Comprehensive Genome-Wide Analysis of Histone Acetylation Genes in Roses and Expression Analyses in Response to Heat Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Heat Stress and Plant Materials Collection
2.2. Identification of the HDAC Genes and HAT Genes in Rose
2.3. Conserved Sequence and Structure Model Analysis
2.4. Physicochemical Properties of HDACs and HATs
2.5. Phylogenetic Analysis of HDACs and HATs
2.6. Chromosomal Location, Homologous Genes, and Synteny
2.7. Transcriptome Analysis of HDACs and HATs
2.8. RNA Preparation and qRT-PCR Analysis
3. Results
3.1. The Identification of HDACs and HATs
3.2. Gene Structure and Conserved Domains
3.3. Physicochemical Properties of HDACs and HATs
3.4. Phylogenetic Relationships of HDACs and HATs
3.4.1. RPD3/HDA1 Subfamily
3.4.2. HD2 Subfamily
3.4.3. SIR2 Subfamily
3.4.4. HAT Family
3.5. Chromosomal Location, Homologous Genes, and Synteny
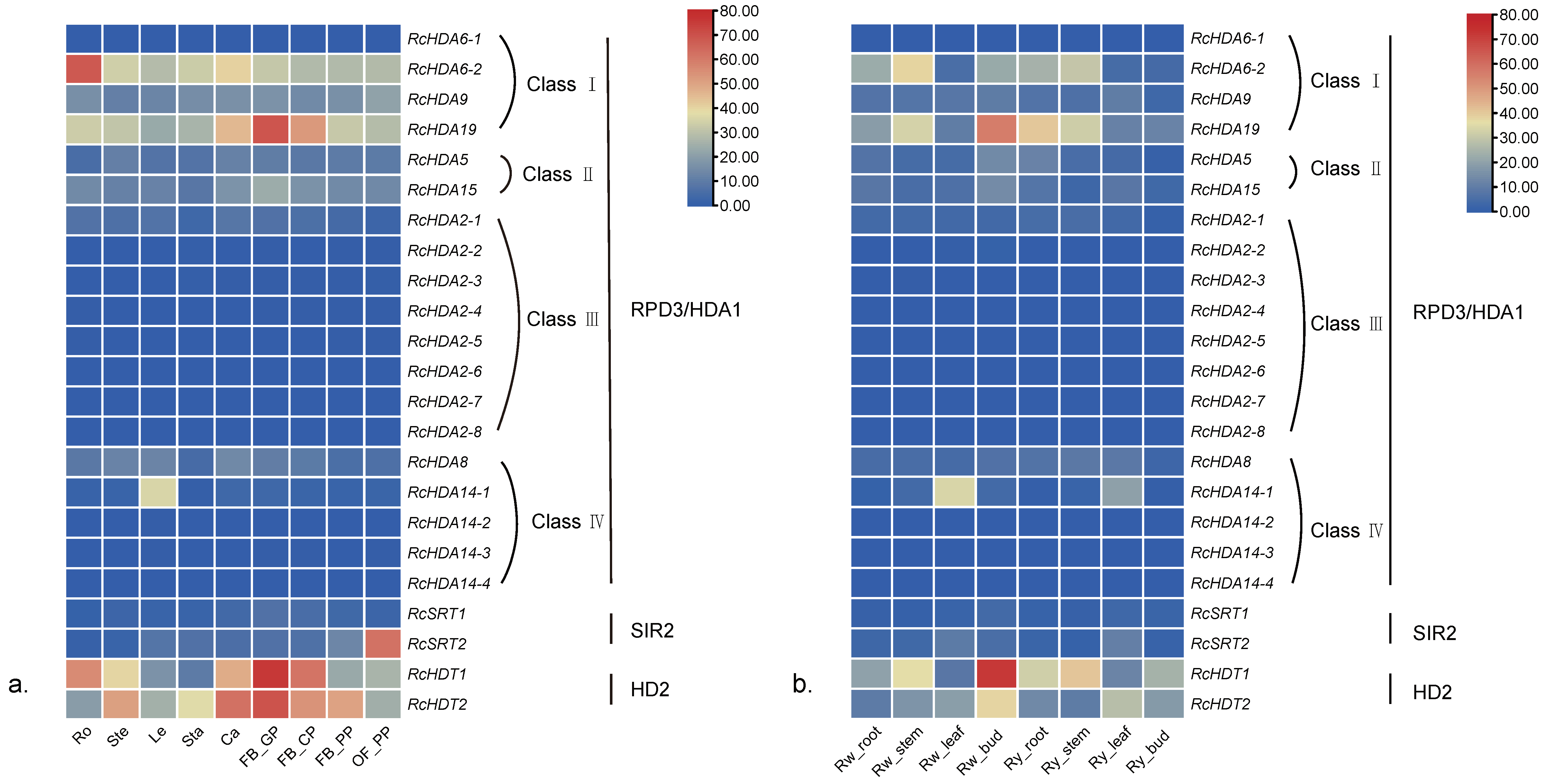
3.6. Expression of HDACs and HATs in Different Organs of Three Roses
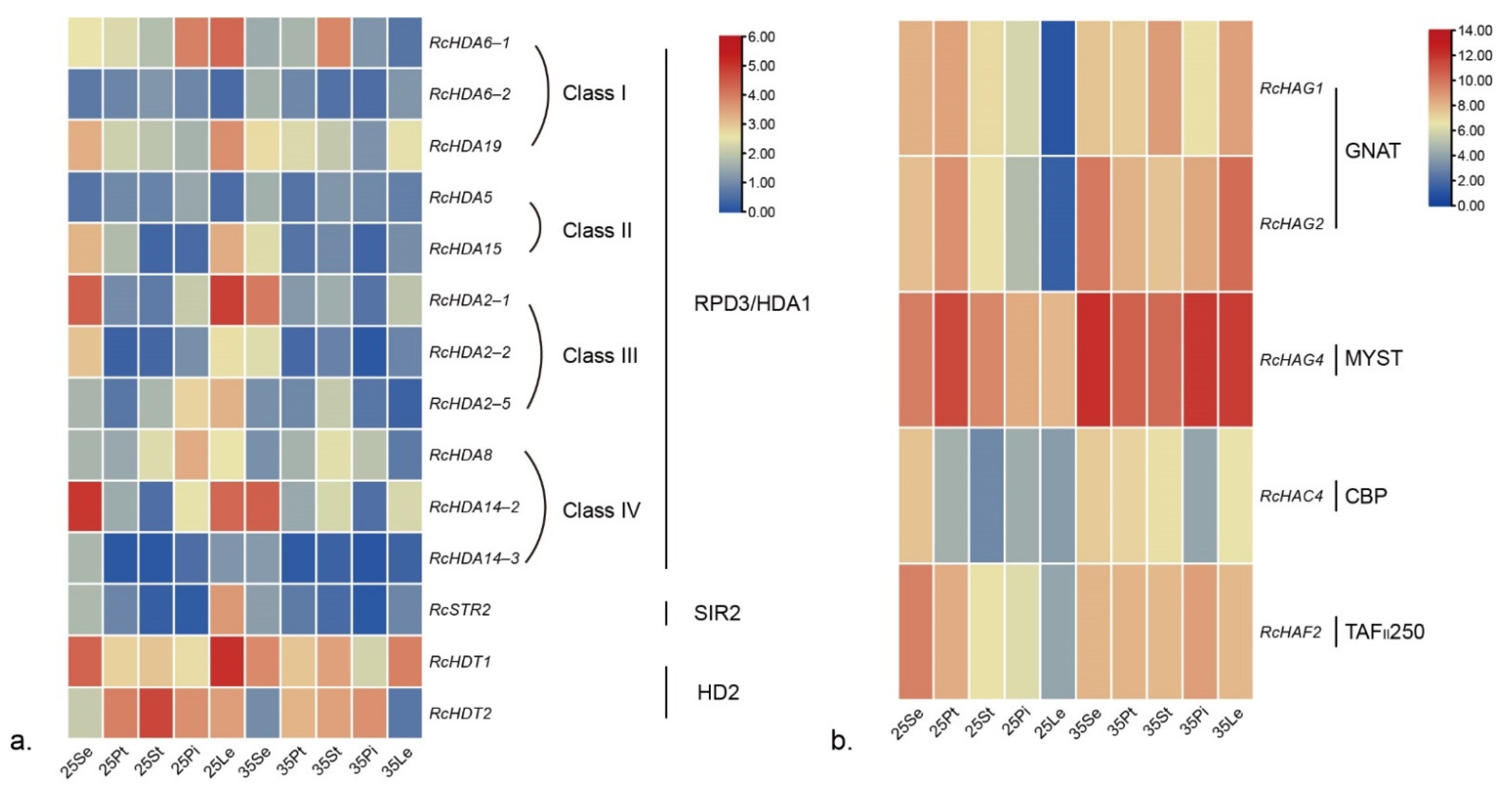
3.7. Responses of RcHDACs and RcHATs to High Temperature
4. Discussion
4.1. The Expression Specificity of HDACs and HATs
4.2. The Antagonistic Expression Patterns of HDACs and HATs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kornet, N.; Scheres, B. Members of the GCN5 Histone Acetyltransferase Complex Regulate PLETHORA-Mediated Root Stem Cell Niche Maintenance and Transit Amplifying Cell Proliferation in Arabidopsis. Plant Cell 2009, 21, 1070–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.K.; Song, J.D.; Noh, Y.S.; Noh, B. Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J. 2007, 49, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Latrasse, D.; Benhamed, M.; Henry, Y.; Domenichini, S.; Kim, W.; Zhou, D.-X.; Delarue, M. The MYST histone acetyltransferases are essential for gametophyte development in Arabidopsis. BMC Plant Biol. 2008, 8, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.; Liu, X.; Luo, M.; Yang, S.; Wu, K. Involvement of histone modifications in plant abiotic stress responses. J. Integr. Plant Biol. 2013, 55, 892–901. [Google Scholar] [CrossRef]
- Bharti, K.; von Koskull-Döring, P.; Bharti, S.; Kumar, P.; Tintschl-Körbitzer, A.; Treuter, E.; Nover, L. Tomato Heat Stress Transcription Factor HsfB1 Represents a Novel Type of General Transcription Coactivator with a Histone-Like Motif Interacting with the Plant CREB Binding Protein Ortholog HAC1. Plant Cell 2004, 16, 1521–1535. [Google Scholar] [CrossRef] [Green Version]
