Identification and Regulatory Network Analysis of Genes Related to Reproductive Performance in the Hypothalamus and Pituitary of Angus Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Tissue Collection
2.2. RNA Extraction and cDNA Library Preparation
2.3. Data Analysis
2.3.1. Quality Control
2.3.2. Gene Functional Annotation
2.3.3. SNP (Single Nucleotide Polymorphisms) Calling
2.3.4. Variable Splicing Event Prediction
2.3.5. Differential Expression Analysis
2.3.6. Quantification of Gene Expression Levels
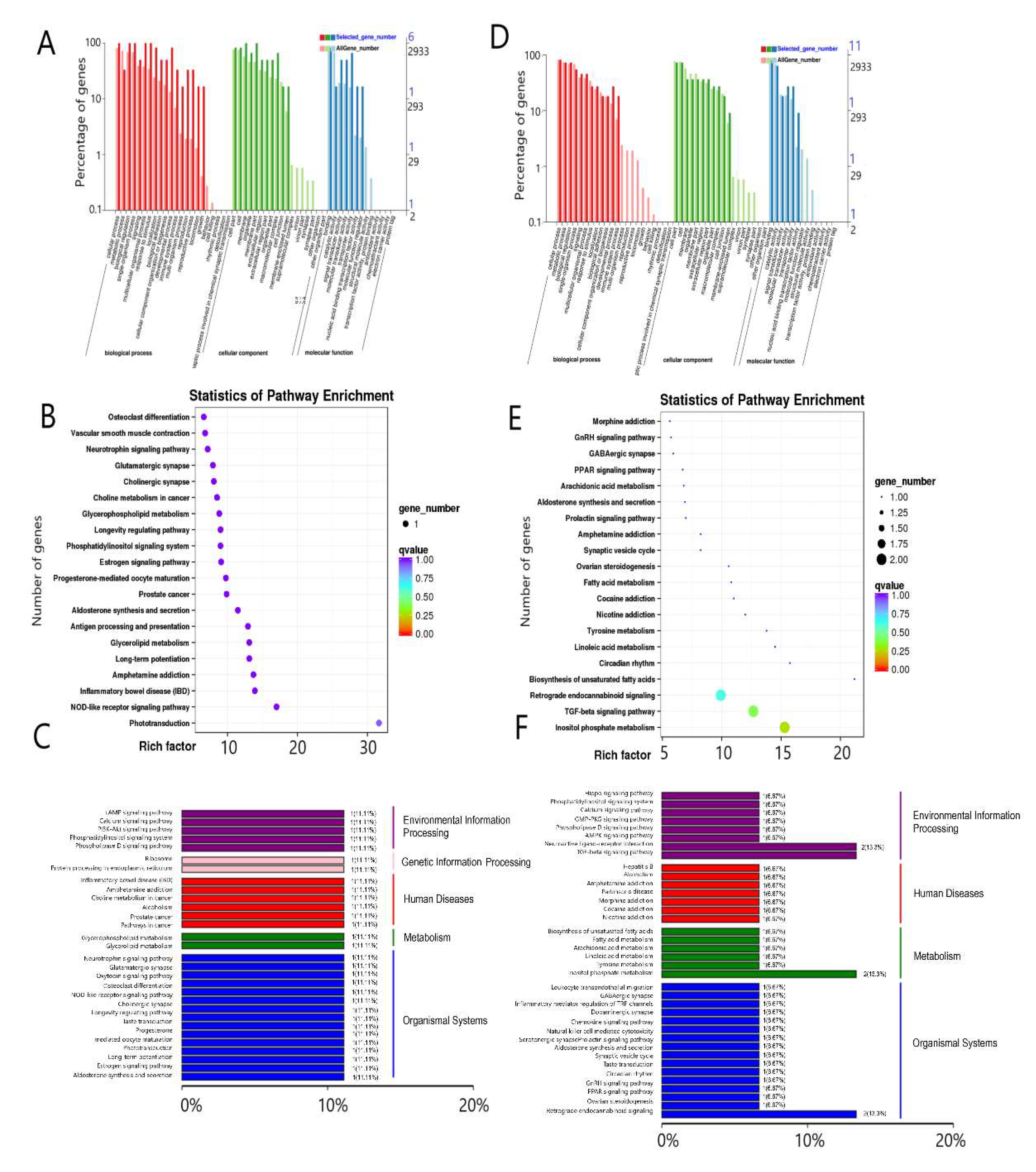
2.3.7. GO Enrichment Analysis
2.3.8. KEGG Pathway Enrichment Analysis
2.3.9. PPI (Protein–Protein Interaction)
2.4. Real-Time RT-PCR and Statistical Analysis
3. Results
3.1. Overview of Reads
3.2. SNP/Indel (Insertion and Deletion) Analysis
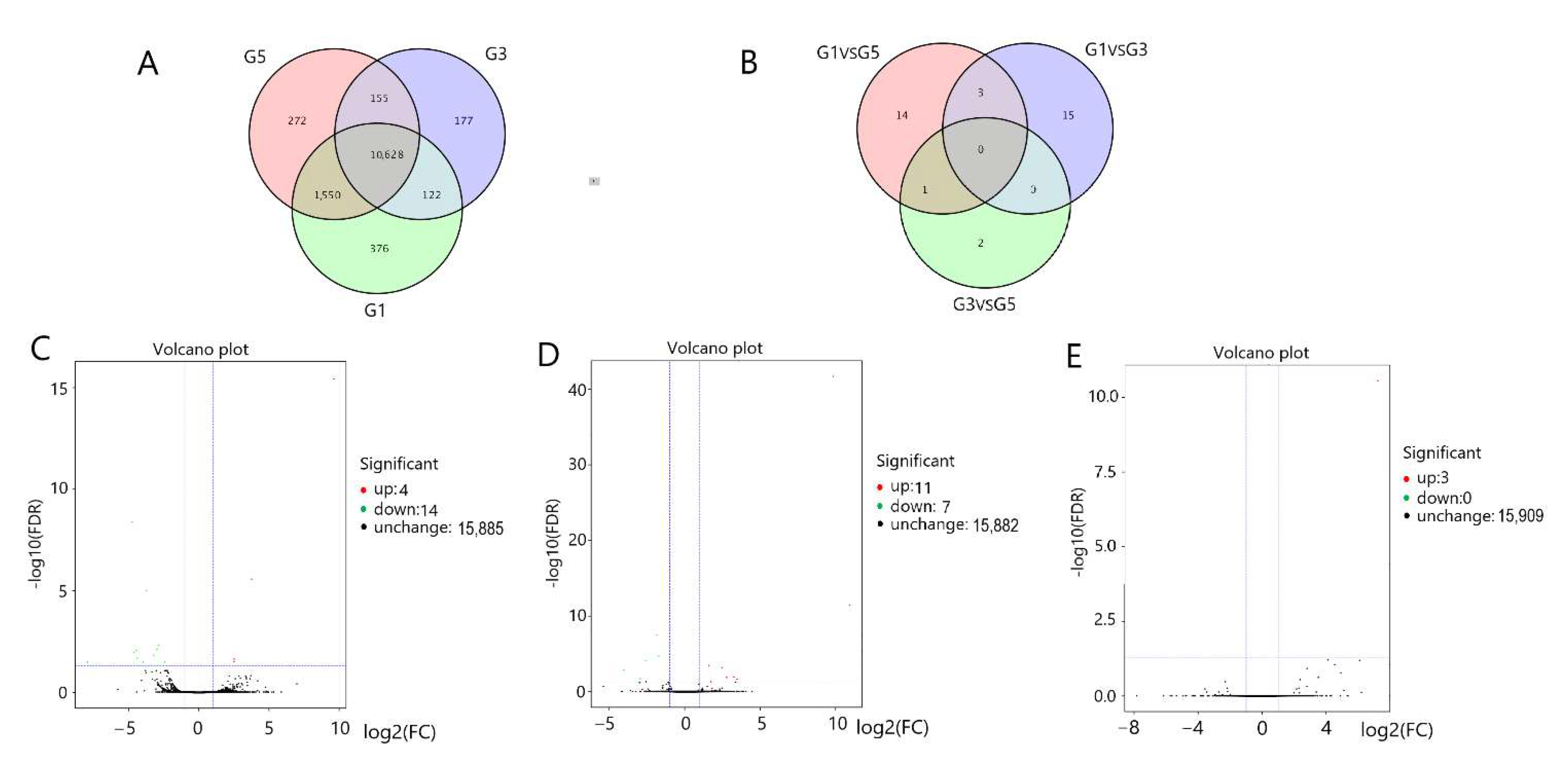
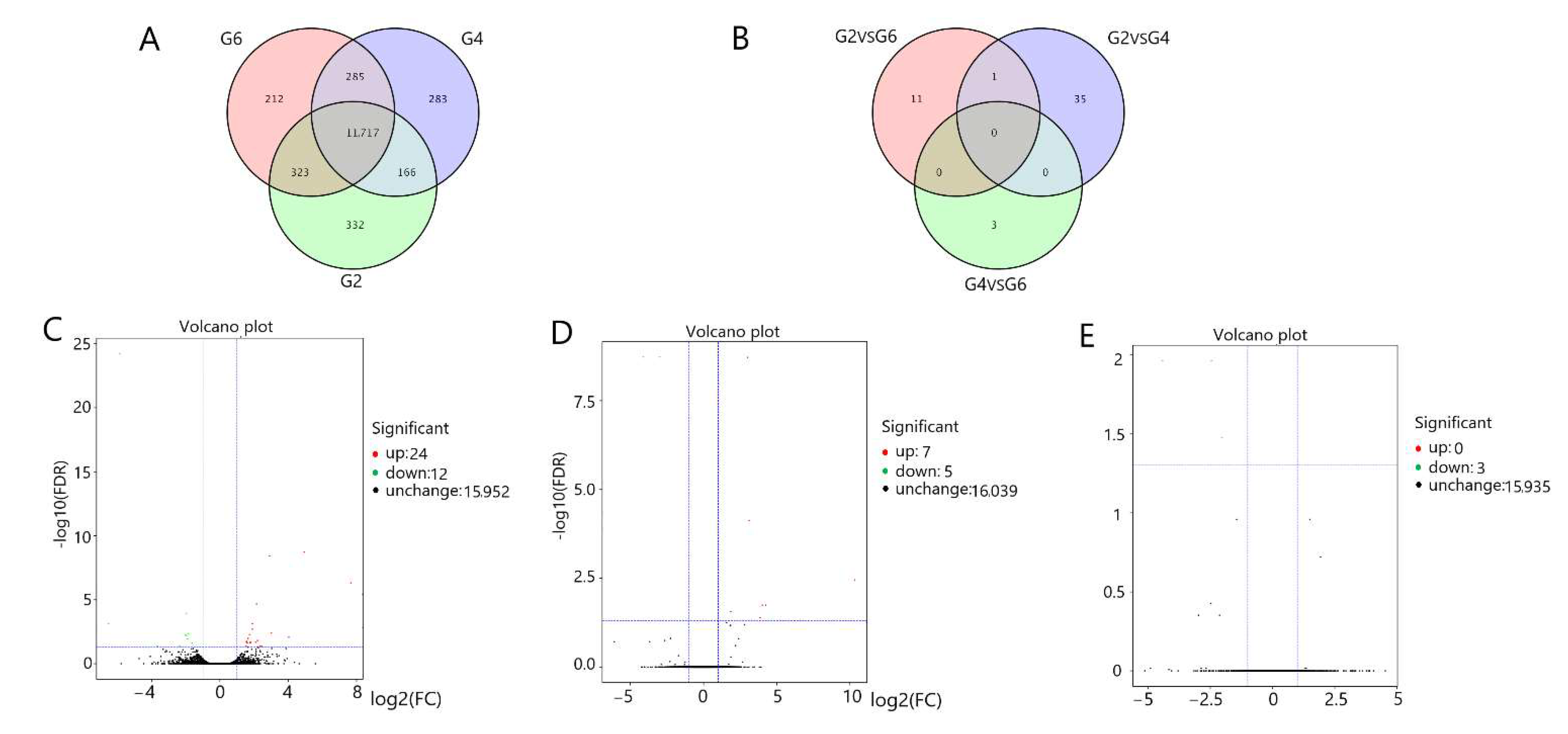
3.3. Screening of Gene Expression in the Pituitary Gland and Hypothalamus
3.4. New Gene Annotation Information in the Pituitary Gland and Hypothalamus
3.5. Known and Novel Transcript Expression Patterns in the Bovine Pituitary Gland and Hypothalamus
3.6. Functional Identification of Differentially Expressed Genes in the Pituitary Gland and Hypothalamus
3.7. The Major Genes of the Pituitary Gland and Hypothalamus
3.8. The Results of RT-qPCR
3.9. Protein Interaction Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riley, D.G.; Chase, C.C., Jr.; Coleman, S.W.; Olson, T.A. Evaluation of the Criollo breed Romosinuano as purebred and crossbred cows with Brahman and Angus in Florida: I. Reproduction and parturition. J. Anim. Sci. 2014, 92, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- López Valiente, S.; Maresca, S.; Rodríguez, A.M.; Palladino, R.A.; Lacau-Mengido, I.M.; Long, N.M.; Quintans, G. Effect of protein restriction of Angus cows during late gestation: Subsequent reproductive performance and milk yield. Prof. Anim. Sci. 2018, 34, 261–268. [Google Scholar] [CrossRef]
