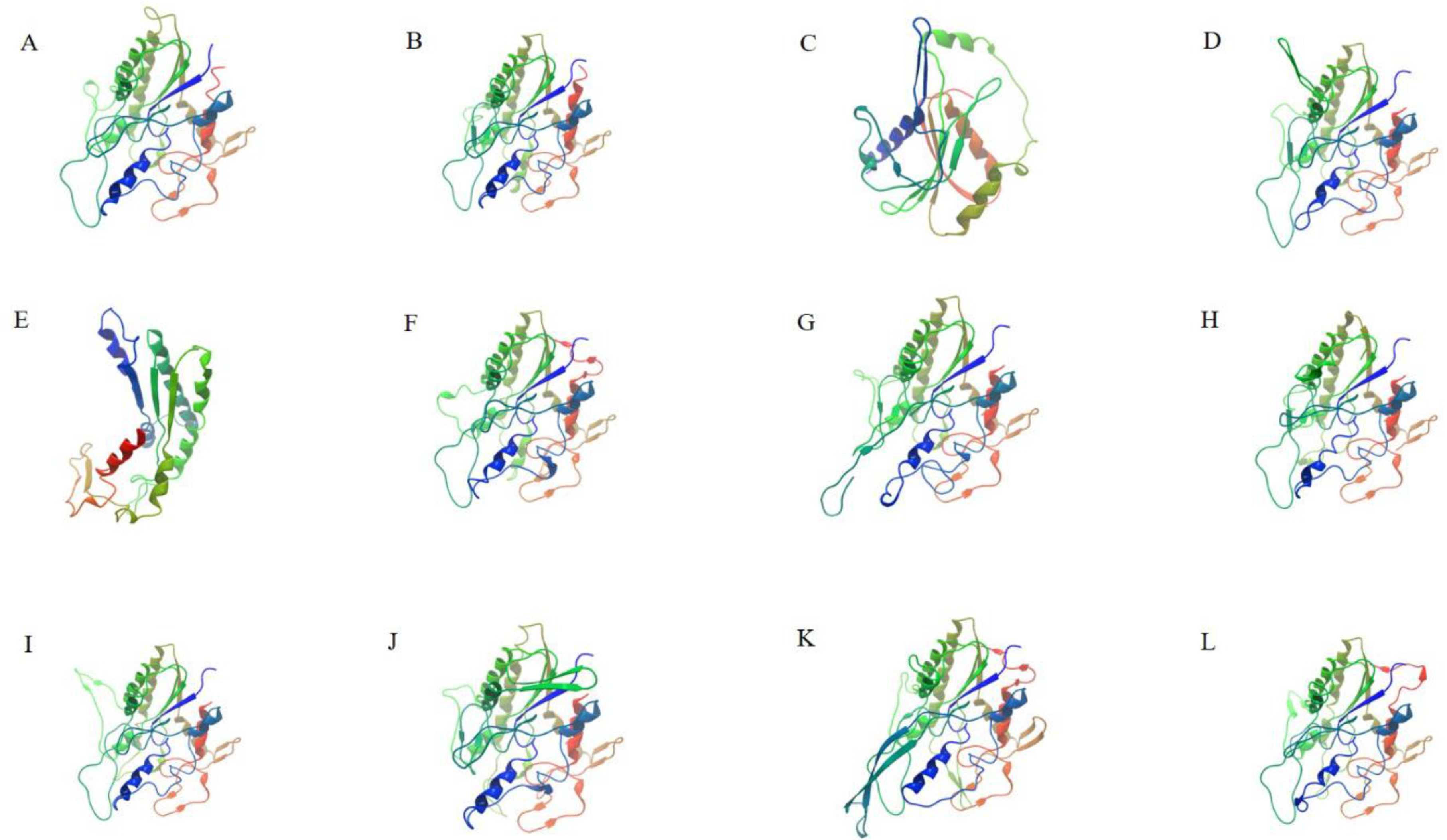
3.2. Phylogenetic Analysis and Gene Structure of 87 GDSLs
3.3. Detection of Cis-Acting Elements in Promoter Regions of CilGDSLs
3.4. Expression Profiles of GDSL Genes and iTRAQ Analysis in Response to Salt Stress
3.5. Weighted Gene Co-Expression Network Analysis (WGCNA) of GDSLs in Pecan under Salt Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lai, C.P.; Huang, L.M.; Chen, L.F.O.; Chan, M.T.; Shaw, J.F. Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis. Plant Mol. Biol. 2017, 95, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Zhao, J.; Zuo, K.; Qiu, C.; Yao, H.; Qin, J.; Sun, X.; Tang, K. Isolation and expression analysis of a GDSL-like lipase gene from Brassica napus L. J. Biochem. Mol. Biol. 2006, 39, 297. [Google Scholar] [CrossRef] [PubMed]
- Chepyshko, H.; Lai, C.P.; Huang, L.M.; Liu, J.H.; Shaw, J.F. Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa L. japonica) genome: New insights from bioinformatics analysis. BMC Genom. 2012, 13, 309. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, R.; Dong, J.; Xu, Z.; Gao, X. Analysis of GDSL lipase (GLIP) family genes in rice (Oryza sativa). Plant Omics 2012, 5, 351–358. [Google Scholar]
- Gao, M.; Yin, X.; Yang, W.; Lam, S.M.; Tong, X.; Liu, J.; Wang, X.; Li, Q.; Shui, G.H.; He, Z.H. GDSL lipases modulate immunity through lipid homeostasis in rice. PLoS Pathog. 2017, 13, e1006724. [Google Scholar] [CrossRef]
- Ding, L.N.; Li, M.; Wang, W.J.; Cao, J.; Wang, Z.; Zhu, K.M.; Yang, Y.H.; Li, Y.L.; Tan, X.L. Advances in plant GDSL lipases: From sequences to functional mechanisms. Acta Physiol. Plant. 2019, 41, 1–11. [Google Scholar] [CrossRef]
- Ling, H. Sequence analysis of GDSL lipase gene family in Arabidopsis thaliana. Pak. J. Biol. Sci. 2008, 11, 763–767. [Google Scholar] [CrossRef]
- Clauß, K.; Baumert, A.; Nimtz, M.; Milkowski, C.; Strack, D. Role of a GDSL lipase-like protein as sinapine esterase in Brassicaceae. Plant J. 2008, 53, 802–813. [Google Scholar] [CrossRef]
- Ni, P.Y.; Ji, X.R.; Guo, D.L. Genome-wide identification, characterization, and expression analysis of GDSL-type esterases/lipases gene family in relation to grape berry ripening. Sci. Hortic. 2020, 264, 109162. [Google Scholar] [CrossRef]
- Dong, X.; Yi, H.; Han, C.T.; Nou, I.S.; Hur, Y. GDSL esterase/lipase genes in Brassica rapa L.: Genome-wide identification and expression analysis. Mol. Genet. Genom. 2016, 291, 531–542. [Google Scholar] [CrossRef]