- Berger, S.L. The complex language of chromatin regulation during transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef]
- Shen, Y.; Wei, W.; Zhou, D.X. Histone Acetylation Enzymes Coordinate Metabolism and Gene Expression. Trends Plant Sci. 2015, 20, 614–621. [Google Scholar] [CrossRef]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [Green Version]
- Lusser, A.; Brosch, G.; Loidl, A.; Haas, H.; Loidl, P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 1997, 277, 88–91. [Google Scholar] [CrossRef]
- Sterner, D.E.; Berger, S.L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef] [Green Version]
- Kuo, M.H.; Allis, C.D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 1998, 20, 615–626. [Google Scholar] [CrossRef]
- Tian, L.; Chen, Z.J. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 2001, 98, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Wang, J.; Fong, M.P.; Chen, M.; Cao, H.; Gelvin, S.B.; Chen, Z.J. Genetic Control of Developmental Changes Induced by Disruption of Arabidopsis Histone Deacetylase 1 (AtHD1) Expression. Genetics 2003, 165, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tan, B.; Luo, M.; Li, Y.; Liu, C.; Chen, C.; Yu, C.-W.; Yang, S.; Dong, S.; Ruan, J.; et al. HISTONE DEACETYLASE19 Interacts with HSL1 and Participates in the Repression of Seed Maturation Genes in Arabidopsis Seedlings. Plant Cell 2013, 25, 134–148. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.; Latrasse, D.; Servet, C.; Zhou, D.-X. Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 2013, 432, 394–398. [Google Scholar] [CrossRef]
- Krogan, N.T.; Hogan, K.; Long, J.A. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 2012, 139, 4180–4190. [Google Scholar] [CrossRef] [Green Version]
- Rossi, V.; Locatelli, S.; Varotto, S.; Donn, G.; Pirona, R.; Henderson, D.A.; Hartings, H.; Motto, M. Maize Histone Deacetylase hda101 Is Involved in Plant Development, Gene Transcription, and Sequence-Specific Modulation of Histone Modification of Genes and Repeats. Plant Cell 2007, 19, 1145–1162. [Google Scholar] [CrossRef] [Green Version]
- Servet, C.; Conde e Silva, N.; Zhou, D.X. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 2010, 3, 670–677. [Google Scholar] [CrossRef]
- Zhou, C.; Labbe, H.; Sridha, S.; Wang, L.; Tian, L.; Latoszek-Green, M.; Yang, Z.; Brown, D.; Miki, B.; Wu, K. Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 2004, 38, 715–724. [Google Scholar] [CrossRef]