- Price, D.M.; Arellano, K.K.; Irsik, M.; Rae, D.O.; Yelich, J.V.; Mjoun, K.; Hersom, M.J. Effects of trace mineral supplement source during gestation and lactation in Angus and Brangus cows and subsequent calf immunoglobulin concentrations, growth, and development. Prof. Anim. Sci. 2017, 33, 194–204. [Google Scholar] [CrossRef]
- Hansen, L.B. Consequences of selection for milk yield from a geneticist’s viewpoint. J. Dairy Sci. 2000, 83, 1145–1150. [Google Scholar] [CrossRef]
- Lake, S.L.; Scholljegerdes, E.J.; Atkinson, R.L.; Nayigihugu, V.; Paisley, S.I.; Rule, D.C.; Moss, G.E.; Robinson, T.J.; Hess, B.W. Body condition score at parturition and postpartum supplemental fat effects on cow and calf performance. J. Anim. Sci. 2005, 83, 2908–2917. [Google Scholar] [CrossRef]
- Wang, K.; Liu, Y.; Tian, J.; Huang, K.; Shi, T.; Dai, X.; Zhang, W. Transcriptional Profiling and Identification of Heat-Responsive Genes in Perennial Ryegrass by RNA-Sequencing. Front. Plant Sci. 2017, 8, 1032. [Google Scholar] [CrossRef]
- Dong, Y.; Xie, M.; Jiang, Y.; Xiao, N.; Du, X.; Zhang, W.; Tosser-Klopp, G.; Wang, J.; Yang, S.; Liang, J.; et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 2013, 31, 135–141. [Google Scholar] [CrossRef]
- Canovas, A.; Reverter, A.; DeAtley, K.L.; Ashley, R.L.; Colgrave, M.L.; Fortes, M.R.; Islas-Trejo, A.; Lehnert, S.; Porto-Neto, L.; Rincon, G.; et al. Multi-tissue omics analyses reveal molecular regulatory networks for puberty in composite beef cattle. PLoS ONE 2014, 9, e102551. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.P.; Chen, Y.W.; He, F.C. Integrated nr Database in Protein Annotation System and Its Localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Whitacre, L.K.; Tizioto, P.C.; Kim, J.; Sonstegard, T.S.; Schroeder, S.G.; Alexander, L.J.; Medrano, J.F.; Schnabel, R.D.; Taylor, J.F.; Decker, J.E. What’s in your next-generation sequence data? An exploration of unmapped DNA and RNA sequence reads from the bovine reference individual. BMC Genomics 2015, 16, 1114. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A. The COG database: A tool for genome scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Stringtie Pertea, M.; Kim, D.; Pertea, G.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Li, Z.; Zhao, Y.; Wang, X.; Chen, L.; Zhao, Z.; Cao, M.; Chen, T.; Iqbal, T.; Zhang, B.; et al. Follicular fluid exosomes: Important modulator in proliferation and steroid synthesis of porcine granulosa cells. FASEB J. 2021, 35, e21610. [Google Scholar] [CrossRef] [PubMed]
- Bovine Genome, S.; Analysis, C.; Elsik, C.G.; Tellam, R.L.; Worley, K.C.; Gibbs, R.A.; Muzny, D.M.; Weinstock, G.M.; Adelson, D.L.; Eichler, E.E.; et al. The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science 2009, 324, 522–528. [Google Scholar]
- Seymour, A.J.; Scott, V.; Augustine, R.A.; Bouwer, G.T.; Campbell, R.E.; Brown, C.H. Development of an excitatory kisspeptin projection to the oxytocin system in late pregnancy. J. Physiol. 2017, 595, 825–838. [Google Scholar] [CrossRef]
- Sharma, D.; Kinsey, W.H. PYK2: A calcium-sensitive protein tyrosine kinase activated in response to fertilization of the zebrafish oocyte. Dev. Biol. 2013, 373, 130–140. [Google Scholar] [CrossRef][Green Version]
- Luo, J.; McGinnis, L.K.; Carlton, C.; Beggs, H.E.; Kinsey, W.H. PTK2b function during fertilization of the mouse oocyte. Biochem. Biophys. Res. Commun. 2014, 450, 1212–1217. [Google Scholar] [CrossRef]
- Deckel, A.W.; Elder, R.; Fuhrer, G. Biphasic developmental changes in Ca2+/calmodulin-dependent proteins in R6/2 Huntington’s disease mice. Neuroreport 2002, 13, 707–711. [Google Scholar] [CrossRef]
- Zhang, Y.; Hood, W.R. Current versus future reproduction and longevity: A re-evaluation of predictions and mechanisms. J. Exp. Biol. 2016, 219, 3177–3189. [Google Scholar] [CrossRef]
- Seibert, J.T.; Adur, M.K.; Schultz, R.B.; Thomas, P.Q.; Kiefer, Z.E.; Keating, A.F.; Baumgard, L.H.; Ross, J.W. Differentiating between the effects of heat stress and lipopolysaccharide on the porcine ovarian heat shock protein response1. J. Anim. Sci. 2019, 97, 4965–4973. [Google Scholar] [CrossRef]
