A Formative Study of the Implementation of Whole Genome Sequencing in Northern Ireland
Abstract
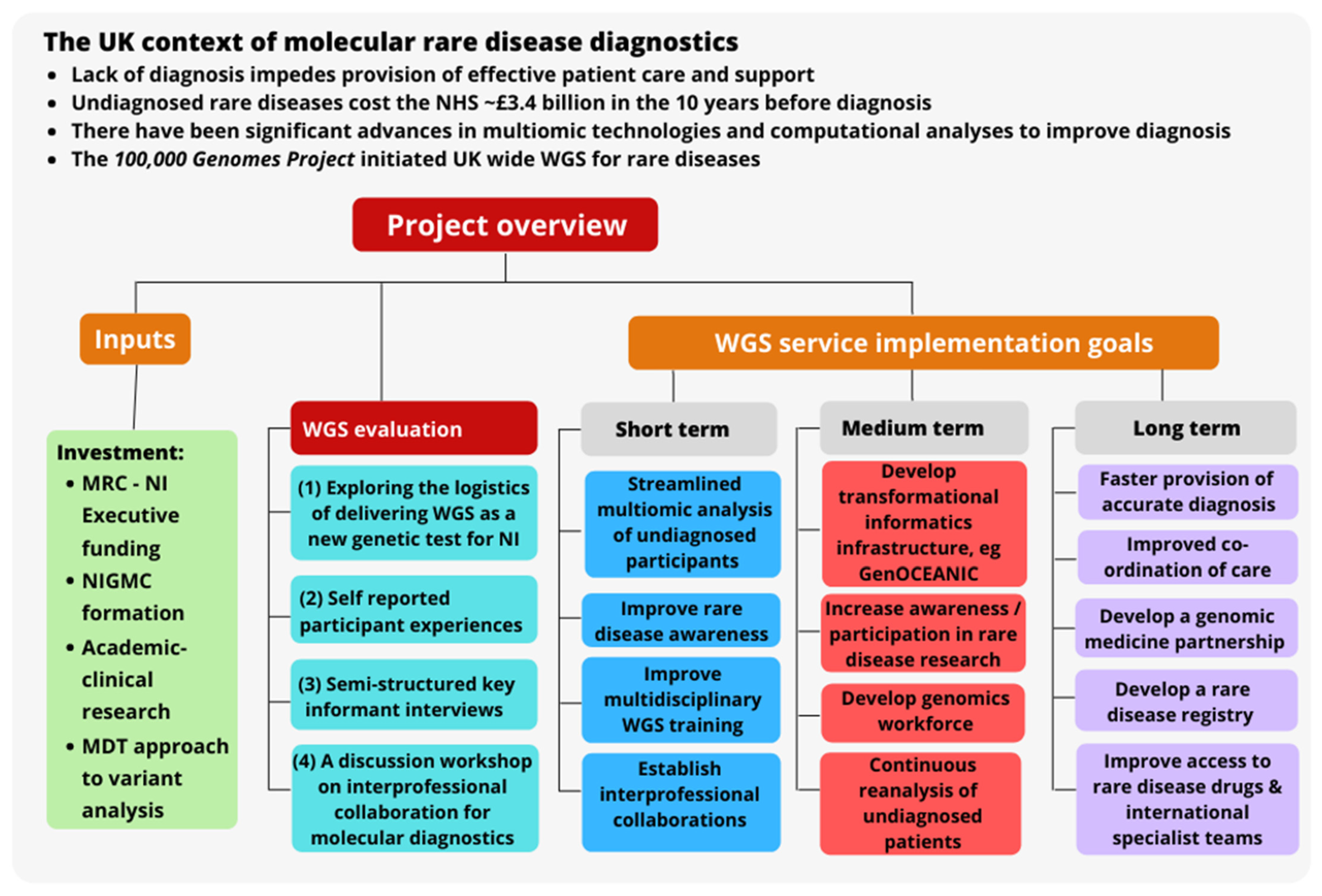
:1. Introduction
2. Materials and Methods
2.1. Quantitative Analysis of the Implementation and Delivery of WGS as a New Genetic Test Service
2.2. Participant Self-Reported Views and Experiences
2.3. Semi-Structured Key Informant Interviews
2.4. Workshop about Interprofessional Collaboration for Molecular Diagnostics
- Benefits of interprofessional collaboration on molecular diagnostics and research.