- Naranjo, M.A.; Forment, J.; RoldÁN, M.; Serrano, R.; Vicente, O. Overexpression of Arabidopsis thaliana LTL1, a salt-induced gene encoding a GDSL-motif lipase, increases salt tolerance in yeast and transgenic plants. Plant Cell Environ. 2006, 29, 1890–1900. [Google Scholar] [CrossRef] [PubMed]
- Zumaquero, A.; Martínez-Ferri, E.; Matas, A.J.; Reeksting, B.; Olivier, N.A.; Pliego-Alfaro, F.; Barceló, A.; van den Berg, N.; Pliego, C. Rosellinia necatrix infection induces differential gene expression between tolerant and susceptible avocado rootstocks. PLoS ONE 2019, 14, e0212359. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, Q.; Dai, Z.; Wang, Y.; Zhang, Y.; Li, B.; Zhao, W.; Hao, J. Identification of QTLs for resistance to maize rough dwarf disease using two connected RIL populations in maize. PLoS ONE 2019, 14, e0226700. [Google Scholar] [CrossRef] [PubMed]
- Manghwar, H.; Hussain, A.; Ali, Q.; Liu, F. Brassinosteroids (BRs) role in plant development and coping with different stresses. Int. J. Mol. Sci. 2022, 23, 1012. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhu, X.; Tian, W.; Cheng, W.; Sun, J.; Jin, S.; Zhu, H. Genome-wide identification and expression analysis of polyamine oxidase genes in upland cotton (Gossypium hirsutum L.). Plant Cell Tissue Org. 2017, 129, 237–249. [Google Scholar] [CrossRef]
- Manghwar, H.; Hussain, A. Mechanism of tobacco osmotin gene in plant responses to biotic and abiotic stress tolerance: A brief history. Biocell 2022, 46, 623. [Google Scholar] [CrossRef]
- Zhang, J.; Jiao, Y.; Shen, D.; Hong, C.; Wei, B.; Pan, C. Effects of salt stress on the fruit quality in pecan (Carya illinoensis K. Koch). J. Zhejiang Sci. Tech. 2022, in press. [Google Scholar]
- Shen, G.; Sun, W.; Chen, Z.; Shi, L.; Hong, J.; Shi, J. Plant GDSL esterases/lipases: Evolutionary, physiological and molecular functions in plant development. Plants 2022, 11, 468. [Google Scholar] [CrossRef]
- Xu, Y.; Yan, F.; Zong, Y.; Li, J.; Gao, H.; Liu, Y.; Wang, Y.; Zhu, Y.; Wang, Q. Proteomic and lipidomics analyses of high fatty acid AhDGAT3 transgenic soybean reveals the key lipase gene associated with the lipid internal mechanism. Genome 2022, 65, 153–164. [Google Scholar] [CrossRef]
- Zhang, J.; Jiao, Y.; Sharma, A.; Shen, D.; Wei, B.; Hong, C.; Zheng, B.; Pan, C. Transcriptomic analysis reveals potential pathways associated with salt resistance in pecan (Carya illinoensis K. Koch). J. Biotechnol. 2021, 330, 17–26. [Google Scholar] [CrossRef]