- Liu, H.; Aramaki, M.; Fu, Y.; Forrest, D. Retinoid-Related Orphan Receptor β and Transcriptional Control of Neuronal Differentiation. Curr. Top. Dev. Biol. 2017, 125, 227–255. [Google Scholar] [PubMed]
- Rangel, P.L.; Sharp, P.J.; Gutierrez, C.G. Testosterone antagonist (flutamide) blocks ovulation and preovulatory surges of progesterone, luteinizing hormone and oestradiol in laying hens. Reproduction 2006, 131, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Stephens, S.B.Z.; Rouse, M.L.; Tolson, K.P.; Liaw, R.B.; Parra, R.A.; Chahal, N.; Kauffman, A.S. Effects of Selective Deletion of Tyrosine Hydroxylase from Kisspeptin Cells on Puberty and Reproduction in Male and Female Mice. eNeuro 2017, 4, ENEURO.0150-17.2017. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Eckardt, S.; Leu, N.A.; McLaughlin, K.J.; Wang, P.J. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J. Cell Biol. 2008, 180, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Boroujeni, P.B.; Sabbaghian, M.; Totonchi, M.; Sodeifi, N.; Sarkardeh, H.; Samadian, A.; Sadighi-Gilani, M.A.; Gourabi, H. Expression analysis of genes encoding TEX11, TEX12, TEX14 and TEX15 in testis tissues of men with non-obstructive azoospermia. JBRA Assist. Reprod. 2018, 22, 185–192. [Google Scholar] [CrossRef]
- Mantere, T.; Tervasmaki, A.; Nurmi, A.; Rapakko, K.; Kauppila, S.; Tang, J.; Schleutker, J.; Kallioniemi, A.; Hartikainen, J.M.; Mannermaa, A.; et al. Case-control analysis of truncating mutations in DNA damage response genes connects TEX15 and FANCD2 with hereditary breast cancer susceptibility. Sci. Rep. 2017, 7, 681. [Google Scholar] [CrossRef]


| Gene | Full Name | Gene | Full Name |
|---|---|---|---|
| TBX21 | T-box 21 | TBR1 | T-box, brain 1 factor |
| SHANK2 | SH3 and multiple ankyrin repeat domains 2 | HSPB7 | Hsp20/α crystallin family |
| PPP1R14A | Protein phosphatase 1 regulatory inhibitor subunit 14A | NOG | Noggin |
