Retrogene Duplication and Expression Patterns Shaped by the Evolution of Sex Chromosomes in Malaria Mosquitoes
Highlights
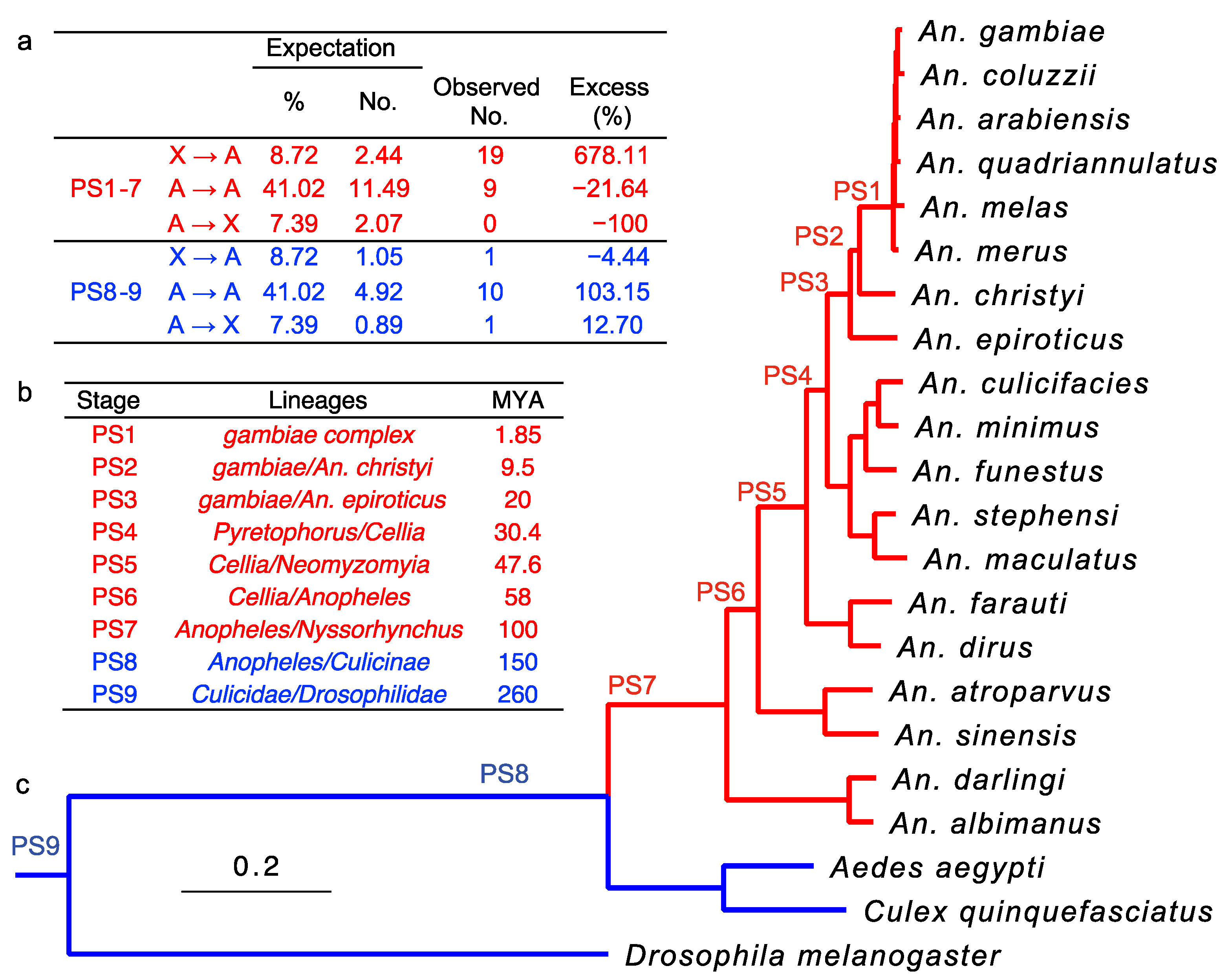
- We discovered a significant excess of young retrogenes duplicated from the X chromosome to autosomes in malaria mosquitoes.
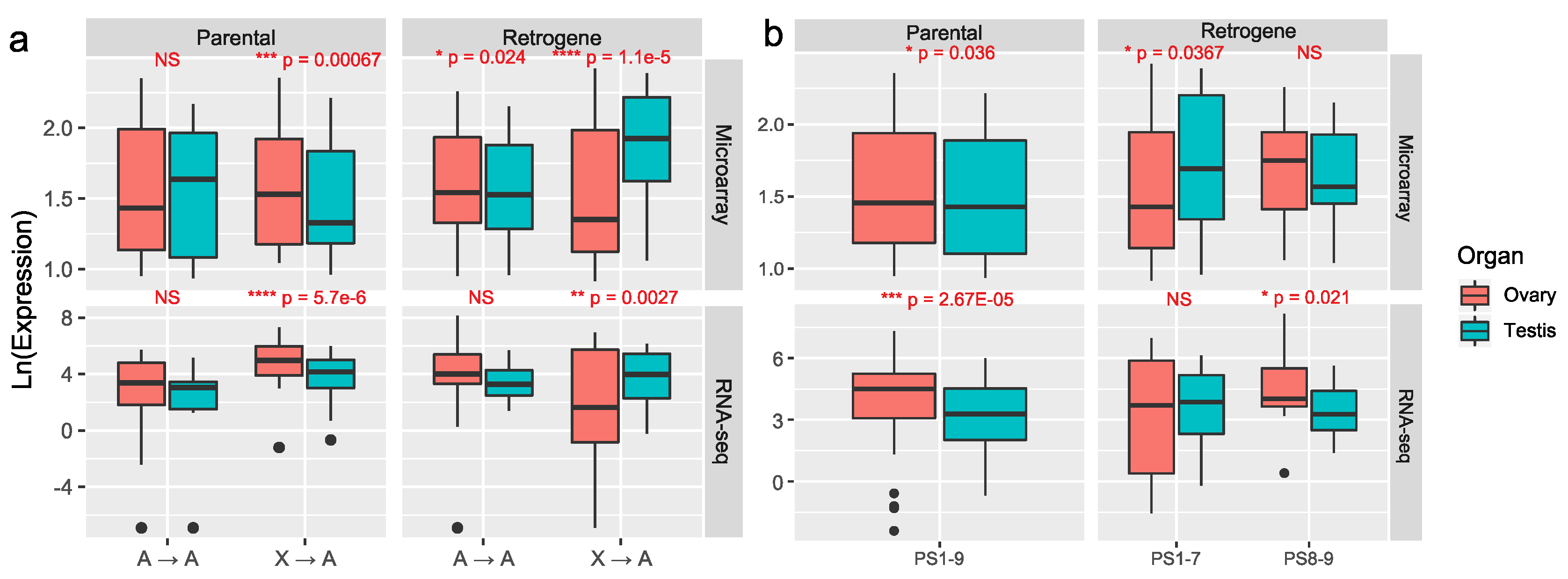
- We demonstrated that these young retrogenes are enriched with testis-biased expression and mitochondrial function.