- Huang, Y.; Xiao, L.; Zhang, Z.; Zhang, R.; Wang, Z.; Huang, C.; Huang, R.; Luan, Y.; Fan, T.; Wang, J.; et al. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. GigaScience 2019, 8, giz036. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liao, L.; Nie, M.; Peng, W.; Zhang, M.; Lei, J.; Zhong, Y.; Liao, H.; Chen, Z. A VIT-like transporter facilitates iron transport into nodule symbiosomes for nitrogen fixation in soybean. New Phytol. 2020, 226, 1413–1428. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Du, H.; Wang, Y.; Wang, H.; Yang, S.; Li, C.; Chen, N.; Yang, H.; Zhang, Y.; Zhu, Y.; et al. The calcium-dependent protein kinase ZmCDPK7 functions in heatstress tolerance in maize. J. Integr. Plant Biol. 2021, 63, 510–527. [Google Scholar] [CrossRef] [PubMed]
- Kramer, L.D.; Ciota, A.T.; Kilpatrick, A.M. Introduction, spread, and establishment of West Nile virus in the Americas. J. Med. Entomol. 2019, 56, 1448–1455. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Pee, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Rychlik, W. OLIGO 7 primer analysis software. In PCR Primer Design; Humana Press: Totowa, NJ, USA, 2007; pp. 35–59. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Liu, A.Y.; Zhang, W.; Lv, S.J.; Ren, K. The effect of pecan oil on hippocampal cells apoptosis of ovariectomized rats. Procedia Eng. 2011, 18, 375–380. [Google Scholar] [CrossRef][Green Version]
- Wang, L.; Zhang, L.; Chen, J.; Huang, D.; Zhang, Y. Physiological analysis and transcriptome comparison of two muskmelon (Cucumis melo L.) cultivars in response to salt stress. Genet. Mol. Res. 2016, 15, 1–18. [Google Scholar] [CrossRef]
- Jiao, Y.; Zhang, J.; Pan, C. Integrated physiological, proteomic, and metabolomic analyses of pecan cultivar ‘Pawnee’adaptation to salt stress. Sci. Rep. 2022, 12, 1841. [Google Scholar] [CrossRef]
- Muralidharan, M.; Buss, K.; Larrimore, K.E.; Segerson, N.A.; Kannan, L.; Mor, T.S. The Arabidopsis thaliana ortholog of a purported maize cholinesterase gene encodes a GDSL-lipase. Plant Mol. Biol. 2013, 81, 565–576. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.J.L.; Au, A.Y.; Ritchie, W.; Rasko, J.E. Intron retention in mRNA: No longer nonsense: Known and putative roles of intron retention in normal and disease biology. Bioessays 2016, 38, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.; Cheong, H.; Park, O. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Kim, B.K.; Kwon, S.J.; Jin, H.C.; Park, O.K. Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 2009, 379, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Yang, X.; Xu, J.; Zhang, M.; Liu, G.; Yao, X. Genome-wide identification and analysis of GDSL-type esterases/lipases in watermelon (Citrullus lanatus). Sci. Hortic. 2019, 289, 110461. [Google Scholar] [CrossRef]