| HAPLN2 | Hyaluronan and proteoglycan link protein | PTK2B | Protein tyrosine kinase 2 β |
| TEX15 | Testis expressed 15 | AGO3 | Argonaute RISC catalytic component 3 |
| CAMK4 | calcium/calmodulin dependent protein kinase IV | ORM1 | Orosomucoid 1 |
| ANGPTL7 | Angiopoietin like 7 | RLF | RLF zinc finger |
| ORM1 | Orosomucoid 1 | CENPA | Centromere protein A |
| HSP90AB1 | Heat shock protein 90 alpha family class B member 1 | LOC506989 | Mitochondrial ribosomal protein L17-like |
| RORB | RAR-related orphan receptor B | ZNF282 | Zinc finger protein 282 |
| Group | Gene | FPKM | FPKM | FDR | log2FC | Regulated | Description |
|---|---|---|---|---|---|---|---|
| G5 vs. G1 | CAMK4 | 14.772 | 40.334 | 0.000 | −1.851 | Down | Calcium/calmodulin dependent protein kinase IV |
| G5 vs. G3 | HSP90AB1 | 27.882 | 0.170 | 0.000 | 7.228 | Up | Hsp90 protein Heat shock protein 90 alphafamily class B member 1 |
| G4 vs. G2 | TH | 7.633 | 27.451 | 0.005 | −2.016 | Down | Tyrosine hydroxylase |
| LOC511936 | 11.263 | 5.386 | 0.022 | 2.100 | Up | Cytochrome P450, family 2, subfamily J | |
| G6 vs. G2 | PTK2B | 7.633 | 31.988 | 0.005 | −2.016 | Down | alpha |
| Group | Gene Name | G1 | G3 | G5 | FDR | log2FC | Regulated | Description |
|---|---|---|---|---|---|---|---|---|
| G3 vs. G1 | DGKH | 2.859 | 0.679 | 1.872 | 0.004 | −2.972 | Down | Diacylglycerol kinase eta |
| LOC101904138 | 0.666 | 0.032 | 0.339 | 0.007 | −4.454 | Down | Uncharacterized LOC101904138 | |
| TPD52 | 2.334 | 33.140 | 8.349 | 0.009 | 3.775 | Up | Tumor protein D52 like 3 | |
| ZNF547 | 2.426 | 0.536 | 1.622 | 0.023 | −3.205 | Down | Zinc finger protein 547 | |
| TBX21 | 1.996 | 0.083 | 2.905 | 0.015 | −4.601 | Down | T-box 21 | |
| LOC101902860 | 38.591 | 1.351 | 25.463 | 0.010 | −4.734 | Down | Ribosomal L15 | |
| SHISA3 | 4.420 | 0.273 | 8.643 | 0.032 | −3.961 | Down | Shisa family member 3 | |
| HAPLN2 | 14.618 | 77.452 | 18.494 | 0.000 | 2.520 | Up | Hyaluronan and proteoglycan link protein | |