- Buszewicz, D.; Archacki, R.; Palusiński, A.; Kotliński, M.; Fogtman, A.; Iwanicka-Nowicka, R.; Sosnowska, K.; Kuciński, J.; Pupel, P.; Olędzki, J.; et al. HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 2016, 39, 2108–2122. [Google Scholar] [CrossRef]
- Hou, J.; Ren, R.; Xiao, H.; Chen, Z.; Yu, J.; Zhang, H.; Shi, Q.; Hou, H.; He, S.; Li, L. Characteristic and evolution of HAT and HDAC genes in Gramineae genomes and their expression analysis under diverse stress in Oryza sativa. Planta 2021, 253, 72. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yan, S.; Zhao, L.; Tan, J.; Zhang, Q.; Gao, F.; Wang, P.; Hou, H.; Li, L. Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 2014, 14, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, H.; Liu, X.; Thorn, G.; Duan, J.; Tian, L. Expression analysis of histone acetyltransferases in rice under drought stress. Biochem. Biophys. Res. Commun. 2014, 443, 400–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, F.; Su, L.; Hu, S.; Xue, J.Y.; Liu, H.; Liu, G.; Jiang, Y.; Du, J.; Qiao, Y.; Fan, Y.; et al. A chromosome-level genome assembly of rugged rose (Rosa rugosa) provides insights into its evolution, ecology, and floral characteristics. Hortic Res. 2021, 8, 141. [Google Scholar] [CrossRef]
- Nakamura, N.; Hirakawa, H.; Sato, S.; Otagaki, S.; Matsumoto, S.; Tabata, S.; Tanaka, Y. Genome structure of Rosa multiflora, a wild ancestor of cultivated roses. DNA Res. 2018, 25, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Pi, M.; Gao, Q.; Liu, Z.; Kang, C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic. Res. 2019, 6, 61. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Wan, H.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis. Sci. Rep. 2017, 7, 43382. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Yu, J.; Zhao, T.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. Dissecting the Genome-Wide Evolution and Function of R2R3-MYB Transcription Factor Family in Rosa chinensis. Genes 2019, 10, 823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, A.; Remay, A.; Raymond, O.; Balzergue, S.; Chauvet, A.; Maene, M.; Pécrix, Y.; Yang, S.H.; Jeauffre, J.; Thouroude, T.; et al. Genomic approach to study floral development genes in Rosa sp. PLoS ONE 2011, 6, e28455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, A.; Raymond, O.; Maene, M.; Baudino, S.; Langlade, N.B.; Boltz, V.; Vergne, P.; Bendahmane, M. Tinkering with the C-function: A molecular frame for the selection of double flowers in cultivated roses. PLoS ONE 2010, 5, e9288. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zang, F.; Ma, Y.; Tu, X.; Huang, P.; Wu, Q.; Li, Z.; Liu, T.; Lin, F.; Pei, S.; Zang, D.; et al. A high-quality chromosome-level genome of wild Rosa rugosa. DNA Res. 2021, 28, dsab017. [Google Scholar] [CrossRef]
