Exome Sequencing Identifies Genetic Variants Associated with Extreme Manifestations of the Cardiovascular Phenotype in Marfan Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Exome Sequencing and Variant Discovery
2.2. VAAST Analysis
2.3. Variant Prioritization
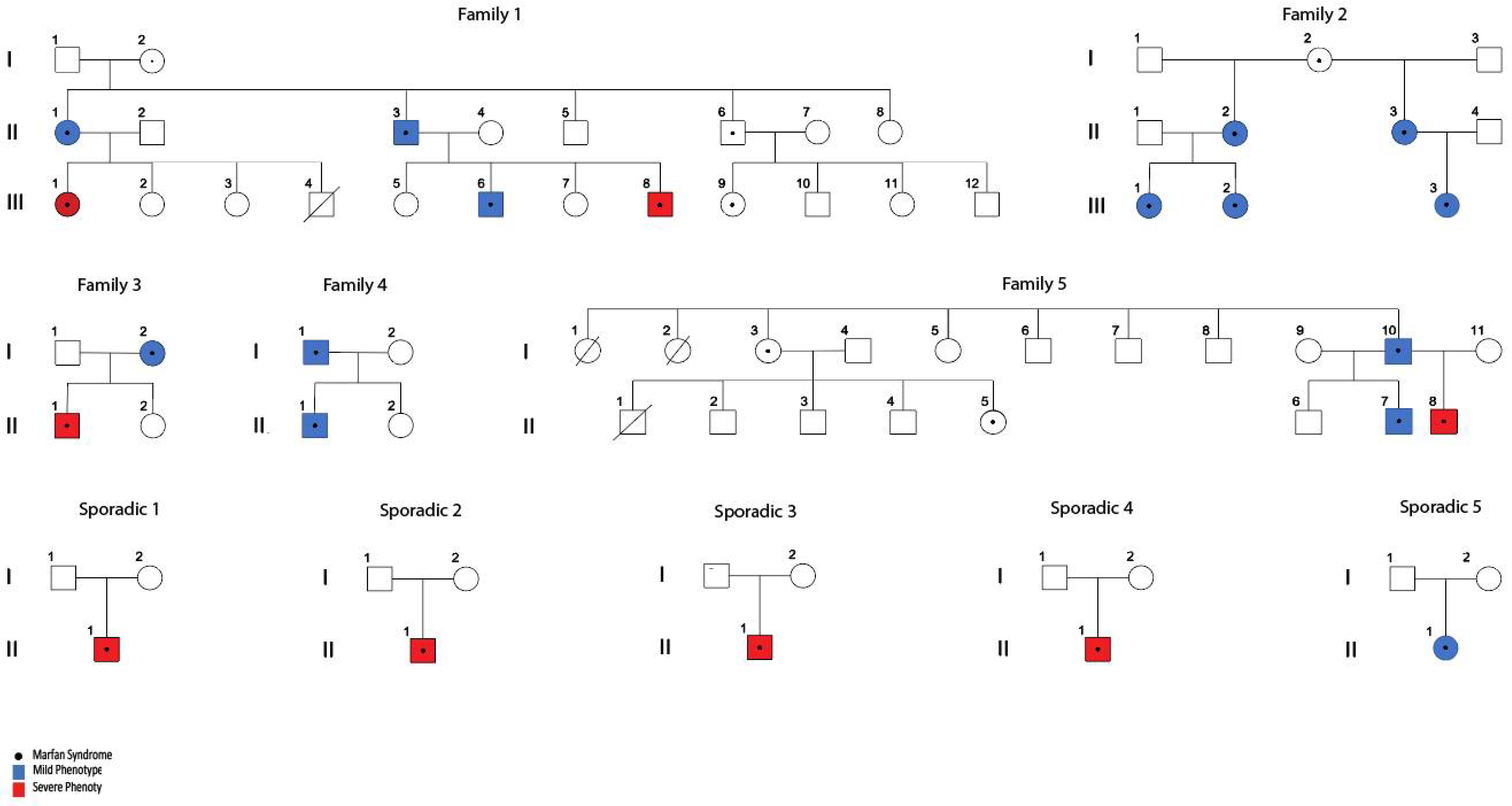
3. Results
3.1. Subjects
| Family or Sporadic Cases | Patient ID | FBN1 Variants HGVS Nomenclature (NM_000138.5) | Variant Type | Clinical Significance (SIFT) | Protein Functional Domain | Aortic Phenotype Classification |
|---|---|---|---|---|---|---|
| Fam 1 | CAS-01-001 | c.7339G>A | Missense | Pathogenic | EGF-like calcium-binding domain | Severe |
| CAS-01-002 | Mild | |||||
| CAS-01-003 | Mild | |||||
| CAS-01-007 | Mild | |||||
| CAS-01-005 | Severe | |||||
| Fam 2 | CAS-01-019 | c.1090C>T | Nonsense (stop gained) | Pathogenic | TB domain | Mild |
| CAS-01-020 | Mild | |||||
| CAS-01-021 | Mild | |||||
| CAS-01-022 | Mild | |||||
| CAS-01-023 | Mild | |||||
| Fam 3 | CAS-01-024 | c.7180C>T | Nonsense (stop gained) | Pathogenic | - | Mild |
| CAS-01-026 | Severe | |||||
| Fam 4 | CAS-01-035 | c.7204+1G>A | splice region | Likely_pathogenic | - | Mild |
| CAS-01-036 | Mild | |||||
| Fam 5 | CAS-01-045 | c.4196_4197insA | frameshift insertion | Pathogenic | EGF-like calcium-binding domain | Severe |
| CAS-01-046 | Mild | |||||
| CAS-01-048 | Mild | |||||
| Sporadic 1 | CAS-01-016 | c.8562delC | frameshift deletion | Pathogenic | - | Severe |
| Sporadic 2 | CAS-01-027 | c.1090C>T | Nonsense | Pathogenic | TB domain | Severe |
| Sporadic 3 | CAS-01-031 | c.5788+5G>T | splice region | Pathogenic | - | Severe |
| Sporadic 4 | CAS-01-043 | c.A2673G | Regulatory region | Likely_pathogenic | TB domain | Severe |
| Sporadic 5 | CAS-01-044 | c.236dupA | frameshift insertion | Pathogenic | N-terminal domain | Mild |
3.2. VAAST Analysis
3.3. Variant Prioritization and Candidate Modifier Genes
3.3.1. Candidate Genes Associated with the Severe Cardiovascular Phenotype in MFS
3.3.2. Candidate Genes Associated with the Mild Cardiovascular Phenotype in MFS
3.3.3. In Silico Analysis of the Effect of Variants in Protein Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Robinson, P.N.; Arteaga-Solis, E.; Baldock, C.; Collod-Béroud, G.; Booms, P.; De Paepe, A.; Dietz, H.C.; Guo, G.; Handford, P.A.; Judge, D.P.; et al. The molecular genetics of Marfan syndrome and related disorders. J. Med. Genet. 2006, 43, 769–787. [Google Scholar] [CrossRef] [PubMed]