- Hegebarth, D.; Jetter, R. Cuticular waxes of Arabidopsis thaliana shoots: Cell-type-specific composition and biosynthesis. Plants 2017, 6, 27. [Google Scholar] [CrossRef]
- Tang, J.; Yang, X.; Xiao, C.; Li, J.; Chen, Y.; Li, R.; Li, S.; Lü, S.; Hu, H. GDSL lipase occluded stomatal pore 1 is required for wax biosynthesis and stomatal cuticular ledge formation. New Phytol. 2020, 228, 1880–1896. [Google Scholar] [CrossRef]
- Ayaz, A.; Huang, H.; Zheng, M.; Zaman, W.; Li, D.; Saqib, S.; Zhao, H.; Lü, S. Molecular cloning and functional analysis of GmLACS2-3 reveals its involvement in cutin and suberin biosynthesis along with abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 9175. [Google Scholar] [CrossRef]
- Ayaz, A.; Saqib, S.; Huang, H.; Zaman, W.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, M.; Li, Y.; Yan, W.; Chang, Z.; Ni, H.; Chen, Z.; Wu, J.; Xu, C.; Deng, X.; et al. GDSL esterase/lipases OsGELP34 and OsGELP110/OsGELP115 are essential for rice pollen development. J. Integr. Plant Biol. 2020, 62, 1574–1593. [Google Scholar] [CrossRef]
- Li, H.; Han, X.; Qiu, W.; Xu, D.; Wang, Y.; Yu, M.; Hu, X.; Zhuo, R. Identification and expression analysis of the GDSL esterase/lipase family genes, and the characterization of SaGLIP8 in Sedum alfredii Hance under cadmium stress. PeerJ 2019, 7, e6741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zou, X.; Tang, W.; Zheng, Y. Revelation on early response and molecular mechanism of submergence tolerance in maize roots by microarray and suppression subtractive hybridization. Environ. Exp. Bot. 2006, 58, 53–63. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Shankar, R.; Jain, M. Whole genome sequence analysis of rice genotypes with contrasting response to salinity stress. Sci. Rep. 2020, 10, 21259. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Zhang, X.; Wang, T.; Wei, W.; Wang, Y.; Chen, J.; Zhou, Y.; Chen, M.; Ma, Y.; Xu, Z.; et al. Genome-Wide Identification, Evolution, and Expression of GDSL-Type Esterase/Lipase Gene Family in Soybean. Front. Plant Sci. 2020, 11, 726. [Google Scholar] [CrossRef]
- Cao, D.; Cheng, H.; Wu, W.; Soo, H.M.; Peng, J. Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 2006, 142, 509–525. [Google Scholar] [CrossRef]
- Guo, Y.; Xiao, P.; Lei, S.; Deng, F.; Xiao, G.G.; Liu, Y.; Chen, X.; Li, L.; Wu, S.; Chen, Y.; et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim. Biophys. Sin. 2008, 40, 426–436. [Google Scholar] [CrossRef]