- The biased retro-duplication of genes from the X chromosome to autosomes indicates that this process is shaped by selection pressure. The testis-biased expression and mitochondrial-related functions of these young retrogenes are suggestive of their potential roles in spermatogenesis.
- Meiotic sex chromosome inactivation (MSCI) in males with heteromorphic sex chromosomes limits the functionality of genes located on the X chromosome during meiosis. As a result, the duplication of such genes to autosomes may be selectively favored, particularly when they are essential for meiosis and spermatogenesis in males.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identifying and Dating Gene Retroposition Events
2.2. Microarray and RNA-Seq Data
2.3. Mosquito Strain and Dissection
2.4. Primer Design, RNA Extraction, and cDNA Synthesis for RT-PCR
2.5. Reverse-Transcription PCR
2.6. Selection Analyses for Molecular Evolution of Parental Genes and Retrogenes
3. Results
3.1. Retroduplications Occurred Predominantly from the X Chromosome to Autosomes after the Evolution of Differentiated Sex Chromosomes
3.2. Most Retrogenes Originated after the Evolution of Sex Chromosomes Evolved Male-Biased Expression in Reproductive Organs whereas Older Retrogenes Acquired Female-Biased Expression
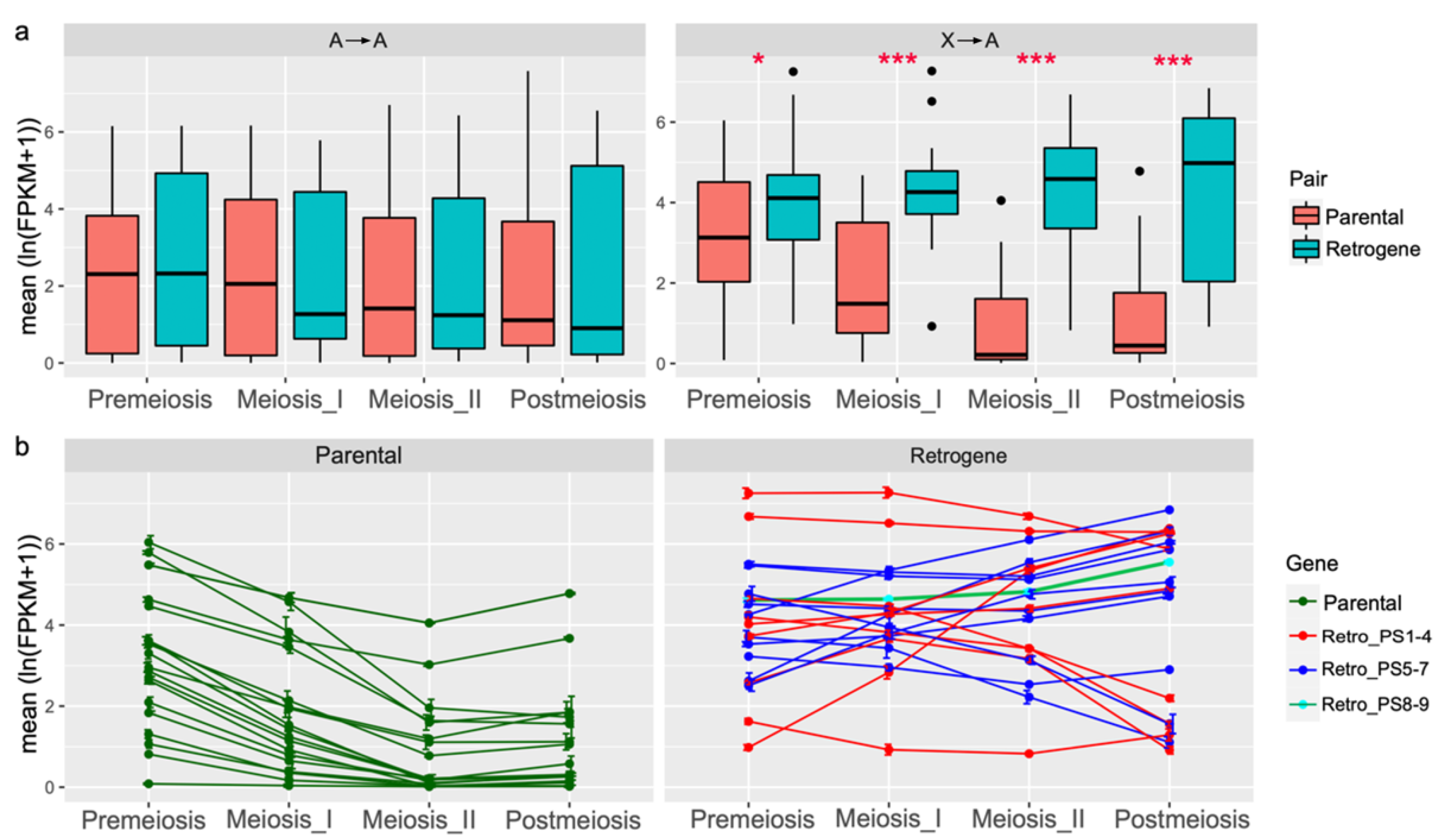
3.3. Autosomal Retrogenes with Increased Meiotic or Postmeiotic Expression Tend to Be Male Biased
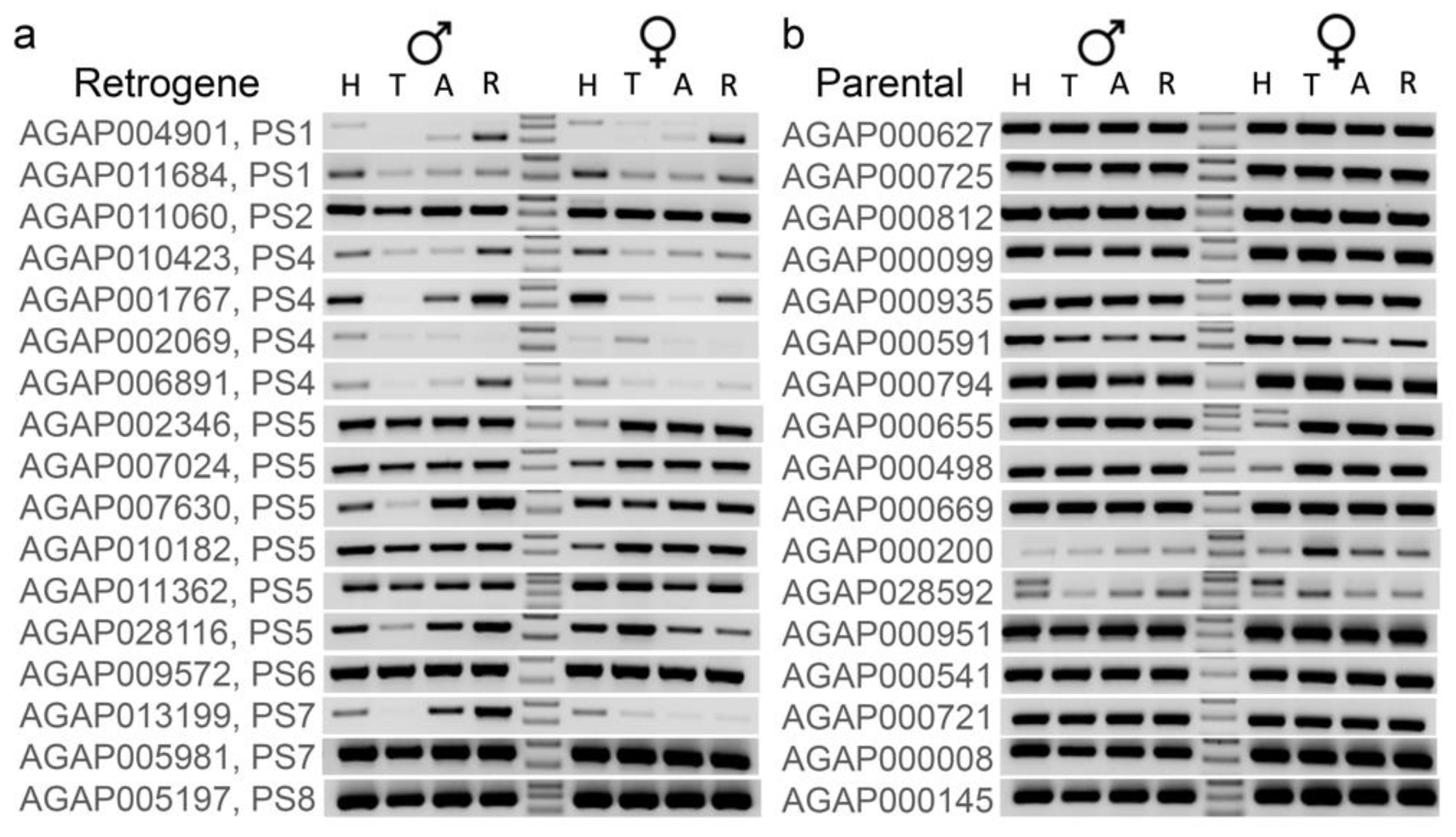
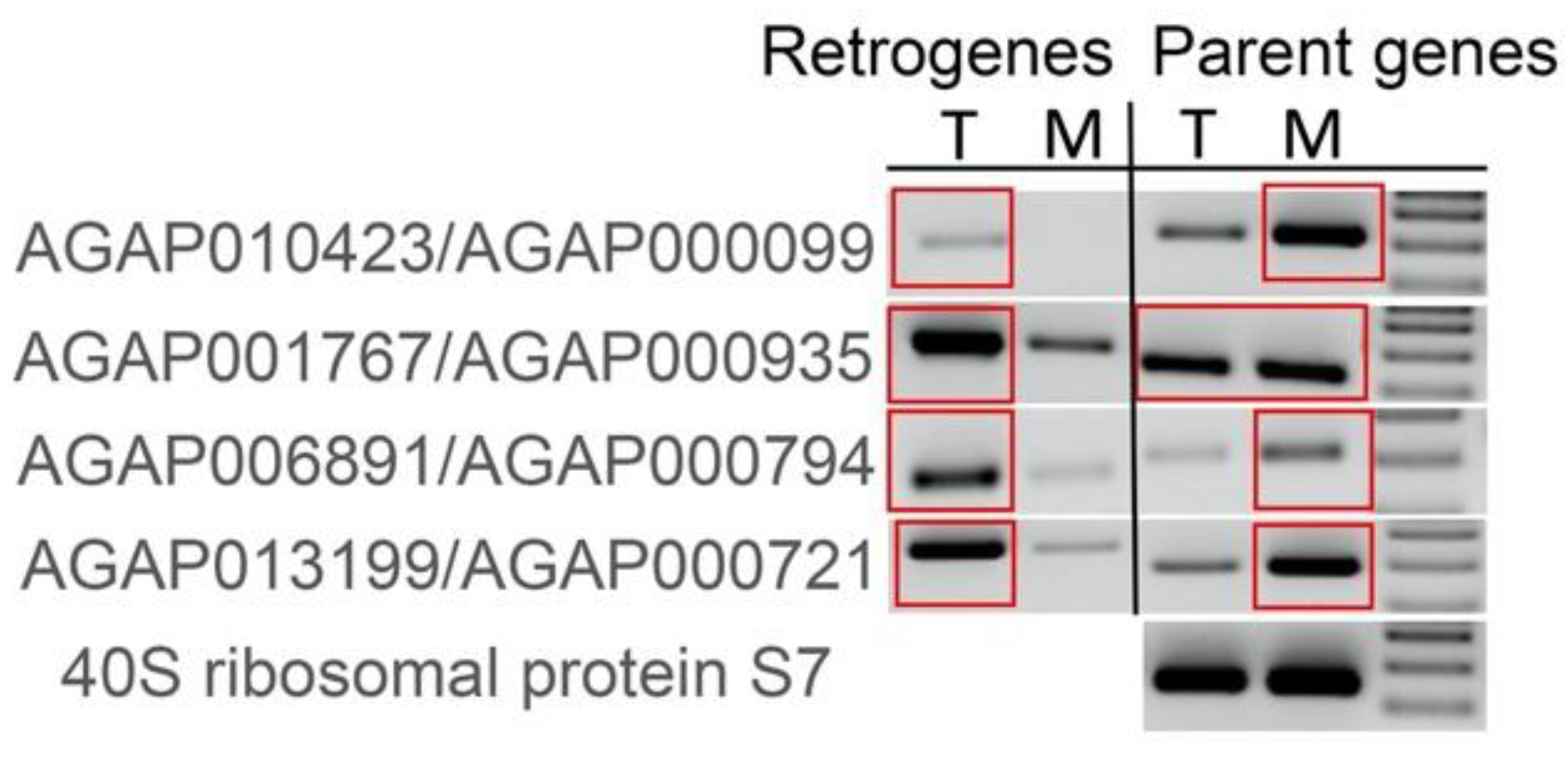
3.4. Younger Retrogenes Have Nonuniform Expression Patterns across Body Parts and Sexes
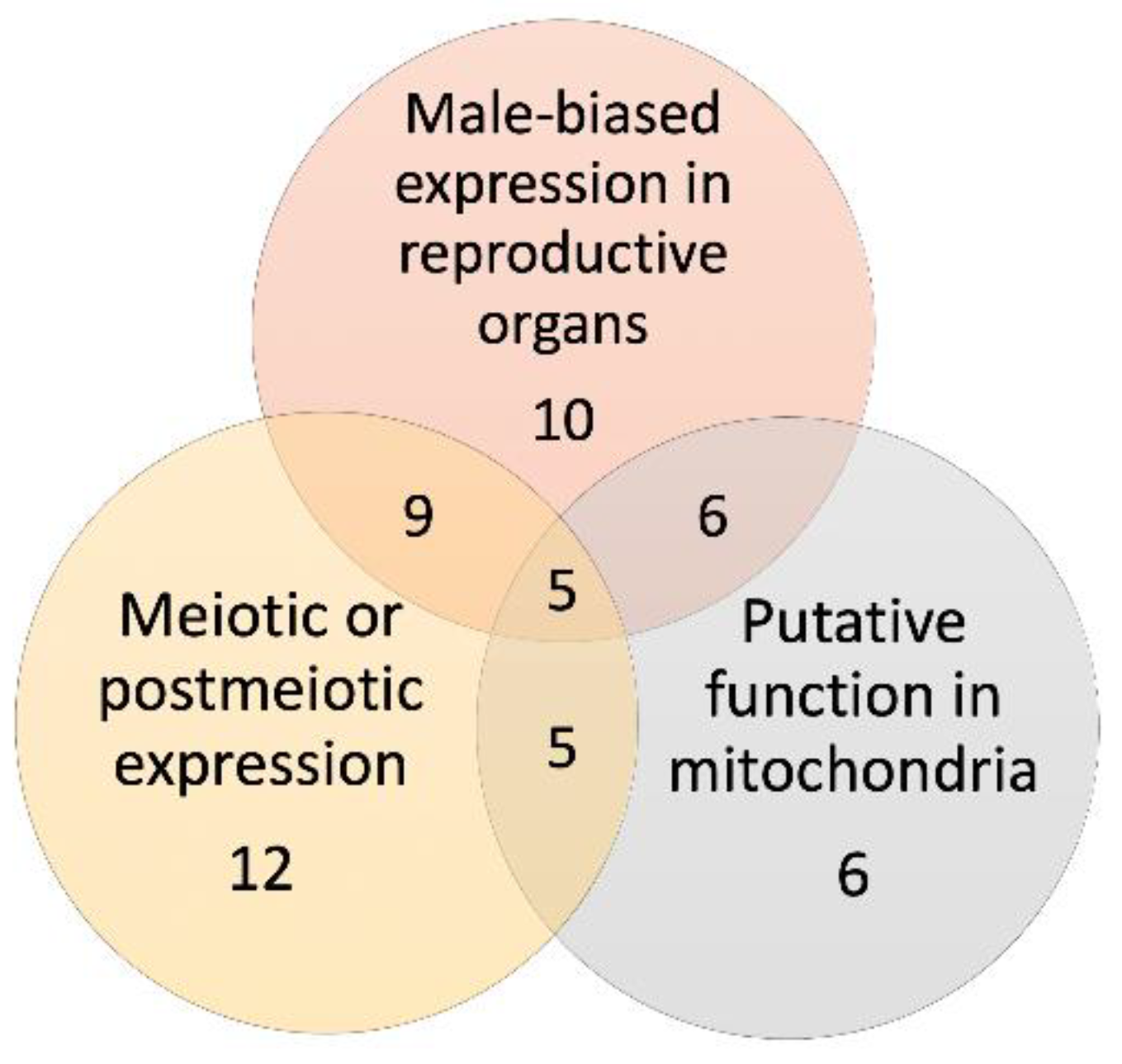
3.5. Mitochondrial Function Is a Distinctive Feature of the Young Retrogenes
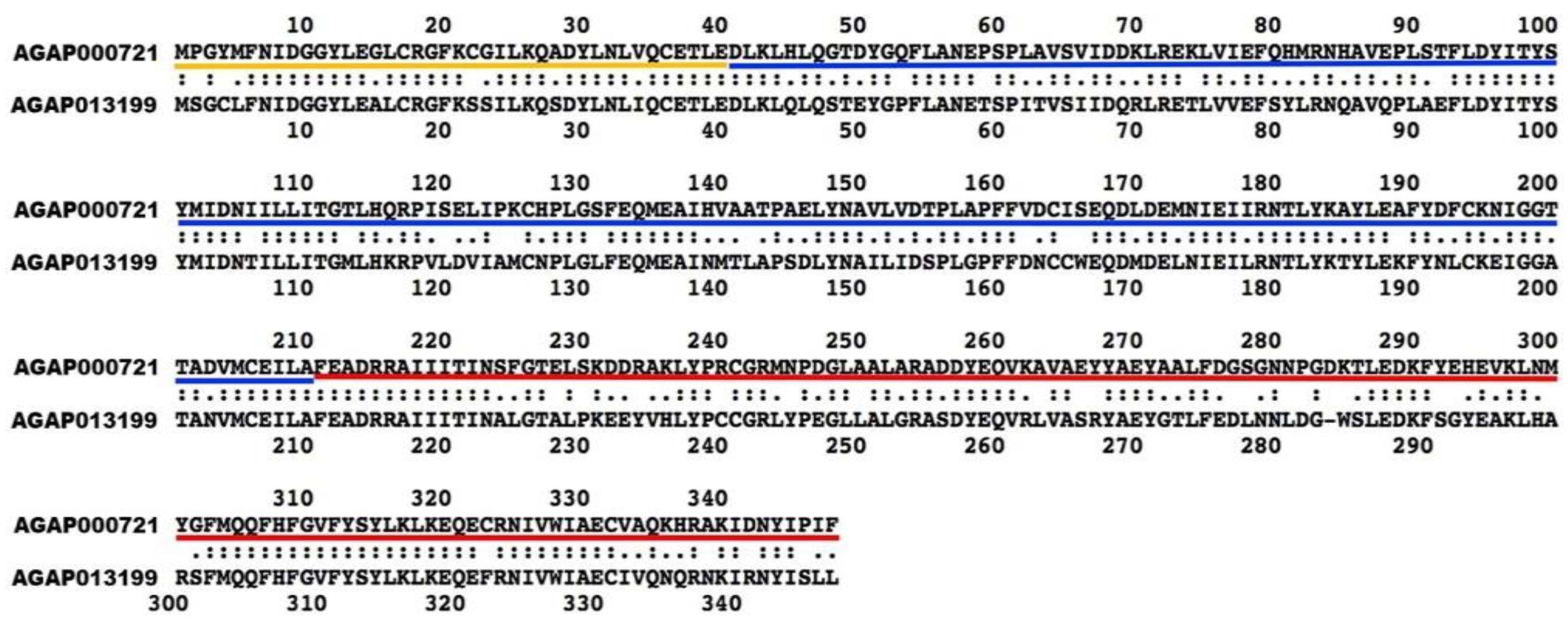
3.6. Most of the An. gambiae Retrogenes Show Signatures of Purifying Selection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casola, C.; Betran, E. The Genomic Impact of Gene Retrocopies: What Have We Learned from Comparative Genomics, Population Genomics, and Transcriptomic Analyses? Genome Biol. Evol. 2017, 9, 1351–1373. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.Z.; Yoshimatsu, T.F.; Long, M.Y. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene 2006, 385, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Vanin, E.F. Processed pseudogenes: Characteristics and evolution. Annu. Rev. Genet. 1985, 19, 253–272. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, A.; Skene, B.; Swift, S.; Lovell-Badge, R. Zfa is an expressed retroposon derived from an alternative transcript of the Zfx gene. EMBO J. 1990, 9, 1529–1534. [Google Scholar] [CrossRef]
- Long, M.; Langley, C.H. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 1993, 260, 91–95. [Google Scholar] [CrossRef]