- Keane, M.G.; Pyeritz, R.E. Medical management of marfan syndrome. Circulation 2008, 117, 2802–2813. [Google Scholar] [CrossRef] [PubMed]
- Sakai, L.Y.; Keene, D.R.; Renard, M.; De Backer, J. FBN1: The disease-causing gene for Marfan syndrome and other genetic disorders. Gene 2016, 592, 279–291. [Google Scholar] [CrossRef]
- Dietz, H.C.; Cutting, C.R.; Pyeritz, R.E.; Maslen, C.L.; Sakai, L.Y.; Corson, G.M.; Puffenberger, E.G.; Hamosh, A.; Nanthakumar, E.J.; Curristin, S.M.; et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature 1991, 352, 337–339. [Google Scholar] [CrossRef]
- Li, Y.; Xu, J.; Chen, M.; Du, B.; Li, Q.; Xing, Q.; Zhang, Y. A FBN1 mutation association with different phenotypes of Marfan syndrome in a Chinese family. Clin. Chim. Acta 2016, 460, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Takeda, N.; Inuzuka, R.; Maemura, S.; Morita, H.; Nawata, K.; Fujita, D.; Taniguchi, Y.; Yamauchi, H.; Yagi, H.; Kato, M.; et al. Impact of Pathogenic FBN1 Variant Types on the Progression of Aortic Disease in Patients With Marfan Syndrome. Circ. Genom. Precis. Med. 2018, 11, e002058. [Google Scholar] [CrossRef] [PubMed]
- Booher, A.M.; Eagle, K.A. Diagnosis and management issues in thoracic aortic aneurysm. Am. Heart J. 2011, 162, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Díaz de Bustamante, A.; Ruiz-Casares, E.; Darnaude, M.T.; Perucho, T.; Martínez-Quesada, G. Phenotypic Variability in Marfan Syndrome in a Family With a Novel Nonsense FBN1 Gene Mutation. Rev. Esp. Cardiol. 2012, 65, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, S.; Furger, A.; Halliday, D.; Judge, D.P.; Jefferson, A.; Dietz, H.C.; Firth, H.; Handford, P.A. Allelic variation in normal human FBN1 expression in a family with Marfan syndrome: A potential modifier of phenotype? Hum. Mol. Genet. 2003, 12, 2269–2276. [Google Scholar] [CrossRef] [PubMed]
- Aubart, M.; Gazal, S.; Arnaud, P.; Benarroch, L.; Gross, M.S.; Buratti, J.; Boland, A.; Meyer, V.; Zouali, H.; Hanna, N.; et al. Association of modifiers and other genetic factors explain Marfan syndrome clinical variability. Eur. J. Hum. Genet. 2018, 26, 1759–1772. [Google Scholar] [CrossRef]