- Barriers to interprofessional collaboration on molecular diagnostics and research.
- What should change about collaboration on molecular diagnostics and research.
3. Results
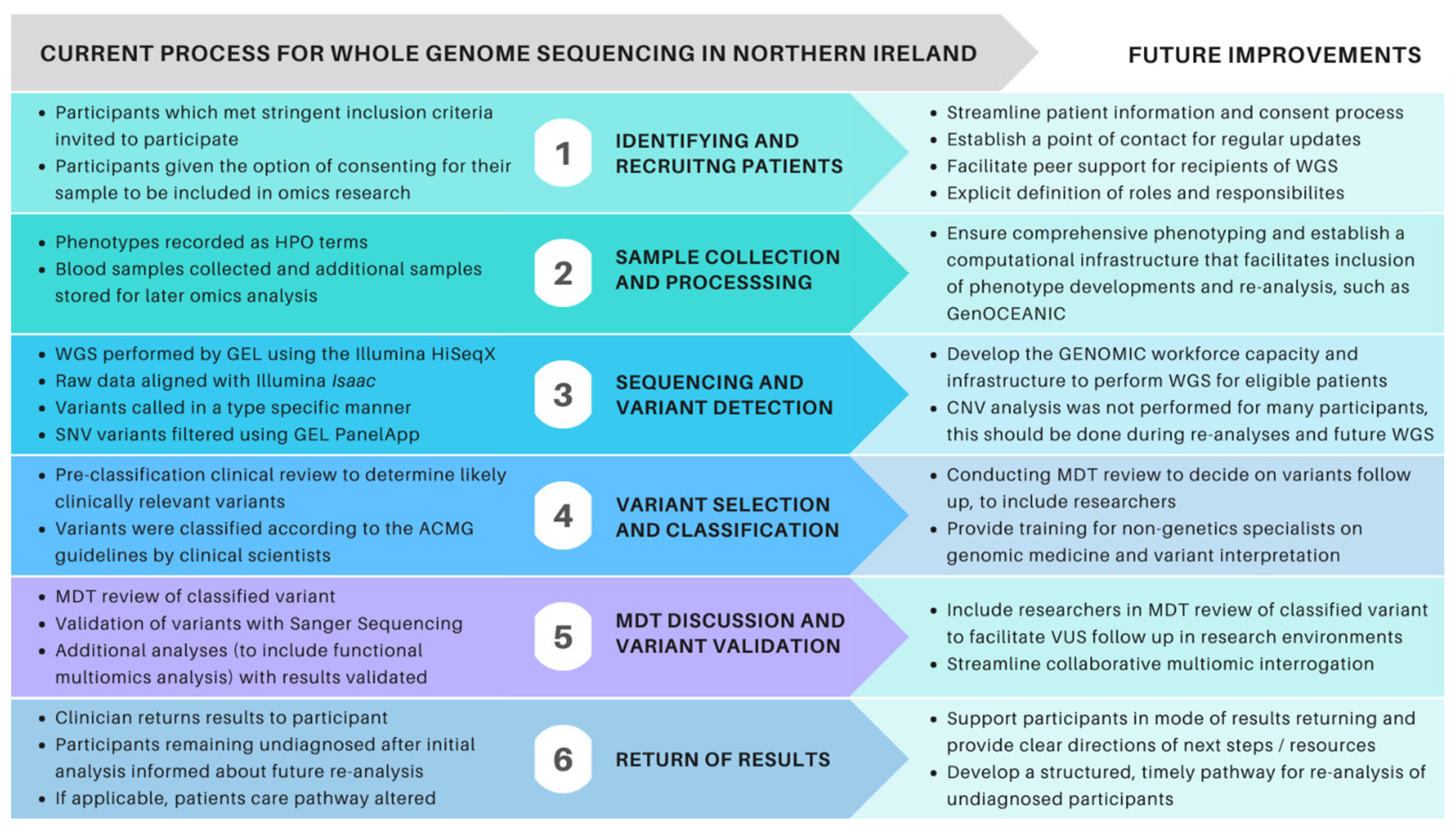
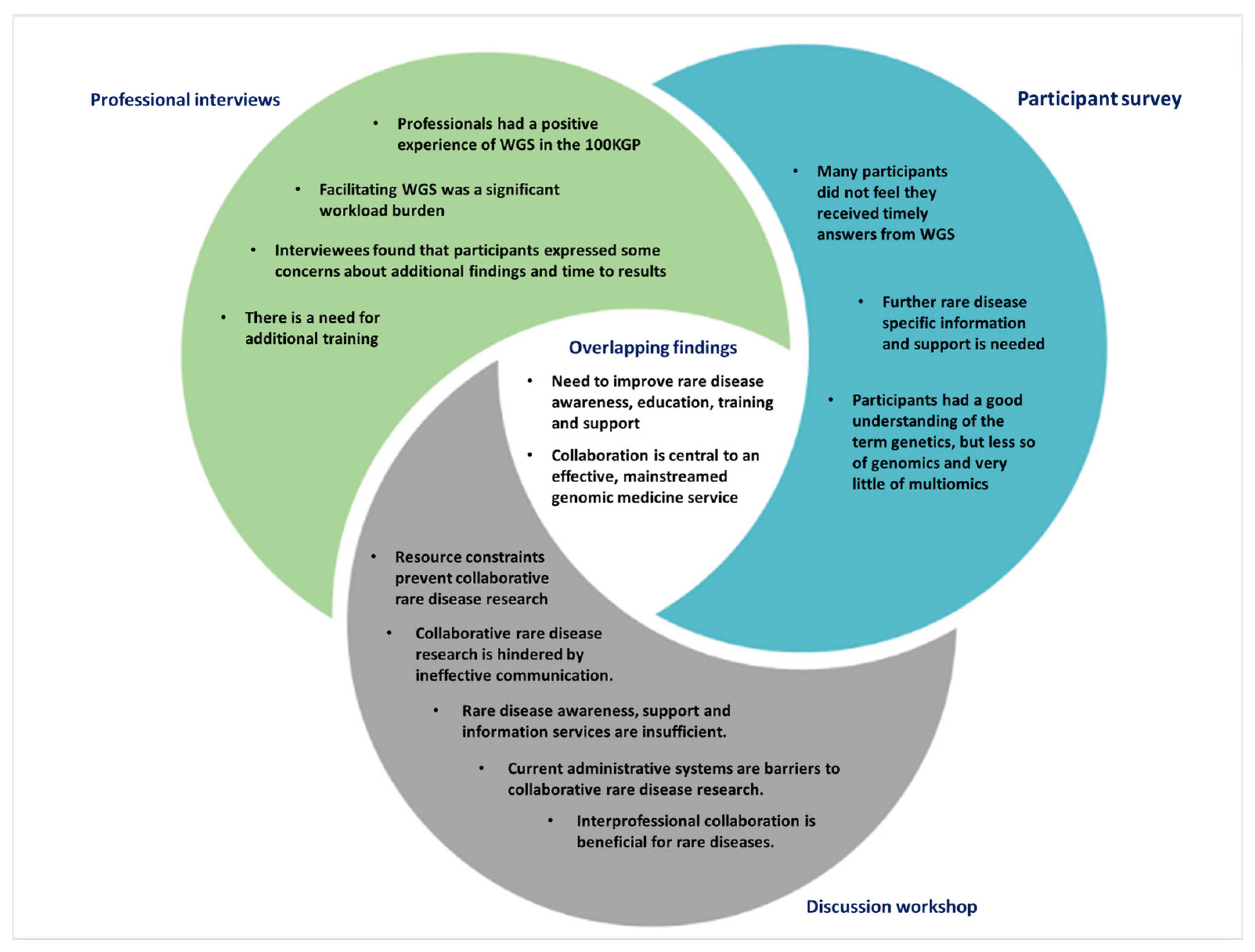
3.1. Exploring the Logistics of Delivering WGS as a New Genetic Test for NI
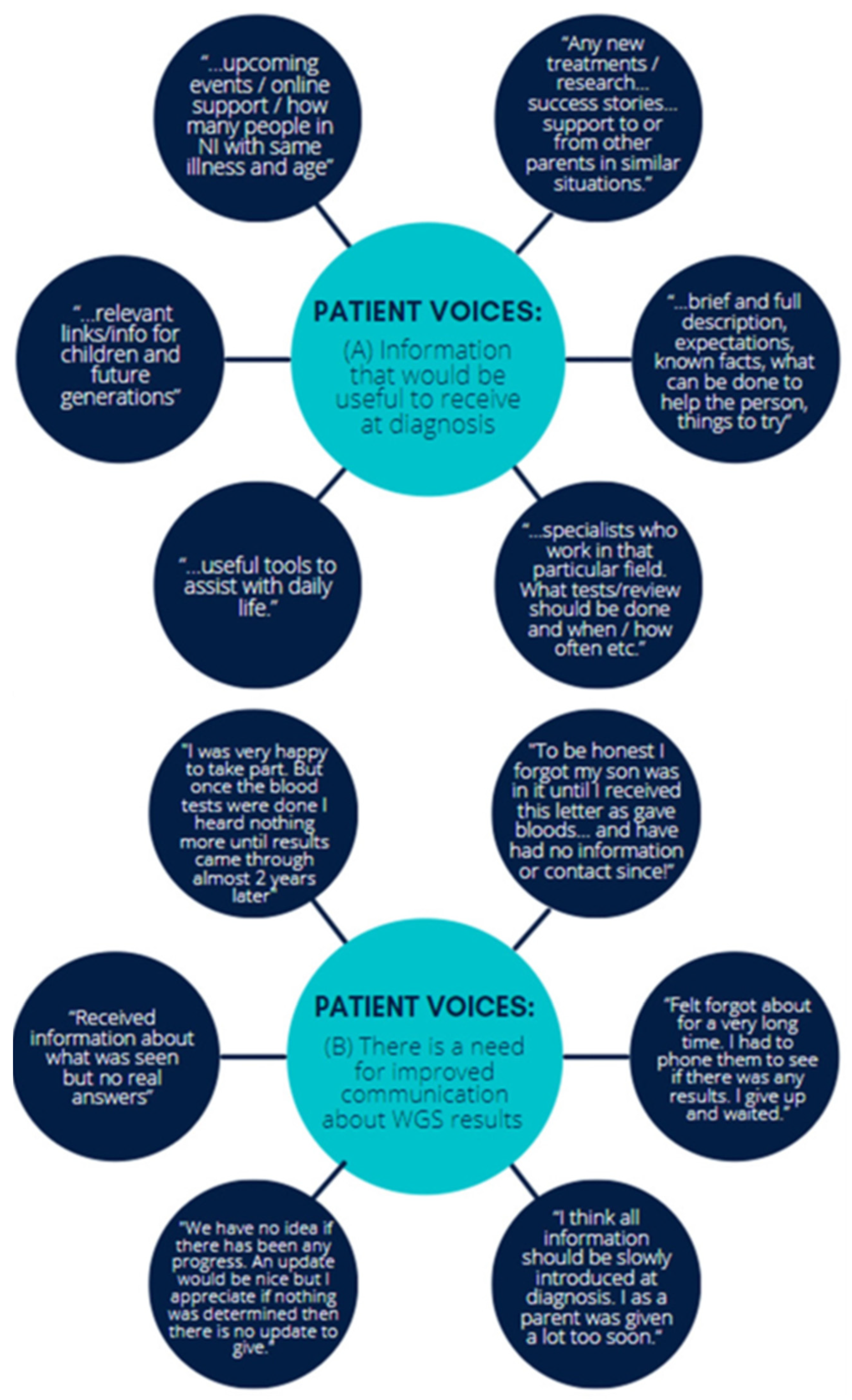
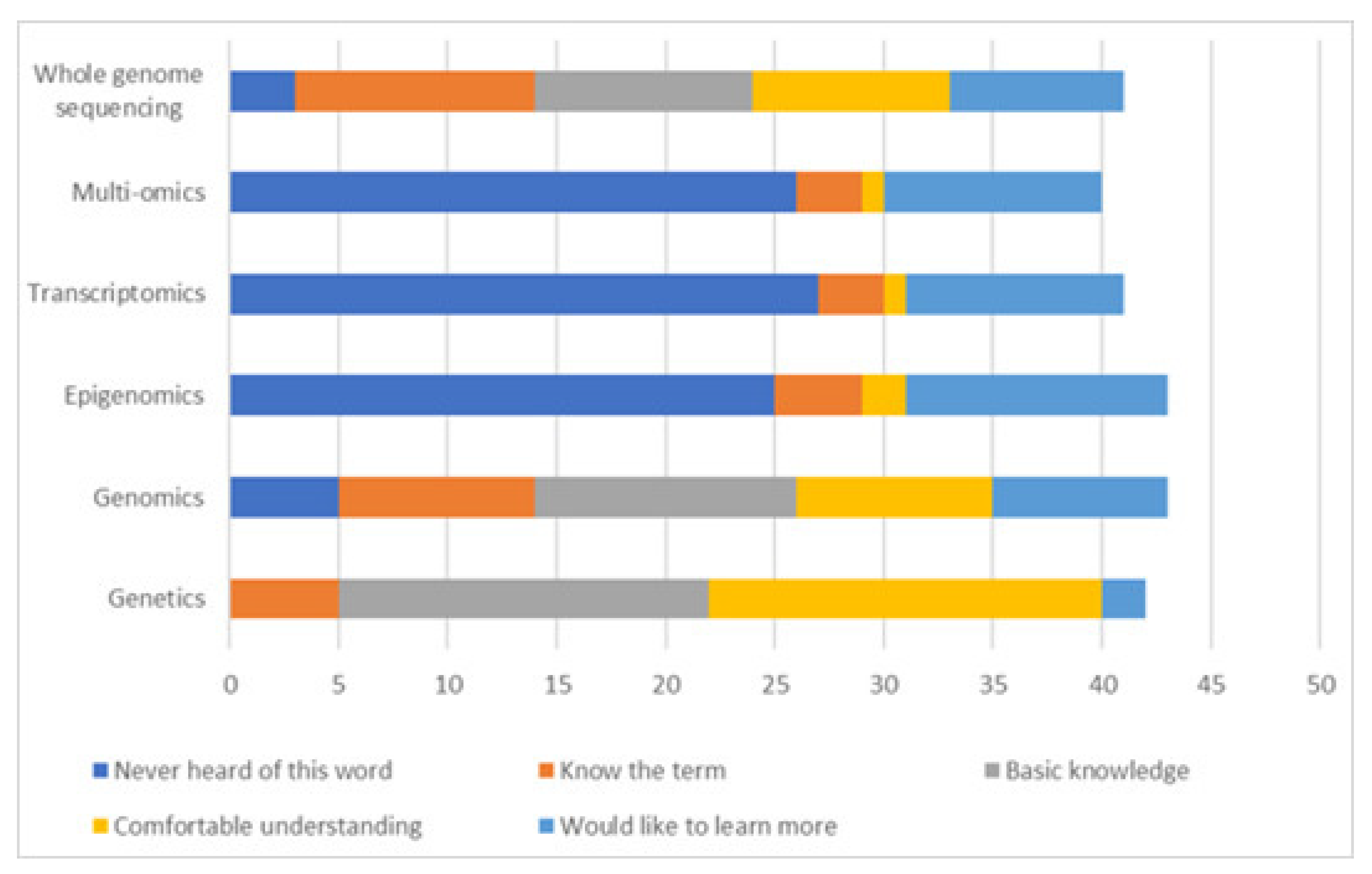
3.2. Participant Self-Reported Views and Experiences