- Fitzgerald, J.; Wilcox, S.A.; Graves, J.A.; Dahl, H.H. A eutherian X-linked gene, PDHA1, is autosomal in marsupials: A model for the evolution of a second, testis-specific variant in eutherian mammals. Genomics 1993, 18, 636–642. [Google Scholar] [CrossRef]
- Dahl, H.H.; Brown, R.M.; Hutchison, W.M.; Maragos, C.; Brown, G.K. A testis-specific form of the human pyruvate dehydrogenase E1 alpha subunit is coded for by an intronless gene on chromosome 4. Genomics 1990, 8, 225–232. [Google Scholar] [CrossRef]
- Betran, E.; Thornton, K.; Long, M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002, 12, 1854–1859. [Google Scholar] [CrossRef]
- Emerson, J.J.; Kaessmann, H.; Betran, E.; Long, M. Extensive gene traffic on the mammalian X chromosome. Science 2004, 303, 537–540. [Google Scholar] [CrossRef]
- Vibranovski, M.D.; Zhang, Y.; Long, M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009, 19, 897–903. [Google Scholar] [CrossRef] [Green Version]
- Kaessmann, H.; Vinckenbosch, N.; Long, M. RNA-based gene duplication: Mechanistic and evolutionary insights. Nat. Rev. Genet. 2009, 10, 19–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parisi, M.; Nuttall, R.; Naiman, D.; Bouffard, G.; Malley, J.; Andrews, J.; Eastman, S.; Oliver, B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 2003, 299, 697–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.J. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol. Metab. 2004, 15, 79–83. [Google Scholar] [CrossRef] [PubMed]
- VanKuren, N.W.; Long, M. Gene duplicates resolving sexual conflict rapidly evolved essential gametogenesis functions. Nat. Ecol. Evol. 2018, 2, 705–712. [Google Scholar] [CrossRef]
- Wyman, M.J.; Cutter, A.D.; Rowe, L. Gene duplication in the evolution of sexual dimorphism. Evolution 2012, 66, 1556–1566. [Google Scholar] [CrossRef]
- Gallach, M.; Domingues, S.; Betran, E. Gene duplication and the genome distribution of sex-biased genes. Int. J. Evol. Biol. 2011, 2011, 989438. [Google Scholar] [CrossRef] [Green Version]
- Gallach, M.; Betran, E. Intralocus sexual conflict resolved through gene duplication. Trends Ecol. Evol. 2011, 26, 222–228. [Google Scholar] [CrossRef] [Green Version]
- Vicoso, B.; Charlesworth, B. Evolution on the X chromosome: Unusual patterns and processes. Nat. Rev. Genet. 2006, 7, 645–653. [Google Scholar] [CrossRef]
- Turner, J.M. Meiotic sex chromosome inactivation. Development 2007, 134, 1823–1831. [Google Scholar] [CrossRef] [Green Version]
- Cloutier, J.M.; Turner, J.M. Meiotic sex chromosome inactivation. Curr. Biol. 2010, 20, R962–R963. [Google Scholar] [CrossRef] [Green Version]