- Fu, W.; Wu, K.; Duan, J. Sequence and expression analysis of histone deacetylases in rice. Biochem. Biophys. Res. Commun. 2007, 356, 843–850. [Google Scholar] [CrossRef]
- Liu, X.; Luo, M.; Zhang, W.; Zhao, J.; Zhang, J.; Wu, K.; Tian, L.; Duan, J. Histone acetyltransferases in rice (Oryza sativa L.): Phylogenetic analysis, subcellular localization and expression. BMC Plant Biol. 2012, 12, 145. [Google Scholar] [CrossRef] [Green Version]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Aiese Cigliano, R.; Sanseverino, W.; Cremona, G.; Ercolano, M.R.; Conicella, C.; Consiglio, F.M. Genome-wide analysis of histone modifiers in tomato: Gaining an insight into their developmental roles. BMC Genom. 2013, 14, 57. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Zhang, L.; Zhou, C.; Yu, C.-W.; Chaikam, V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 2008, 59, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, L.; Ko, E.E.; Shao, K.; Qiao, H. Histone Deacetylases SRT1 and SRT2 Interact with ENAP1 to Mediate Ethylene-Induced Transcriptional Repression. Plant Cell 2018, 30, 153–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, H.; Liu, X.; Ma, N.; Xue, J.; Lu, W.; Bai, J.; Gao, J. Ethylene-influenced flower opening and expression of genes encoding Etrs, Ctrs, and Ein3s in two cut rose cultivars. Postharvest Biol. Technol. 2006, 40, 97–105. [Google Scholar] [CrossRef]
- Wu, K.; Tian, L.; Zhou, C.; Brown, D.; Miki, B. Repression of gene expression by Arabidopsis HD2 histone deacetylases. Plant J. 2003, 34, 241–247. [Google Scholar] [CrossRef]
- Kuang, J.F.; Chen, J.Y.; Luo, M.; Wu, K.Q.; Sun, W.; Jiang, Y.M.; Lu, W.J. Histone deacetylase HD2 interacts with ERF1 and is involved in longan fruit senescence. J. Exp. Bot. 2012, 63, 441–454. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Huang, L.; Xu, C.; Zhao, Y.; Zhou, D.X. Altered levels of histone deacetylase OsHDT1 affect differential gene expression patterns in hybrid rice. PLoS ONE 2011, 6, e21789. [Google Scholar] [CrossRef]