“… gain a broader knowledge and was able to see that this project would have such positive outcomes for the future and the identification and treatment of rare diseases.”[p22]
“Obviously COVID-19 knocked things all about. Our child is very precious so to receive first a letter with all the medical diagnosis and language was very unnerving. A phone call or appointment would have been more appropriate, but I know it is strange times… It was very nerve wrecking until I got speaking to [the doctors].”[p26]
“It was an excellent opportunity…Getting the results during COVID19 via letter was very disheartening.”[p26]
“Would be great to have information instead of google…”[p20]
“I believe the more information and knowledge I have to help my child, helps more than not knowing at all and only working on scraps of information”[p28]
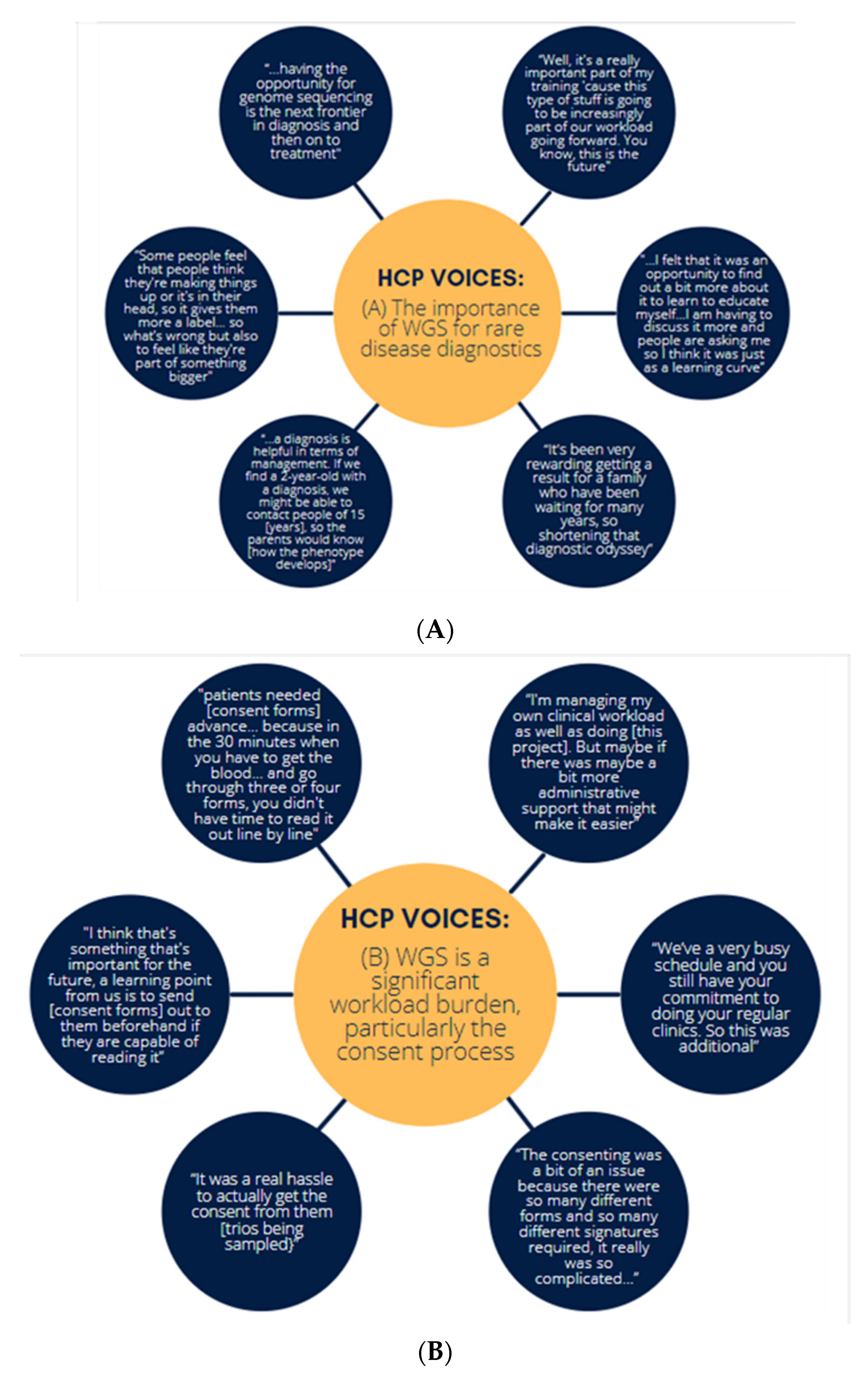
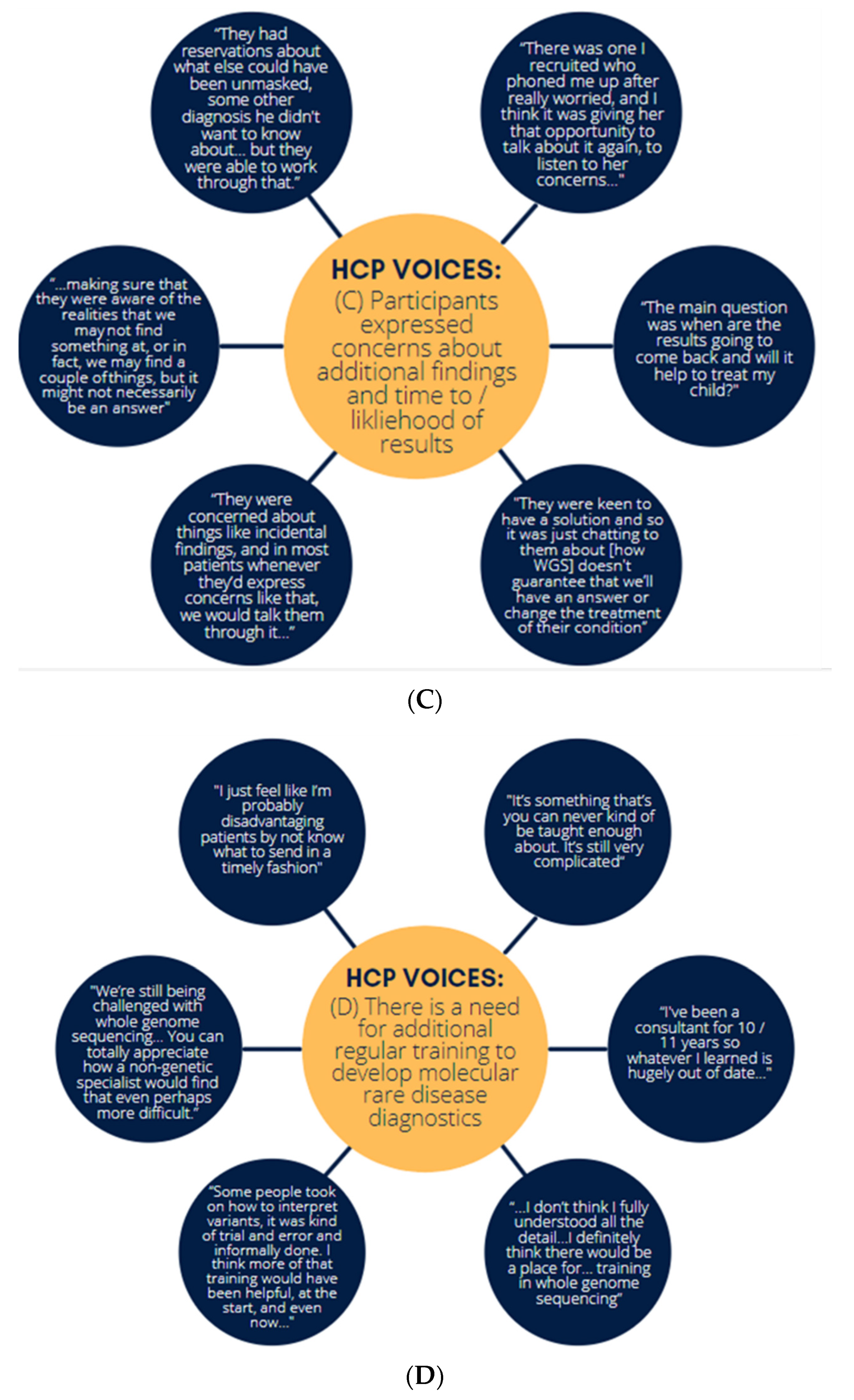
3.3. Semi-Structured Interviews of Healthcare Professionals Who Participated in WGS
- Healthcare professionals had a positive experience with WGS.