- Richardson, S.R.; Salvador-Palomeque, C.; Faulkner, G.J. Diversity through duplication: Whole-genome sequencing reveals novel gene retrocopies in the human population. Bioessays 2014, 36, 475–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinka, M.E.; Bangs, M.J.; Manguin, S.; Rubio-Palis, Y.; Chareonviriyaphap, T.; Coetzee, M.; Mbogo, C.M.; Hemingway, J.; Patil, A.P.; Temperley, W.H.; et al. A global map of dominant malaria vectors. Parasites Vectors 2012, 5, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N. Insecticide resistance in mosquitoes: Impact, mechanisms, and research directions. Annu. Rev. Entomol. 2015, 60, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.S. A comparative study of mosquito karyotypes. Ann. Entomol. Soc. Am. 1963, 56, 160–170. [Google Scholar] [CrossRef]
- Toups, M.A.; Hahn, M.W. Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes. Genetics 2010, 186, 763–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, D.A.; Russell, S. Role of testis-specific gene expression in sex-chromosome evolution of Anopheles gambiae. Genetics 2011, 189, 1117–1120. [Google Scholar] [CrossRef] [Green Version]
- Magnusson, K.; Lycett, G.J.; Mendes, A.M.; Lynd, A.; Papathanos, P.A.; Crisanti, A.; Windbichler, N. Demasculinization of the Anopheles gambiae X chromosome. BMC Evol. Biol. 2012, 12, 69. [Google Scholar] [CrossRef] [Green Version]
- Taxiarchi, C.; Kranjc, N.; Kriezis, A.; Kyrou, K.; Bernardini, F.; Russell, S.; Nolan, T.; Crisanti, A.; Galizi, R. High-resolution transcriptional profiling of Anopheles gambiae spermatogenesis reveals mechanisms of sex chromosome regulation. Sci. Rep. 2019, 9, 14841. [Google Scholar] [CrossRef]
- Connallon, T.; Clark, A.G. Evolutionary inevitability of sexual antagonism. Proc. Biol. Sci. 2014, 281, 20132123. [Google Scholar] [CrossRef]
- Connallon, T.; Clark, A.G. Sex-differential selection and the evolution of X inactivation strategies. PLoS Genet. 2013, 9, e1003440. [Google Scholar] [CrossRef]
- Sharakhova, M.V.; Hammond, M.P.; Lobo, N.F.; Krzywinski, J.; Unger, M.F.; Hillenmeyer, M.E.; Bruggner, R.V.; Birney, E.; Collins, F.H. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 2007, 8, R5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearson, W.R. Finding protein and nucleotide similarities with FASTA. Curr. Protoc. Bioinform. 2016, 53, 3.9.1–3.9.25. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, G.; Zhang, Y.; Xu, S.; Zhao, R.; Zhan, Z.; Li, X.; Ding, Y.; Yang, S.; Wang, W. On the origin of new genes in Drosophila. Genome Res. 2008, 18, 1446–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neafsey, D.E.; Waterhouse, R.M.; Abai, M.R.; Aganezov, S.S.; Alekseyev, M.A.; Allen, J.E.; Amon, J.; Arcà, B.; Arensburger, P.; Artemov, G.; et al. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science 2015, 347, 1258522. [Google Scholar] [CrossRef] [Green Version]