- Cohen, R.; Schocken, J.; Kaldis, A.; Vlachonasios, K.E.; Hark, A.T.; McCain, E.R. The histone acetyltransferase GCN5 affects the inflorescence meristem and stamen development in Arabidopsis. Planta 2009, 230, 1207–1221. [Google Scholar] [CrossRef]
- Bertrand, C.; Bergounioux, C.; Domenichini, S.; Delarue, M.; Zhou, D.X. Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 2003, 278, 28246–28251. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Liu, C.; Pei, Y.; Deng, X.; Niu, L.; Cao, X. Involvement of the Histone Acetyltransferase AtHAC1 in the Regulation of Flowering Time via Repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiol. 2007, 143, 1660–1668. [Google Scholar] [CrossRef]
- Li, C.; Xu, J.; Li, J.; Li, Q.; Yang, H. Involvement of Arabidopsis HAC family genes in pleiotropic developmental processes. Plant Signal. Behav. 2014, 9, e28173. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-T.; Luo, M.; Wang, Y.-Y.; Wu, K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010, 61, 3345–3353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sridha, S.; Wu, K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006, 46, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Tian, L.; Malik, K.; Brown, D.; Miki, B. Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 2000, 22, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Pécrix, Y.; Rallo, G.; Folzer, H.; Cigna, M.; Gudin, S.; Le Bris, M. Polyploidization mechanisms: Temperature environment can induce diploid gamete formation in Rosa sp. J. Exp. Bot. 2011, 62, 3587–3597. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [Green Version]






| Subfamily | Name | Size(aa) 1 | PI | MW | Ii | Classifies | GRAVY |
|---|---|---|---|---|---|---|---|
| RPD3/HDA1 | RcHDA6-1 | 297 | 6.05 | 33,956.99 | 44.54 | unstable | −0.191 |
| RcHDA6-2 | 466 | 5.33 | 52,454.87 | 45.42 | unstable | −0.514 | |
| RcHDA8 | 377 | 5.63 | 41,217.82 | 29.37 | stable | −0.166 | |
| RcHDA15 | 575 | 5.97 | 62,865.81 | 37.96 | stable | −0.328 | |
| RcHDA14-1 | 445 | 5.96 | 48,359.75 | 37.04 | stable | −0.137 | |
| RcHDA2-1 | 397 | 9.04 | 44,059.8 | 43.81 | unstable | −0.038 | |
| RcHDA19 | 492 | 5.14 | 55,455.22 | 41.09 | unstable | −0.531 | |
| RcHDA9 | 432 | 5.38 | 49,400.64 | 37.07 | stable | −0.404 | |
| RcHDA5 | 661 | 5.29 | 73,513.29 | 40.76 | unstable | −0.291 | |
| RcHDA2-2 | 274 | 8.93 | 31,459.6 | 44.02 | unstable | −0.131 | |
| RcHDA14-2 | 83 | 5.83 | 9359.63 | 16.39 | stable | 0.294 | |
| RcHDA2-3 | 198 | 7.61 | 22,437.29 | 38.39 | stable | 0.432 | |
| RcHDA14-3 | 75 | 5.12 | 8463.73 | 14.47 | stable | 0.611 | |