- Facilitating WGS was a significant workload burden.
- Interviewees found that participants expressed some concerns about additional findings and time to results.
- There is a need for additional training.
“I think that the multi-disciplinary team meetings with the lab were brilliant… I do think that kind of lab liaison is helpful… both in terms of understanding the variant interpretation and being able to explain that to patients. It was helpful for the whole clinical process and from an educational point of view, and even just people working together better.”[p14]
“We can’t multiply people overnight… One of the great outcomes [for the NIGMC] is the upskilling and the education and training of their staff… so there’s a lot of good, but … there’s a lot of unknowns that should be flagged up in the future for any major new service.”[p9]
…” I do wonder if the consent form and our discussion with patients will truly have prepared them for the incidental findings… to be honest, I don’t know how well they did understand what they were signing up for. But as a doctor, I think we saw that it was overwhelmingly positive and worth it for them. And the only really realistic, next step in terms of testing.”[p4]
“There were some patients who were like, “I don’t really care what I’m consenting for but I just desperately want an answer for my child.” I was having to say, “but I need to go through this with you.” A very typical comment would have been, “Oh, anything I can do to help your research doctor.” And yes, there could be fruitful answers out of it, hopefully. Sometimes they were just so desperate to be eligible. I think for me the biggest thing was managing people’s expectations of a diagnosis at the end of the project.”[p10]
“…having something you can actually go back to refresh your memory could be really helpful”.[p18]
“Sometimes you get, you get everything’s taught at the same time and then it’s gonna be ages before you actually get to use that information. And then you kind of forget.”[p14]
“I think it’s a tier three that’s probably more niggling to myself and my colleagues, because we realise a large portion of our patients are waiting from results specifically out of tier 3 and we haven’t got the resources or man power yet to address those, and I think that is where the problem potentially lies.”[p16]
“We have found that whatever way the algorithm’s working, that a fairly high percentage of the time our actual results, that causative result, is in Tier 3. OK, definitely not the majority of the time, or else in the un-tiered, but it could be as high as 5–10% of the time.”[p4]
“[Patients] had to come up to Belfast for appointments, which was probably a problem for the patients, especially if they had any disabilities or children, then just getting them up for travel. And the appointment times started at 9:00 AM in the morning, if they had other children to get out to school or anything.”[p12]
- A simple approach to rare disease information that both non-specialists and health care professionals could access easily.
- Financial entitlements that patients are entitled to with relevant links.
- Online consenting for research, perhaps via a video call.
- Signposting facility to avoid the website being too heavy on information, e.g., to patient support groups.
- Disease specific information.
- Educational resources regarding genetic/genomic testing.
- Contact details of willing clinicians working in particular areas of rare disease.
3.4. A Discussion Workshop on Interprofessional Collaboration for Molecular Diagnostics
- Resource constraints prevent collaborative rare disease research.
- Collaborative rare disease research is hindered by ineffective communication.
- Rare disease awareness, support and information services are insufficient.
- Current administrative systems are barriers to collaborative rare disease research.
- Interprofessional collaboration is beneficial for rare diseases.
“Venues need to be accessible to all. …[often] accessible facilities aren’t truly accessible.”[p1]
“Research is not encouraged in NI clinical practice and a lot of people in areas of rare disease have lost their opportunity to be involved in research. It’s difficult to motivate training clinicians to be involved in research.”[p2]
“We need to not only encourage people to do research but also show them a pathway to make this research applicable to their career, e.g., is there funding for a post that then continues [their research]? How does this augment my career?”[p3]
“It’s very difficult to bring in money to do this type of research, money is so competitive.”[p1]
“We need more rare disease researchers and more investment in rare disease research in NI”[p4]
“[Rare disease awareness] starts from primary school, if you have somebody who has a rare disease, if you have teachers that are made aware of it, it can grow and mushroom in the community.”[p1]
“There is a lot of these workshops and surveys but there isn’t a great amount of progress from it, what happens from this research?”[p1]
“My only experience of a rare disease research project was as a patient, signing a consent form prior to a surgery and then never hearing anything else from it. I want to know what comes out of the research projects!”[p3]
“You don’t hear any of the results that’s coming out of [research projects], especially true in rural areas…We need more information to be posted on social media. You don’t hear about project progress either.”[p4]
“You can’t take part if you don’t know about research”[p1]
“Seeing the consultant is great and getting the letter is great, but if you have no idea how to contact them back, people fall apart. Having some way of being able to contact them back and [knowing] what do we need to do next is crucial.”[p3]
“We don’t have a [rare disease] centre, it would be nice to have this so that families or people that are affected could meet and discuss mental health issues.”[p1]
“Emotional support for this type of research is really crucial, need for reassurance, there may be anxiety about taking part in studies, so there needs to be somewhere they can go ask questions, could possibly work with charities to do this.”[p6]
“It’s not the healthcare professionals’ fault that they don’t always know about all rare diseases, we need to increase awareness amongst them too.”[p1]
“Still waiting on approval for minor [ethics] revisions (sitting on it for 3 months). With [University] institutions you can easily monitor the progress of ethics applications, but with the hospitals they go into a black hole. It’s very hard to get feedback.”[p3]
“… the biggest challenge is the ethical approval, it takes significantly longer than any other region…”[p2]
“In England they had a collaborative approach, we were able to produce research mostly by systematic reviews which was able to shape the R21 pathway. It was really nice to be part of a team that translated [research] into progress.”[p2]
“A multidisciplinary approach was beneficial, (including scientists, midwifes, clinical geneticists) to discuss the case and interpret the evidence.”[p2]
“Maybe there’s one person in NI who has a rare disease, that person may only be able to go to America to find out something about their rare disease. Therefore, benefits of collaborative research do outweigh the challenges.”[p1]
“Rather than trying to reinvent the wheel, especially North and South of the [Irish] border, we only have a certain number of specialists in this field (particularly in genomic specialist researchers), so there needs to be better collaboration.”[p2]
4. Discussion
4.1. Administrative Considerations for WGS Rare Disease Diagnostics in a Regional Setting
4.2. Analytical Considerations for WGS
4.3. Educational Considerations for WGS
4.4. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferreira, C.R. The burden of rare diseases. Am. J. Med. Genet. A 2019, 179, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Worldometer. Available online: https://www.worldometers.info/ (accessed on 20 April 2022).