- Franken, R.; Teixido-Tura, G.; Brion, M.; Forteza, A.; Rodriguez-Palomares, J.; Gutierrez, L.; Garcia Dorado, D.; Pals, G.; Mulder, B.J.; Evangelista, A. Relationship between fibrillin-1 genotype and severity of cardiovascular involvement in Marfan syndrome. Heart 2017, 103, 1795–1799. [Google Scholar] [CrossRef]
- Fernandes, G.R.; Massironi, S.M.G.; Pereira, L.V. Identification of Loci Modulating the Cardiovascular and Skeletal Phenotypes of Marfan Syndrome in Mice. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Gerdes Gyuricza, I.; Barbosa de Souza, R.; Farinha-Arcieri, L.E.; Ribeiro Fernandes, G.; Veiga Pereira, L. Is HSPG2 a modifier gene for Marfan syndrome? Eur. J. Hum. Genet. 2020, 28, 1292–1296. [Google Scholar] [CrossRef]
- Lima, B.L.; Santos, E.J.C.; Fernandes, G.R.; Merkel, C.; Mello, M.R.B.; Gomes, J.P.A.; Soukoyan, M.; Kerkis, A.; Massironi, S.M.G.; Visintin, J.A.; et al. A new mouse model for marfan syndrome presents phenotypic variability associated with the genetic background and overall levels of Fbn1 expression. PLoS ONE 2010, 5, e14136. [Google Scholar] [CrossRef]
- Michel, J.-B.; Jondeau, G.; Milewicz, D.M. From genetics to response to injury: Vascular smooth muscle cells in aneurysms and dissections of the ascending aorta. Cardiovasc. Res. 2018, 114, 578–589. [Google Scholar] [CrossRef]
- Bolar, N.; Van Laer, L.; Loeys, B.L. Marfan syndrome. Curr. Opin. Pediatr. 2012, 24, 498–504. [Google Scholar] [CrossRef]
- Castellano, J.M.; Kovacic, J.C.; Sanz, J.; Fuster, V. Are we ignoring the dilated thoracic aorta? Ann. N. Y. Acad. Sci. 2012, 1254, 164–174. [Google Scholar] [CrossRef]
- Habashi, J.P.; Judge, D.P.; Holm, T.M.; Cohn, R.D.; Loeys, B.L.; Cooper, T.K.; Myers, L.; Klein, E.C.; Liu, G.; Calvi, C.; et al. Losartan, an AT1 Antagonist, Prevents Aortic Aneurysm in a Mouse Model of Marfan Syndrome. Science 2006, 312, 117–121. [Google Scholar] [CrossRef]
- Loeys, B.L.; Dietz, H.C.; Braverman, A.C.; Callewaert, B.L.; De Backer, J.; Devereux, R.B.; Hilhorst-Hofstee, Y.; Jondeau, G.; Faivre, L.; Milewicz, D.M.; et al. The revised Ghent nosology for the Marfan syndrome. J. Med. Genet. 2010, 47, 476–485. [Google Scholar] [CrossRef]
- Depristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Picard Tools—By Broad Institute. Available online: http://broadinstitute.github.io/picard/ (accessed on 3 June 2022).