- Fontaine, M.C.; Pease, J.B.; Steele, A.; Waterhouse, R.M.; Neafsey, D.E.; Sharakhov, I.V.; Jiang, X.F.; Hall, A.B.; Catteruccia, F.; Kakani, E.; et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 2015, 347, 42. [Google Scholar] [CrossRef] [Green Version]
- Kamali, M.; Marek, P.E.; Peery, A.; Antonio-Nkondjio, C.; Ndo, C.; Tu, Z.; Simard, F.; Sharakhov, I.V. Multigene phylogenetics reveals temporal diversification of major african malaria vectors. PLoS ONE 2014, 9, e93580. [Google Scholar] [CrossRef] [Green Version]
- Clark, A.G.; Eisen, M.B.; Smith, D.R.; Bergman, C.M.; Oliver, B.; Markow, T.A.; Kaufman, T.C.; Kellis, M.; Gelbart, W.; Iyer, V.N.; et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature 2007, 450, 203–218. [Google Scholar] [CrossRef]
- Timoshevskiy, V.A.; Kinney, N.A.; deBruyn, B.S.; Mao, C.; Tu, Z.; Severson, D.W.; Sharakhov, I.V.; Sharakhova, M.V. Genomic composition and evolution of Aedes aegypti chromosomes revealed by the analysis of physically mapped supercontigs. BMC Biol. 2014, 12, 27. [Google Scholar] [CrossRef] [Green Version]
- Baker, D.A.; Nolan, T.; Fischer, B.; Pinder, A.; Crisanti, A.; Russell, S. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genom. 2011, 12, 296. [Google Scholar] [CrossRef] [Green Version]
- Papa, F.; Windbichler, N.; Waterhouse, R.M.; Cagnetti, A.; D’Amato, R.; Persampieri, T.; Lawniczak, M.K.N.; Nolan, T.; Papathanos, P.A. Rapid evolution of female-biased genes among four species of Anopheles malaria mosquitoes. Genome Res. 2017, 27, 1536–1548. [Google Scholar] [CrossRef] [Green Version]
- Giraldo-Calderon, G.I.; Harb, O.S.; Kelly, S.A.; Rund, S.S.; Roos, D.S.; McDowell, M.A. VectorBase.org updates: Bioinformatic resources for invertebrate vectors of human pathogens and related organisms. Curr. Opin. Insect Sci. 2021, 50, 100860. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Wang, J.; Xie, W.; Zhou, G.; Long, M.; Zhang, Q. Dynamic programming procedure for searching optimal models to estimate substitution rates based on the maximum-likelihood method. Proc. Natl. Acad. Sci. USA 2011, 108, 7860–7865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Dos Reis, M. Statistical properties of the branch-site test of positive selection. Mol. Biol. Evol. 2010, 28, 1217–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, G.; Krzywinska, E.; Kim, J.; Revuelta, L.; Ferretti, L.; Krzywinski, J. Dosage compensation in the african malaria mosquito Anopheles gambiae. Genome Biol. Evol. 2016, 8, 411–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicoso, B.; Bachtrog, D. Numerous Transitions of Sex Chromosomes in Diptera. PLoS Biol. 2015, 13, e1002078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betran, E. The “Life Histories” of Genes. J. Mol. Evol. 2015, 80, 186–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assis, R. Out of the testis, into the ovary: Biased outcomes of gene duplication and deletion in Drosophila. Evolution 2019, 73, 1850–1862. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yuan, Z.; Ma, Z.; Song, J.; Xie, X.; Chen, Y. KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol. Biosyst. 2014, 10, 2441–2447. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Rottschaefer, S.M.; Crawford, J.E.; Riehle, M.M.; Guelbeogo, W.M.; Gneme, A.; Sagnon, N.; Vernick, K.D.; Lazzaro, B.P. Population genetics of Anopheles coluzzii immune pathways and genes. G3 (Bethesda) 2014, 5, 329–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barillas-Mury, C.; Han, Y.S.; Seeley, D.; Kafatos, F.C. Anopheles gambiae Ag-STAT, a new insect member of the STAT family, is activated in response to bacterial infection. EMBO J. 1999, 18, 959–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, L.; Molina-Cruz, A.; Kumar, S.; Rodrigues, J.; Dixit, R.; Zamora, R.E.; Barillas-Mury, C. The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe 2009, 5, 498–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Lavorgna, A.; Bowman, M.; Hiscott, J.; Harhaj, E.W. Aryl hydrocarbon receptor interacting protein targets irf7 to suppress antiviral signaling and the induction of type i interferon. J. Biol. Chem. 2015, 290, 14729–14739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, M.W.; Lanzaro, G.C. Female-biased gene expression in the malaria mosquito Anopheles gambiae. Curr. Biol. 2005, 15, R192–R193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richler, C.; Soreq, H.; Wahrman, J. X inactivation in mammalian testis is correlated with inactive X-specific transcription. Nat. Genet. 1992, 2, 192–195. [Google Scholar] [CrossRef]
- Otto, S.P.; Scott, M.F.; Immler, S. Evolution of haploid selection in predominantly diploid organisms. Proc. Natl. Acad. Sci. USA 2015, 112, 15952–15957. [Google Scholar] [CrossRef] [Green Version]
- Raices, J.B.; Otto, P.A.; Vibranovski, M.D. Haploid selection drives new gene male germline expression. Genome Res. 2019, 29, 1115–1122. [Google Scholar] [CrossRef] [Green Version]
- Su, Q.; He, H.; Zhou, Q. On the origin and evolution of Drosophila new genes during spermatogenesis. Genes 2021, 12, 1796. [Google Scholar] [CrossRef]
- Fontaine, A.; Filipovic, L.; Fansiri, T.; Hoffmann, A.A.; Cheng, C.; Kirkpatrick, M.; Rasic, G.; Lambrechts, L. Extensive genetic differentiation between homomorphic sex chromosomes in the mosquito vector, Aedes aegypti. Genome Biol. Evol. 2017, 9, 2322–2335. [Google Scholar] [CrossRef] [Green Version]