- Badreldin, A.A.; Bagheri, L.; Zhang, B.; Larson, A.N.; van Wijnen, A.J. Relative mRNA and protein stability of epigenetic regulators in musculoskeletal cell culture models. Gene 2021, 766, 145032. [Google Scholar] [CrossRef]



| No. | Gene | Protein Symbol | Description * | Log2 (Fold Change) | Subgroup | ||
|---|---|---|---|---|---|---|---|
| 0.3% | 0.6% | 0.9% | |||||
| 1 | CilGDSL41.11 | LOC108993637 | GDSL-like Lipase, At5g33370-like | 1.28 | 1.37 | 1.27 | L |
| 2 | CilGDSL39.84 | LOC108993591 | Li-tolerant lipase 1, LTL1-like | 1.20 | 1.39 | 1.38 | L |
| 3 | CilGDSL31.75 | LOC108981353 | GDSL-like Lipase, At5g03610-like | 1.28 | 1.27 | 1.45 | F |
| 4 | CilGDSL38.59 | LOC108983791 | GDSL esterase/lipase APG | - | 1.38 | 1.37 | A |
| 5 | CilGDSL42.57 | LOC108982502 | GDSL-like, AtFXG1-homologous | - | 1.41 | 1.67 | D |
| 6 | CilGDSL40.34 | LOC108998072 | GDSL-like Lipase, At5g45670-like | 1.21 | 1.40 | 1.43 | I |
| 7 | CilGDSL27.88 | LOC108983860 | GDSL-like Lipase, At5g03610-like | 1.32 | 1.21 | 1.34 | F |
| 8 | CilGDSL40.78 | LOC108988817 | GDSL-like Lipase, At5g33370-like | 0.83 | 0.94 | 0.80 | L |
| 9 | CilGDSL31.4 | LOC109012662 | GDSL-like Lipase, At4g28780-like | - | 1.29 | 1.35 | K |
| 10 | CilGDSL27.35 | LOC108981001 | SGNH hydrolase-type esterase, At5g62930-like | - | 1.23 | 1.43 | C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Zhang, J.; Pan, C. Genome-Wide Analysis of the GDSL Genes in Pecan (Carya illinoensis K. Koch): Phylogeny, Structure, Promoter Cis-Elements, Co-Expression Networks, and Response to Salt Stresses. Genes 2022, 13, 1103. https://doi.org/10.3390/genes13071103
Jiao Y, Zhang J, Pan C. Genome-Wide Analysis of the GDSL Genes in Pecan (Carya illinoensis K. Koch): Phylogeny, Structure, Promoter Cis-Elements, Co-Expression Networks, and Response to Salt Stresses. Genes. 2022; 13(7):1103. https://doi.org/10.3390/genes13071103
Chicago/Turabian StyleJiao, Yun, Jianhong Zhang, and Cunde Pan. 2022. "Genome-Wide Analysis of the GDSL Genes in Pecan (Carya illinoensis K. Koch): Phylogeny, Structure, Promoter Cis-Elements, Co-Expression Networks, and Response to Salt Stresses" Genes 13, no. 7: 1103. https://doi.org/10.3390/genes13071103
APA StyleJiao, Y., Zhang, J., & Pan, C. (2022). Genome-Wide Analysis of the GDSL Genes in Pecan (Carya illinoensis K. Koch): Phylogeny, Structure, Promoter Cis-Elements, Co-Expression Networks, and Response to Salt Stresses. Genes, 13(7), 1103. https://doi.org/10.3390/genes13071103