| SHANK2 | 14.665 | 2.981 | 6.003 | 0.030 | −2.428 | Down | SH3 and multiple ankyrin repeat domains 2 | |
| PPP1R14A | 9.619 | 117.037 | 9.727 | 0.000 | 2.525 | Up | Protein phosphatase 1 regulatory inhibitor subunit 14A | |
| LOC101904796 | 0.014 | 17.472 | 21.182 | 0.032 | 9.622 | Up | Heterogeneous nuclear ribonucleoproteins A2/B1 pseudogene | |
| LOC534155 | 14.330 | 1.879 | 13.502 | 0.000 | −2.859 | Down | Immunoglobulin (CD79A) binding protein 1-like | |
| G5 vs. G1 | ORM1 | 0.013 | 22.558 | 35.752 | 0.000 | 10.936 | Up | Orosomucoid 1 |
| LOC101906580 | 1.021 | 0.188 | 0.060 | 0.001 | −4.021 | Down | Uncharacterized LOC101906580 | |
| GPATCH2L | 5.110 | 7.325 | 9.942 | 0.000 | 1.600 | Up | Phospholipase A2 group IVE | |
| CCDC168 | 0.000 | 0.166 | 0.383 | 0.000 | Inf | Up | Coiled-coil domain containing 168 | |
| ANGPTL7 | 0.628 | 2.566 | 4.627 | 0.000 | 2.788 | Up | Angiopoietin like 7 | |
| TEX15 | 2.016 | 0.541 | 0.484 | 0.012 | −2.572 | Down | Testis expressed 15 | |
| GNAT2 | 0.000 | 0.648 | 0.689 | 0.000 | Inf | Up | G protein subunit alpha transducin 2 | |
| LYRM7 | 15.766 | 7.591 | 5.663 | 0.021 | −1.716 | Down | LYR motif containing 7 | |
| PCDHGA7 | 0.215 | 0.910 | 2.116 | 0.000 | 3.262 | Up | Protocadherin gammasubfamily A, 7 | |
| LOC101904796 | 0.014 | 17.472 | 21.182 | 0.011 | 9.828 | Up | Heterogeneous nuclear ribonucleoproteins A2/B1 pseudogene | |
| LOC100848679 | 3.066 | 2.119 | 0.967 | 0.000 | −2.940 | Down | DEAD-box ATP-dependent RNA helicase 30-like |
| Group | Gene Name | G2 | G4 | G6 | FDR | log2FC | Regulated | Description |
|---|---|---|---|---|---|---|---|---|
| G4 vs. G2 | RORB | 2.771 | 0.887 | 1.385 | 0.041 | −2.354 | Down | RAR related orphan receptor B |
| PCP4 | 199.707 | 645.127 | 419.042 | 0.010 | 1.654 | Up | Purkinje cell protein 4 | |
| LOC101904985 | 1.690 | 0.000 | 0.188 | 0.002 | -- | Down | Regulating synaptic membrane exocytosis protein 1-like | |
| SYNJ2 | 10.812 | 3.234 | 6.590 | 0.005 | −1.855 | Down | synaptojanin 2 | |