- Ramalho-Santos, J.; Varum, S.; Amaral, S.; Mota, P.C.; Sousa, A.P.; Amaral, A. Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 2009, 15, 553–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piomboni, P.; Focarelli, R.; Stendardi, A.; Ferramosca, A.; Zara, V. The role of mitochondria in energy production for human sperm motility. Int. J. Androl. 2012, 35, 109–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eslamieh, M.; Mirsalehi, A.; Markova, D.N.; Betran, E. COX4-like, a nuclear-encoded mitochondrial gene duplicate, is essential for male fertility in Drosophila melanogaster. Genes 2022, 13, 424. [Google Scholar] [CrossRef] [PubMed]
- Waris, G.; Ahsan, H. Reactive oxygen species: Role in the development of cancer and various chronic conditions. J. Carcinog. 2006, 5, 14. [Google Scholar] [CrossRef]
- Gallach, M.; Chandrasekaran, C.; Betran, E. Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol. Evol. 2010, 2, 835–850. [Google Scholar] [CrossRef] [Green Version]
- Rand, D.M. Mitochondrial genetics of aging: Intergenomic conflict resolution. Sci. Aging Knowl. Environ. 2005, 2005, re5. [Google Scholar] [CrossRef]
- Allen, J.F. Separate sexes and the mitochondrial theory of ageing. J. Theor. Biol. 1996, 180, 135–140. [Google Scholar] [CrossRef]
- Han, M.V.; Hahn, M.W. Inferring the history of interchromosomal gene transposition in Drosophila using n-dimensional parsimony. Genetics 2012, 190, 813–825. [Google Scholar] [CrossRef] [Green Version]
- Phadnis, N.; Hsieh, E.; Malik, H.S. Birth, death, and replacement of karyopherins in Drosophila. Mol. Biol. Evol. 2012, 29, 1429–1440. [Google Scholar] [CrossRef] [Green Version]
- Rohozinski, J.; Lamb, D.J.; Bishop, C.E. UTP14c is a recently acquired retrogene associated with spermatogenesis and fertility in man. Biol. Reprod. 2006, 74, 644–651. [Google Scholar] [CrossRef]
- Vemuganti, S.A.; de Villena, F.P.; O’Brien, D.A. Frequent and recent retrotransposition of orthologous genes plays a role in the evolution of sperm glycolytic enzymes. BMC Genom. 2010, 11, 285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eslamieh, M.; Williford, A.; Betran, E. Few Nuclear-Encoded Mitochondrial Gene Duplicates Contribute to Male Germline-Specific Functions in Humans. Genome Biol. Evol. 2017, 9, 2782–2790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeidler, M.P.; Bach, E.A.; Perrimon, N. The roles of the Drosophila JAK/STAT pathway. Oncogene 2000, 19, 2598–2606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lachance, C.; Leclerc, P. Mediators of the Jak/STAT signaling pathway in human spermatozoa. Biol. Reprod. 2011, 85, 1222–1231. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Wawersik, M.; Matunis, E.L. The Jak-STAT target Chinmo prevents sex transformation of adult stem cells in the Drosophila testis niche. Dev. Cell 2014, 31, 474–486. [Google Scholar] [CrossRef] [Green Version]