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Kennedy, B.; Kronenberg, Z.; Hu, H.; Moore, B.; Flygare, S.; Reese, M.G.; Jorde, L.B.; Yandell, M.; Huff, C. Using VAAST to Identify Disease-Associated Variants in Next-Generation Sequencing Data. Curr. Protoc. Hum. Genet. 2014, 81, 6–14. [Google Scholar] [CrossRef]
- Yandell, M.; Huff, C.; Hu, H.; Singleton, M.; Moore, B.; Xing, J.; Jorde, L.B.; Reese, M.G. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011, 21, 1529–1542. [Google Scholar] [CrossRef] [PubMed]
- Bruse, S.; Moreau, M.; Bromberg, Y.; Jang, J.H.; Wang, N.; Ha, H.; Picchi, M.; Lin, Y.; Langley, R.J.; Qualls, C.; et al. Whole exome sequencing identifies novel candidate genes that modify chronic obstructive pulmonary disease susceptibility. Hum. Genom. 2016, 10, 1–13. [Google Scholar] [CrossRef]
- Pinnaro, C.T.; Henry, T.; Major, H.J.; Parida, M.; DesJardin, L.E.; Manak, J.R.; Darbro, B.W. Candidate modifier genes for immune function in 22q11.2 deletion syndrome. Mol. Genet. Genom. Med. 2020, 8, 1–10. [Google Scholar] [CrossRef]
- Génin, E.; Feingold, J.; Clerget-Darpoux, F. Identifying modifier genes of monogenic disease: Strategies and difficulties. Hum. Genet. 2008, 124, 357–368. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef]
- Ardlie, K.G.; DeLuca, D.S.; Segrè, A.V.; Sullivan, T.J.; Young, T.R.; Gelfand, E.T.; Trowbridge, C.A.; Maller, J.B.; Tukiainen, T.; Lek, M.; et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Kim, H.J.; Ham, S.A.; Kim, S.U.; Hwang, J.Y.; Kim, J.H.; Chang, K.C.; Yabe-Nishimura, C.; Kim, J.H.; Seo, H.G. Transforming growth factor-β1 is a molecular target for the peroxisome proliferator-activated receptor δ. Circ. Res. 2008, 102, 193–200. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, M.Y.; Jin, H.; Kim, H.J.; Kang, S.S.; Kim, H.J.; Lee, J.H.; Chang, K.C.; Hwang, J.Y.; Yabe-Nishimura, C.; et al. Peroxisome proliferator-Activated receptor δ regulates extracellular matrix and apoptosis of vascular smooth muscle cells through the activation of transforming growth factor-β1/Smad3. Circ. Res. 2009, 105, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Payne, T.J.; Mohanty, D.K. Effects of Slow, Sustained, and Rate-Tunable Nitric Oxide Donors on Human Aortic Smooth Muscle Cells Proliferation. Chem. Biol. Drug Des. 2011, 78, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Orr, A.W.; Hastings, N.E.; Blackman, B.R.; Wamhoff, B.R. Complex regulation and function of the inflammatory smooth muscle cell phenotype in atherosclerosis. J. Vasc. Res. 2010, 47, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Petit, C.; Karkhaneh Yousefi, A.A.; Ben Moussa, O.; Michel, J.B.; Guignandon, A.; Avril, S. Regulation of SMC traction forces in human aortic thoracic aneurysms. Biomech. Model. Mechanobiol. 2021, 20, 717–731. [Google Scholar] [CrossRef]
- Li, S.; Yang, B.; Du, Y.; Lin, Y.; Liu, J.; Huang, S.; Zhang, A.; Jia, Z.; Zhang, Y. Targeting PPARα for the Treatment and Understanding of Cardiovascular Diseases. Cell. Physiol. Biochem. 2018, 51, 2760–2775. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Li, X.; Li, Y.; Tang, X.; Xu, J.; Li, R.; Hao, P.; Sun, Y. PPAR δ agonist GW501516 inhibits PDGF-stimulated pulmonary arterial smooth muscle cell function related to pathological vascular remodeling. BioMed Res. Int. 2013, 2013. [Google Scholar] [CrossRef]