| KCNK3 | 2.834 | 8.648 | 6.530 | 0.024 | 1.784 | Up | Potassium two pore domain channel subfamily K member 3 | |
| KCNF1 | 27.825 | 105.381 | 69.183 | 0.001 | 1.907 | Up | Potassium two pore domain channel subfamily K member 1 | |
| TBR1 | 13.041 | 1.779 | 5.804 | 0.044 | −3.671 | Down | T-box, brain 1 factor | |
| NKAIN3 | 8.259 | 2.222 | 5.496 | 0.000 | −1.971 | Down | sodium/potassium transporting ATPase interacting 3 | |
| NPPB | 2.539 | 13.985 | 7.234 | 0.044 | 2.455 | Up | Natriuretic peptide B | |
| PLCH2 | 12.934 | 39.087 | 30.698 | 0.022 | 1.553 | Up | Phospholipase C eta 2 | |
| GABRD | 20.752 | 56.832 | 46.509 | 0.040 | 1.538 | Up | Gamma-aminobutyric acid type A receptor subunit delta | |
| DMKN | 0.110 | 1.857 | 1.536 | 0.009 | 4.041 | Up | Dermokine | |
| NOG | 12.227 | 37.659 | 28.866 | 0.024 | 1.604 | Up | Noggin | |
| KCNAB3 | 6.489 | 25.054 | 17.821 | 0.002 | 1.904 | Up | KN motif and ankyrin repeat domains 3 | |
| PCP4L1 | 67.687 | 310.150 | 217.532 | 0.047 | 2.215 | Up | Purkinje cell protein 4 like 1 | |
| ANKRD29 | 9.223 | 32.066 | 26.675 | 0.022 | 1.788 | Up | Ankyrin repeat domain 29 | |
| LOC101906058 | 50.638 | 13.883 | 37.250 | 0.011 | −1.890 | Down | Acyl-CoA desaturase-like | |
| NHSL2 | 4.488 | 1.153 | 3.759 | 0.007 | −1.961 | Down | NHS like 2 | |
| SLC9A7 | 3.038 | 14.306 | 9.154 | 0.000 | 2.145 | Up | Solute carrier family 9 member A7 | |
| BMP8B | 0.776 | 3.641 | 2.071 | 0.015 | 2.205 | Up | Bone morphogenetic protein 8b | |
| CSMD2 | 4.889 | 1.299 | 3.543 | 0.025 | −1.630 | Down | CUB and Sushi multiple domains 2 | |
| SHISA8 | 5.045 | 34.331 | 20.559 | 0.000 | 2.903 | Up | Shisa family member 8 | |
| CPLX1 | 39.927 | 121.167 | 89.915 | 0.018 | 1.569 | Up | Complexin 1 | |
| HSPB7 | 1.072 | 5.446 | -- | 0.005 | 1.734 | Up | Hsp20/alpha crystallin family | |
| G6 vs. G2 | CHRNA2 | 1.429 | 5.469 | 10.428 | 0.000 | 3.120 | Up | Cholinergic receptor nicotinic alpha 2 subunit |