- Carelli, F.N.; Hayakawa, T.; Go, Y.; Imai, H.; Warnefors, M.; Kaessmann, H. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 2016, 26, 301–314. [Google Scholar] [CrossRef] [Green Version]
- Kaessmann, H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010, 20, 1313–1326. [Google Scholar] [CrossRef] [Green Version]
- Vinckenbosch, N.; Dupanloup, I.; Kaessmann, H. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl. Acad. Sci. USA 2006, 103, 3220–3225. [Google Scholar] [CrossRef] [Green Version]
- Soumillon, M.; Necsulea, A.; Weier, M.; Brawand, D.; Zhang, X.; Gu, H.; Barthes, P.; Kokkinaki, M.; Nef, S.; Gnirke, A.; et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013, 3, 2179–2190. [Google Scholar] [CrossRef]
- Gabrieli, P.; Kakani, E.G.; Mitchell, S.N.; Mameli, E.; Want, E.J.; Mariezcurrena Anton, A.; Serrao, A.; Baldini, F.; Catteruccia, F. Sexual transfer of the steroid hormone 20E induces the postmating switch in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2014, 111, 16353–16358. [Google Scholar] [CrossRef] [Green Version]
- Thailayil, J.; Magnusson, K.; Godfray, H.C.; Crisanti, A.; Catteruccia, F. Spermless males elicit large-scale female responses to mating in the malaria mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2011, 108, 13677–13681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, S.N.; Catteruccia, F. Anopheline Reproductive Biology: Impacts on Vectorial Capacity and Potential Avenues for Malaria Control. Cold Spring Harb. Perspect. Med. 2017, 7, a025593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.T.; Hodge, J.M.; Sharakhov, I.V. Asymmetric phenotypes of sterile hybrid males from reciprocal crosses between species of the Anopheles gambiae complex. Front. Ecol. Evol. 2021, 9, 1–17. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, D.; Chen, J.; Liang, J.; Betrán, E.; Long, M.; Sharakhov, I.V. Retrogene Duplication and Expression Patterns Shaped by the Evolution of Sex Chromosomes in Malaria Mosquitoes. Genes 2022, 13, 968. https://doi.org/10.3390/genes13060968
Miller D, Chen J, Liang J, Betrán E, Long M, Sharakhov IV. Retrogene Duplication and Expression Patterns Shaped by the Evolution of Sex Chromosomes in Malaria Mosquitoes. Genes. 2022; 13(6):968. https://doi.org/10.3390/genes13060968
Chicago/Turabian StyleMiller, Duncan, Jianhai Chen, Jiangtao Liang, Esther Betrán, Manyuan Long, and Igor V. Sharakhov. 2022. "Retrogene Duplication and Expression Patterns Shaped by the Evolution of Sex Chromosomes in Malaria Mosquitoes" Genes 13, no. 6: 968. https://doi.org/10.3390/genes13060968
APA StyleMiller, D., Chen, J., Liang, J., Betrán, E., Long, M., & Sharakhov, I. V. (2022). Retrogene Duplication and Expression Patterns Shaped by the Evolution of Sex Chromosomes in Malaria Mosquitoes. Genes, 13(6), 968. https://doi.org/10.3390/genes13060968