- Orphanet Nomenclature. Available online: http://www.orphadata.org/cgi-bin/ORPHAnomenclature.html (accessed on 14 May 2022).
- Momozawa, Y.; Mizukami, K. Unique roles of rare variants in the genetics of complex diseases in humans. J. Hum. Genet. 2021, 66, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Blöß, S.; Klemann, C.; Rother, A.-K.; Mehmecke, S.; Schumacher, U.; Mücke, U.; Mücke, M.; Stieber, C.; Klawonn, F.; Kortum, X.; et al. Diagnostic needs for rare diseases and shared prediagnostic phenomena: Results of a German-wide expert Delphi survey. PLoS ONE 2017, 12, e0172532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spillmann, R.C.; McConkie-Rosell, A.; Pena, L.; Jiang, Y.-H.; Adams, C.J.; Adams, D.R.; Alejandro, M.E.; Allard, P.; Ashley, E.A.; Azamian, M.S.; et al. A window into living with an undiagnosed disease: Illness narratives from the Undiagnosed Diseases Network. Orphanet J. Rare Dis. 2017, 12, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowe, A.L.; McKnight, A.J.; McAneney, H. Communication Needs for Individuals with Rare Diseases within and around the Healthcare System of Northern Ireland. Front. Public Health 2019, 7, 236. [Google Scholar] [CrossRef] [Green Version]
- Bogart, K.R.; Irvin, V.L. Health-related quality of life among adults with diverse rare disorders. Orphanet J. Rare Dis. 2017, 12, 177. [Google Scholar] [CrossRef] [Green Version]
- McMullan, J.; Crowe, A.; Bailie, C.; Moore, K.; McMullan, L.; Shamandi, N.; McAneney, H.; McKnight, A.J. Improvements needed to support people living and working with a rare disease in Northern Ireland: Current rare disease support perceived as inadequate. Orphanet J. Rare Dis. 2020, 15, 315. [Google Scholar] [CrossRef]
- McMullan, J.; Crowe, A.L.; Downes, K.; McAneney, H.; McKnight, A.J. Carer reported experiences: Supporting someone with a rare disease. Health Soc. Care Community 2022, 30, 1097–1108. [Google Scholar] [CrossRef]
- Harrison, K. A preliminary assessment of the potential impact of rare diseases on the NHS. In Imperial College Health Partners; Imperial College Health Partners: London, UK, 2019. [Google Scholar]
- Nisar, H.; Wajid, B.; Shahid, S.; Anwar, F.; Wajid, I.; Khatoon, A.; Sattar, M.U.; Sadaf, S. Whole-genome sequencing as a first-tier diagnostic framework for rare genetic diseases. Exp. Biol. Med. 2021, 246, 2610–2617. [Google Scholar] [CrossRef]
- Bick, D.; Jones, M.; Taylor, S.L.; Taft, R.J.; Belmont, J. Case for genome sequencing in infants and children with rare, undiagnosed or genetic diseases. J. Med. Genet. 2019, 56, 783–791. [Google Scholar] [CrossRef] [Green Version]
- Wu, A.C.; McMahon, P.; Lu, C. Ending the Diagnostic Odyssey-Is Whole-Genome Sequencing the Answer? JAMA Pediatrics 2020, 174, 821–822. [Google Scholar] [CrossRef]
- Murdock, D.R.; Rosenfeld, J.A.; Lee, B. What Has the Undiagnosed Diseases Network Taught Us about the Clinical Applications of Genomic Testing? Annu. Rev. Med. 2022, 73, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhu, L.; Roberts, R.; Tong, W. Toward Clinical Implementation of Next-Generation Sequencing-Based Genetic Testing in Rare Diseases: Where Are We? Trends Genet. 2019, 35, 852–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ontario Health (Quality). Genome-Wide Sequencing for Unexplained Developmental Disabilities or Multiple Congenital Anomalies: A Health Technology Assessment. Ont. Health Technol. Assess. Ser. 2020, 20, 1–178. [Google Scholar]
- Ibañez, K.; Polke, J.; Hagelstrom, R.T.; Dolzhenko, E.; Pasko, D.; Thomas, E.R.A.; Daugherty, L.C.; Kasperaviciute, D.; Smith, K.R.; McDonagh, E.M.; et al. Whole genome sequencing for the diagnosis of neurological repeat expansion disorders in the UK: A retrospective diagnostic accuracy and prospective clinical validation study. Lancet Neurol. 2022, 21, 234–245. [Google Scholar] [CrossRef]
- Northern Ireland Statistics and Research Agency. Available online: https://www.nisra.gov.uk/?msclkid=dad032a2c0ba11eca58369318b3e2434 (accessed on 11 May 2022).
- Crowe, A.; McAneney, H.; Morrison, P.J.; Cupples, M.E.; McKnight, A.J. A quick reference guide for rare disease: Supporting rare disease management in general practice. Br. J. Gen. Pract. 2020, 70, 260–261. [Google Scholar] [CrossRef]
- Eligibility Wheels for the 100,000 Genomes Project. Available online: https://www.genomicseducation.hee.nhs.uk/eligibility-wheels/ (accessed on 26 April 2022).
- Smedley, D.; Smith, K.R.; Martin, A.; Thomas, E.A.; McDonagh, E.M.; Cipriani, V.; Ellingford, J.M.; Arno, G.; Tucci, A.; Vandrovcova, J.; et al. 100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care—Preliminary Report. N. Engl. J. Med. 2021, 385, 1868–1880. [Google Scholar]
- Varvasovszky, Z.; Brugha, R. A stakeholder analysis. Health Policy Plan. 2000, 15, 338–345. [Google Scholar] [CrossRef]
- Nutbeam, D. Evaluation in a Nutshell: A Practical Guide to the Evaluation of Health Promotion Programs/Don Nutbeam, Adrian Bauman; McGraw-Hill: North Ryde, NSW, Australia, 2006. [Google Scholar]
- Neergaard, M.A.; Olesen, F.; Andersen, R.S.; Sondergaard, J. Qualitative description—The poor cousin of health research? BMC Med. Res. Methodol. 2009, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Bailie, C.M.J.; Crowe, A.; Flanaghan, C.; McKee, S.; McKnight, A.J. Patient and Family Reflections on Their Experience of Participating in the 100,000 Genomes Project within the NI Genomic Medicine Centre. [CrossRef]
- McKnight, A. Department of Health, Social Services and Public Safety: Providing High Quality Care for People Affected by Rare Diseases—Recommendation for a Collaborative Centre of Expertise for Rare Diseases in Northern Ireland (CERDNI). Available online: https://pure.qub.ac.uk/en/publications/recommendation-for-a-collaborative-centre-of-expertise-for-rare-d (accessed on 10 May 2022).