| ORM1 | 0.039 | 15.589 | 56.033 | 0.003 | 10.333 | Up | Orosomucoid 1 | |
| VSIG10 | 2.858 | 13.395 | 25.630 | 0.000 | 3.010 | Up | V-set and immunoglobulin domain containing 10 | |
| SOX4 | 0.226 | 2.999 | 3.162 | 0.041 | 3.872 | Up | SRY-box transcription factor 4 | |
| RLF | 3.370 | 4.590 | 6.354 | 0.027 | 1.858 | Up | RLF zinc finger | |
| AGO3 | 6.829 | 4.594 | 1.811 | 0.000 | −2.984 | Down | argonaute RISC catalytic component 3 | |
| G6 vs. G4 | CENPA | 0.774 | 1.052 | 0.035 | 0.011 | −4.415 | Down | Centromere protein A |
| ZNF282 | 2.947 | 5.137 | 1.750 | 0.034 | −2.030 | Down | Zinc finger protein 282 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Yuan, C.; Zhao, Y.; Li, C.; Cao, M.; Li, H.; Zhao, Z.; Sun, A.; Basang, W.; Zhu, Y.; et al. Identification and Regulatory Network Analysis of Genes Related to Reproductive Performance in the Hypothalamus and Pituitary of Angus Cattle. Genes 2022, 13, 965. https://doi.org/10.3390/genes13060965
Huang Y, Yuan C, Zhao Y, Li C, Cao M, Li H, Zhao Z, Sun A, Basang W, Zhu Y, et al. Identification and Regulatory Network Analysis of Genes Related to Reproductive Performance in the Hypothalamus and Pituitary of Angus Cattle. Genes. 2022; 13(6):965. https://doi.org/10.3390/genes13060965
Chicago/Turabian StyleHuang, Yuwen, Chenfeng Yuan, Yun Zhao, Chunjin Li, Maosheng Cao, Haobang Li, Zijiao Zhao, Ao Sun, Wangdui Basang, Yanbin Zhu, and et al. 2022. "Identification and Regulatory Network Analysis of Genes Related to Reproductive Performance in the Hypothalamus and Pituitary of Angus Cattle" Genes 13, no. 6: 965. https://doi.org/10.3390/genes13060965
APA StyleHuang, Y., Yuan, C., Zhao, Y., Li, C., Cao, M., Li, H., Zhao, Z., Sun, A., Basang, W., Zhu, Y., Chen, L., He, F., Huan, C., Zhang, B., Iqbal, T., Wei, Y., Fan, W., Yi, K., & Zhou, X. (2022). Identification and Regulatory Network Analysis of Genes Related to Reproductive Performance in the Hypothalamus and Pituitary of Angus Cattle. Genes, 13(6), 965. https://doi.org/10.3390/genes13060965