- Lobley, A.; Sadowski, M.I.; Jones, D.T. pGenTHREADER and pDomTHREADER: New methods for improved protein fold recognition and superfamily discrimination. Bioinformatics 2009, 25, 1761–1767. [Google Scholar] [CrossRef]
- Volkenstein, M.V. Coding of Polar and Non-polar Amino-acids. Nature 1965, 207, 294–295. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, S.L.; Hartley, T.; Dyment, D.A.; Beaulieu, C.L.; Schwartzentruber, J.; Smith, A.; Bedford, H.M.; Bernard, G.; Bernier, F.P.; Brais, B.; et al. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: Time to address gaps in care. Clin. Genet. 2016, 89, 275–284. [Google Scholar] [CrossRef]
- Fouillade, C.; Monet-Leprêtre, M.; Baron-Menguy, C.; Joutel, A. Notch signalling in smooth muscle cells during development and disease. Cardiovasc. Res. 2012, 95, 138–146. [Google Scholar] [CrossRef]
- Manderfield, L.J.; High, F.A.; Engleka, K.A.; Liu, F.; Li, L.; Rentschler, S.; Epstein, J.A. Notch activation of Jagged1 contributes to the assembly of the arterial wall. Circulation 2012, 125, 314–323. [Google Scholar] [CrossRef] [PubMed]
- De la Pompa, J.L.; Epstein, J.A. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 2013, 22, 244–254. [Google Scholar] [CrossRef]
- Loerakker, S.; Stassen, O.M.J.A.; Ter Huurne, F.M.; Boareto, M.; Bouten, C.V.C.; Sahlgren, C.M. Mechanosensitivity of Jagged–Notch signaling can induce a switch-type behavior in vascular homeostasis. Proc. Natl. Acad. Sci. USA 2018, 115, E3682–E3691. [Google Scholar] [CrossRef]
- Saarinen, J.; Kalkkinen, N.; Welgus, H.G.; Kovanen, P.T. Activation of human interstitial procollagenase through direct cleavage of the Leu83-Thr84 bond by mast cell chymase. J. Biol. Chem. 1994, 269, 18134–18140. [Google Scholar] [CrossRef]
- Tchougounova, E.; Forsberg, E.; Angelborg, G.; Kjellén, L.; Pejler, G. Altered processing of fibronectin in mice lacking heparin. A role for heparin-dependent mast cell chymase in fibronectin degradation. J. Biol. Chem. 2001, 276, 3772–3777. [Google Scholar] [CrossRef] [PubMed]
- Leskinen, M.J.; Lindstedt, K.A.; Wang, Y.; Kovanen, P.T. Mast cell chymase induces smooth muscle cell apoptosis by a mechanism involving fibronectin degradation and disruption of focal adhesions. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 238–243. [Google Scholar] [CrossRef]
- Raman, M.; Chen, W.; Cobb, M.H. Differential regulation and properties of MAPKs. Oncogene 2007, 26, 3100–3112. [Google Scholar] [CrossRef]
- Karin, M.; Gallagher, E. From JNK to pay dirt: Jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life 2005, 57, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Karagyozov, L.; Grozdanov, P.N.; Böhmer, F.D. The translation attenuating arginine-rich sequence in the extended signal peptide of the protein-tyrosine phosphatase PTPRJ/DEP1 is conserved in mammals. PLoS ONE 2020, 15, e0240498. [Google Scholar] [CrossRef]
- Oshikawa, J.; Urao, N.; Kim, H.W.; Kaplan, N.; Razvi, M.; McKinney, R.; Poole, L.B.; Fukai, T.; Ushio-Fukai, M. Extracellular SOD-derived H2O2 promotes VEGF signaling in caveolae/lipid rafts and post-ischemic angiogenesis in mice. PLoS ONE 2010, 5, e10189. [Google Scholar] [CrossRef]
- Fournier, P.; Dussault, S.; Fusco, A.; Rivard, A.; Royal, I. Tyrosine phosphatase PTPRJ/DEP-1 Is an essential promoter of vascular permeability, angiogenesis, and tumor progression. Cancer Res. 2016, 76, 5080–5091. [Google Scholar] [CrossRef]