- Northern Ireland Rare Diseases—Action Plan 2022/23. Available online: https://www.health-ni.gov.uk/publications/northern-ireland-rare-diseases-action-plan-202223 (accessed on 12 May 2022).
- Kingsmore, S.F.; Cakici, J.A.; Clark, M.M.; Gaughran, M.; Feddock, M.; Batalov, S.; Bainbridge, M.N.; Carroll, J.; Caylor, S.A.; Clarke, C.; et al. A Randomized, Controlled Trial of the Analytic and Diagnostic Performance of Singleton and Trio, Rapid Genome and Exome Sequencing in Ill Infants. Am. J. Hum. Genet. 2019, 105, 719–733. [Google Scholar] [CrossRef]
- Hill, M.; Hammond, J.; Lewis, C.; Mellis, R.; Clement, E.; Chitty, L.S. Delivering genome sequencing for rapid genetic diagnosis in critically ill children: Parent and professional views, experiences and challenges. Eur. J. Hum. Genet. 2020, 28, 1529–1540. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.R.; Biesecker, B.B.; Blout, C.L.; Christensen, K.D.; Amendola, L.M.; Bergstrom, K.L.; Biswas, S.; Bowling, K.M.; Brothers, K.B.; Conlin, L.K.; et al. Secondary findings from clinical genomic sequencing: Prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet. Med. 2019, 21, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.S.; Robinson, J.O.; Diamond, P.M.; Bharadwaj, A.; Christensen, K.D.; Lee, K.B.; Green, R.C.; McGuire, A.L.; MedSeq Project Team. Patient understanding of, satisfaction with, and perceived utility of whole-genome sequencing: Findings from the MedSeq Project. Genet. Med. 2018, 20, 1069–1076. [Google Scholar] [CrossRef] [Green Version]
- Insurance. Genomics England. Available online: https://www.genomicsengland.co.uk/patients-participants/taking-part/resources/insurance (accessed on 14 April 2022).
- Powell, S.N.; Byfield, G.; Bennetone, A.; Frantz, A.M.; Harrison, L.K.; James-Crook, E.R.; Osborne, H.; Owens, T.H.; Shaw, J.L.; O’Daniel, J.; et al. Parental Guidance Suggested: Engaging Parents as Partners in Research Studies of Genomic Screening for a Pediatric Population. Front. Genet. 2022, 13, 867030. [Google Scholar] [CrossRef] [PubMed]
- Terry, A.B.; Wylie, A.; Raspa, M.; Vogel, B.; Sanghavi, K.; Djurdjinovic, L.; Caggana, M.; Bodurtha, J. Clinical models of telehealth in genetics: A regional telegenetics landscape. J. Genet. Couns. 2019, 28, 673–691. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Thygesen, J.H.; Shi, T.; Gkoutos, G.V.; Hemingway, H.; Guthrie, B.; Wu, H.; Genomics England Research Consortium. Increased COVID-19 mortality rate in rare disease patients: A retrospective cohort study in participants of the Genomics England 100,000 Genomes project. Orphanet J. Rare Dis. 2022, 17, 166. [Google Scholar] [CrossRef]
- Yu, J.-H.; Appelbaum, P.S.; Brothers, K.B.; Joffe, S.; Kauffman, T.L.; Koenig, B.A.; Prince, A.E.; Scollon, S.; Wolf, S.M.; Bernhardt, B.A.; et al. Consent for clinical genome sequencing: Considerations from the Clinical Sequencing Exploratory Research Consortium. PerMed 2019, 16, 325–333. [Google Scholar] [CrossRef]
- Cuddy, D.G.; Quinn, D.M. Our Digital Future-Encompass: Delivering Care Together. Ulster Med. J. 2019, 88, 4–5. [Google Scholar]
- Schwarze, K.; Buchanan, J.; Fermont, J.M.; Dreau, H.; Tilley, M.W.; Taylor, J.M.; Antoniou, P.; Knight, S.J.L.; Camps, C.; Pentony, M.M.; et al. The complete costs of genome sequencing: A microcosting study in cancer and rare diseases from a single center in the United Kingdom. Genet. Med. 2020, 22, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Son, J.H.; Xie, G.; Yuan, C.; Ena, L.; Li, Z.; Goldstein, A.; Huang, L.; Wang, L.; Shen, F.; Liu, H.; et al. Deep Phenotyping on Electronic Health Records Facilitates Genetic Diagnosis by Clinical Exomes. Am. J. Hum. Genet. 2018, 103, 58–73. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Tang, X.; Yang, K.; Huo, X.; Zhang, H.; Ding, K.; Liao, S. Deep phenotyping and whole-exome sequencing improved the diagnostic yield for nuclear pedigrees with neurodevelopmental disorders. Mol. Genet. Genom. Med. 2022, 10, e1918. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.C.; Yu, H.C.; Martin, R.; Cirulli, E.T.; Schenker-Ahmed, N.M.; Hicks, M.; Cohen, I.V.; Jönsson, T.J.; Heister, R.; Napier, L.; et al. Precision medicine integrating whole-genome sequencing, comprehensive metabolomics, and advanced imaging. Proc. Natl. Acad. Sci. USA 2020, 117, 3053–3062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balachandar, S.; Graves, T.J.; Shimonty, A.; Kerr, K.; Kilner, J.; Xiao, S.; Slade, R.; Sroya, M.; Alikian, M.; Curetean, E.; et al. Identification and validation of a novel pathogenic variant in GDF2 (BMP9) responsible for hereditary hemorrhagic telangiectasia and pulmonary arteriovenous malformations. Am. J. Med. Genet. Part A 2022, 188A, 959–964. [Google Scholar] [CrossRef]
- Brnich, S.E.; Abou Tayoun, A.N.; Couch, F.J.; Cutting, G.R.; Greenblatt, M.S.; Heinen, C.D.; Kanavy, D.M.; Luo, X.; McNulty, S.M.; Starita, L.M.; et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019, 12, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eales, J.M.; Jiang, X.; Xu, X.; Saluja, S.; Akbarov, A.; Cano-Gamez, E.; McNulty, M.T.; Finan, C.; Guo, H.; Wystrychowski, W.; et al. Uncovering genetic mechanisms of hypertension through multi-omic analysis of the kidney. Nat. Genet. 2021, 53, 630–637. [Google Scholar] [CrossRef]
- Ge, S.; Wang, Y.; Song, M.; Li, X.; Yu, X.; Wang, H.; Wang, J.; Zeng, Q.; Wang, W. Type 2 Diabetes Mellitus: Integrative Analysis of Multiomics Data for Biomarker Discovery. Omics 2018, 22, 514–523. [Google Scholar] [CrossRef]
- Marwaha, S.; Knowles, J.W.; Ashley, E.A. A guide for the diagnosis of rare and undiagnosed disease: Beyond the exome. Genome Med. 2022, 14, 23. [Google Scholar] [CrossRef]