| RcHDA2-4 | 84 | 5.40 | 9646.11 | 27.87 | stable | −0.029 | |
| RcHDA2-5 | 46 | 7.66 | 5062.87 | 34.16 | stable | 0.004 | |
| RcHDA14-4 | 162 | 9.11 | 18,202.9 | 37.42 | stable | −0.154 | |
| RcHDA2-6 | 127 | 9.39 | 14,334.89 | 51.03 | unstable | 0.130 | |
| RcHDA2-7 | 61 | 5.81 | 7012.15 | 24.68 | stable | −0.051 | |
| RcHDA2-8 | 261 | 6.54 | 28,953.16 | 40.33 | unstable | −0.182 | |
| SIR2 | RcSRT1 | 469 | 9.10 | 52,092.43 | 46.23 | unstable | −0.191 |
| RcSRT2 | 387 | 9.54 | 42,858.86 | 44.43 | unstable | −0.290 | |
| HD2 | RcHDT1 | 115 | 4.70 | 12,675.34 | 29.21 | stable | 0.050 |
| RcHDT2 | 105 | 9.12 | 11,575.43 | 29.42 | stable | 0.000 | |
| GNAT | RcHAG1 | 547 | 6.27 | 61,085.47 | 46.09 | unstable | −0.597 |
| RcHAG2 | 460 | 5.12 | 51,748.95 | 41.74 | unstable | −0.224 | |
| RcHAG3-1 | 567 | 8.74 | 63,437.87 | 30.76 | stable | −0.333 | |
| RcHAG3-2 | 95 | 10.12 | 10,489.60 | 30.19 | stable | −0.071 | |
| MYST | RcHAG4 | 470 | 6.66 | 54,232.93 | 41.66 | unstable | −0.583 |
| CBP | RcHAC1 | 1377 | 6.48 | 154,650.31 | 57.02 | unstable | −0.477 |
| RcHAC2 | 1717 | 8.46 | 194,137.68 | 57.04 | unstable | −0.753 | |
| RcHAC4 | 955 | 7.22 | 107,704.14 | 44.62 | unstable | −0.413 | |
| TAFII250 | RcHAF1 | 1900 | 5.74 | 214,349.67 | 51.34 | unstable | −0.822 |
| RcHAF2 | 1692 | 5.54 | 191,691.06 | 52.75 | unstable | −0.724 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Huang, Q.; Guan, H.; Zhang, X.; Bao, M.; Bendahmane, M.; Fu, X. Comprehensive Genome-Wide Analysis of Histone Acetylation Genes in Roses and Expression Analyses in Response to Heat Stress. Genes 2022, 13, 980. https://doi.org/10.3390/genes13060980
Wu Q, Huang Q, Guan H, Zhang X, Bao M, Bendahmane M, Fu X. Comprehensive Genome-Wide Analysis of Histone Acetylation Genes in Roses and Expression Analyses in Response to Heat Stress. Genes. 2022; 13(6):980. https://doi.org/10.3390/genes13060980
Chicago/Turabian StyleWu, Quanshu, Qiuyue Huang, Huilin Guan, Xiaoni Zhang, Manzhu Bao, Mohammed Bendahmane, and Xiaopeng Fu. 2022. "Comprehensive Genome-Wide Analysis of Histone Acetylation Genes in Roses and Expression Analyses in Response to Heat Stress" Genes 13, no. 6: 980. https://doi.org/10.3390/genes13060980
APA StyleWu, Q., Huang, Q., Guan, H., Zhang, X., Bao, M., Bendahmane, M., & Fu, X. (2022). Comprehensive Genome-Wide Analysis of Histone Acetylation Genes in Roses and Expression Analyses in Response to Heat Stress. Genes, 13(6), 980. https://doi.org/10.3390/genes13060980