- Kerr, K.; McAneney, H.; Smyth, L.J.; Bailie, C.; McKee, S.; McKnight, A.J. A scoping review and proposed workflow for multi-omic rare disease research. Orphanet J. Rare Dis. 2020, 15, 107. [Google Scholar] [CrossRef]
- Ward, R.A.; Aghaeepour, N.; Bhattacharyya, R.P.; Clish, C.B.; Gaudillière, B.; Hacohen, N.; Mansour, M.K.; Mudd, P.A.; Pasupneti, S.; Presti, R.M.; et al. Harnessing the Potential of Multiomics Studies for Precision Medicine in Infectious Disease. Open Forum Infect. Dis. 2021, 8, ofab483. [Google Scholar] [CrossRef]
- Cipriani, V.; Pontikos, N.; Arno, G.; Sergouniotis, P.I.; Lenassi, E.; Thawong, P.; Danis, D.; Michaelides, M.; Webster, A.R.; Moore, A.T.; et al. An Improved Phenotype-Driven Tool for Rare Mendelian Variant Prioritization: Benchmarking Exomiser on Real Patient Whole-Exome Data. Genes 2020, 11, 460. [Google Scholar] [CrossRef]
- De La Vega, F.M.; Chowdhury, S.; Moore, B.; Frise, E.; McCarthy, J.; Hernandez, E.J.; Wong, T.; James, K.; Guidugli, L.; Agrawal, P.B.; et al. Artificial intelligence enables comprehensive genome interpretation and nomination of candidate diagnoses for rare genetic diseases. Genome Med. 2021, 13, 153. [Google Scholar] [CrossRef]
- Schaefer, J.; Lehne, M.; Schepers, J.; Prasser, F.; Thun, S. The use of machine learning in rare diseases: A scoping review. Orphanet J. Rare Dis. 2020, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- Whitley, K.V.; Tueller, J.A.; Weber, K.S. Genomics Education in the Era of Personal Genomics: Academic, Professional, and Public Considerations. Int. J. Mol. Sci. 2020, 21, 768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, A.; Fonseca, M.J.; Lemos, M.; Lencastre, L.; Tavares, F. Bioinformatics-Based Activities in High School: Fostering Students’ Literacy, Interest, and Attitudes on Gene Regulation, Genomics, and Evolution. Front. Microbiol. 2020, 11, 578099. [Google Scholar] [CrossRef] [PubMed]
- Cornel, M.C. Evidence-Based Genetic Education of Non-Genetic-Expert Physicians: Experiences over Three Decades in Amsterdam. Front. Genet. 2019, 10, 712. [Google Scholar] [CrossRef]
- Grove, M.E.; White, S.; Fisk, D.G.; Rego, S.; Dagan-Rosenfeld, O.; Kohler, J.N.; Reuter, C.M.; Bonner, D.; Wheeler, M.T.; Bernstein, J.A.; et al. Developing a genomics rotation: Practical training around variant interpretation for genetic counseling students. J. Genet. Couns. 2019, 28, 466–476. [Google Scholar] [CrossRef]
- Guilabert, M.; Martínez-García, A.; Sala-González, M.; Solas, O.; Mira, J.J. Results of a Patient Reported Experience Measure (PREM) to measure the rare disease patients and caregivers experience: A Spanish cross-sectional study. Orphanet J. Rare Dis. 2021, 16, 67. [Google Scholar] [CrossRef]
- Whittal, A.; Meregaglia, M.; Nicod, E. The Use of Patient-Reported Outcome Measures in Rare Diseases and Implications for Health Technology Assessment. Patient 2021, 14, 485–503. [Google Scholar] [CrossRef]
- Bootpetch, T.C.; Hafrén, L.; Elling, C.L.; Baschal, E.E.; Manichaikul, A.W.; Pine, H.S.; Szeremeta, W.; Scholes, M.A.; Cass, S.P.; Larson, E.D.; et al. Multi-omic studies on missense PLG variants in families with otitis media. Sci. Rep. 2020, 10, 15035. [Google Scholar] [CrossRef]
- Straub, I.R.; Weraarpachai, W.; Shoubridge, E.A. Multi-OMICS study of a CHCHD10 variant causing ALS demonstrates metabolic rewiring and activation of endoplasmic reticulum and mitochondrial unfolded protein responses. Hum. Mol. Genet. 2021, 30, 687–705. [Google Scholar] [CrossRef]
- Labory, J.; Fierville, M.; Ait-El-Mkadem, S.; Bannwarth, S.; Paquis-Flucklinger, V.; Bottini, S. Multi-Omics Approaches to Improve Mitochondrial Disease Diagnosis: Challenges, Advances, and Perspectives. Front. Mol. Biosci. 2020, 7, 590842. [Google Scholar] [CrossRef] [PubMed]
- Decherchi, S.; Pedrini, E.; Mordenti, M.; Cavalli, A.; Sangiorgi, L. Opportunities and Challenges for Machine Learning in Rare Diseases. Front. Med. 2021, 8, 747612. [Google Scholar] [CrossRef] [PubMed]
| Participant Characteristic | Count (Total n = 442) | Percentage |
|---|---|---|
| 245/197 | 55%/45% |
Age at recruitment (2018)
| 21.4 years (0.5–91) | Not applicable |
Recruiting clinic
| 296 41 23 71 15 | 67.0% 9.3% 5.2% 15.2% 3.4% |
Recruitment type
| 120 51 250 21 | 27.2% 11.5% 56.6% 4.8% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerr, K.; McKenna, C.; Heggarty, S.; Bailie, C.; McMullan, J.; Crowe, A.; Kilner, J.; Donnelly, M.; Boyle, S.; Rea, G.; et al. A Formative Study of the Implementation of Whole Genome Sequencing in Northern Ireland. Genes 2022, 13, 1104. https://doi.org/10.3390/genes13071104
Kerr K, McKenna C, Heggarty S, Bailie C, McMullan J, Crowe A, Kilner J, Donnelly M, Boyle S, Rea G, et al. A Formative Study of the Implementation of Whole Genome Sequencing in Northern Ireland. Genes. 2022; 13(7):1104. https://doi.org/10.3390/genes13071104
Chicago/Turabian StyleKerr, Katie, Caoimhe McKenna, Shirley Heggarty, Caitlin Bailie, Julie McMullan, Ashleen Crowe, Jill Kilner, Michael Donnelly, Saralynne Boyle, Gillian Rea, and et al. 2022. "A Formative Study of the Implementation of Whole Genome Sequencing in Northern Ireland" Genes 13, no. 7: 1104. https://doi.org/10.3390/genes13071104
APA StyleKerr, K., McKenna, C., Heggarty, S., Bailie, C., McMullan, J., Crowe, A., Kilner, J., Donnelly, M., Boyle, S., Rea, G., Flanagan, C., McKee, S., & McKnight, A. J. (2022). A Formative Study of the Implementation of Whole Genome Sequencing in Northern Ireland. Genes, 13(7), 1104. https://doi.org/10.3390/genes13071104