- Sanchez-Castro, M.; Eldjouzi, H.; Charpentier, E.; Busson, P.F.; Hauet, Q.; Lindenbaum, P.; Delasalle-Guyomarch, B.; Baudry, A.; Pichon, O.; Pascal, C.; et al. Search for Rare Copy-Number Variants in Congenital Heart Defects Identifies Novel Candidate Genes and a Potential Role for FOXC1 in Patients with Coarctation of the Aorta. Circ. Cardiovasc. Genet. 2016, 9, 86–94. [Google Scholar] [CrossRef]
- Magouliotis, D.E.; Fergadi, M.P.; Christodoulidis, G.; Svokos, A.A.; Svokos, K.A.; Bareka, M.; Athanasiou, T. In-depth bioinformatic study of the cadherin 5 interactome in patients with thoracic aortic aneurysm unveils 8 novel biomarkers. Eur. J. Cardio-Thorac. Surg. 2021, 61, 11–18. [Google Scholar] [CrossRef]
- Akashi, M.; Higashi, T.; Masuda, S.; Komori, T.; Furuse, M. A coronary artery disease-associated gene product, JCAD/KIAA1462, is a novel component of endothelial cell-cell junctions. Biochem. Biophys. Res. Commun. 2011, 413, 224–229. [Google Scholar] [CrossRef]
- Hara, T.; Monguchi, T.; Iwamoto, N.; Akashi, M.; Mori, K.; Oshita, T.; Okano, M.; Toh, R.; Irino, Y.; Shinohara, M.; et al. Targeted Disruption of JCAD (Junctional Protein Associated with Coronary Artery Disease)/KIAA1462, a Coronary Artery Disease-Associated Gene Product, Inhibits Angiogenic Processes in Vitro and in Vivo. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1667–1673. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Xu, Y.; Liu, P.; Zhang, S.; Liu, H.; Slavin, S.; Kumar, S.; Koroleva, M.; Luo, J.; Wu, X.; et al. The novel coronary artery disease risk gene JCAD/KIAA1462 promotes endothelial dysfunction and atherosclerosis. Eur. Heart J. 2019, 40, 2398–2408. [Google Scholar] [CrossRef]
- Fan, J.; Li, X.; Zhong, L.; Hao-Tong; Di, J.; Liu, F.; Zhao, H.H.; Bai, S.L. MCP-1, ICAM-1 and VCAM-1 are present in early aneurysmal dilatation in experimental rats. Folia Histochem. Cytobiol. 2010, 48, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.D.; Kaiser, M.A.; Ghaderi Najafabadi, M.; Koplev, S.; Zhao, Y.; Douglas, G.; Kyriakou, T.; Andrews, S.; Rajmohan, R.; Watkins, H.; et al. JCAD, a Gene at the 10p11 Coronary Artery Disease Locus, Regulates Hippo Signaling in Endothelial Cells. Arterioscler. Thromb. Vasc. Biol. 2018, 38. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.J.; Ren, W.H.; Liu, X.J.; Liu, Y.; Wu, F.J.; Sun, L.Z.; Lan, F.; Du, J.; Zhang, H.J. Disruption of mechanical stress in extracellular matrix is related to Stanford type A aortic dissection through down-regulation of Yesassociated protein. Aging 2016, 8, 1923–1939. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Xu, J.; Guo, J.-L.; Lin, C.-Y.; Luo, W.-M.; Yuan, Y.; Liu, H.; Zhang, J. YAP1 up-regulation inhibits apoptosis of aortic dissection vascular smooth muscle cells. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4632–4639. [Google Scholar] [PubMed]
- Gao, J.; Wang, S.; Liu, S. The involvement of protein TNFSF18 in promoting p-STAT1 phosphorylation to induce coronary microcirculation disturbance in atherosclerotic mouse model. Drug Dev. Res. 2021, 82, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Blobe, G.C.; Schiemann, W.P.; Pepin, M.-C.; Beauchemin, M.; Moustakas, A.; Lodish, H.F.; O’Connor-McCourt, M.D. Functional Roles for the Cytoplasmic Domain of the Type III Transforming Growth Factor β Receptor in Regulating Transforming Growth Factor β Signaling. J. Biol. Chem. 2001, 276. [Google Scholar] [CrossRef]
- Villarreal, M.M.; Kim, S.K.; Barron, L.; Kodali, R.; Baardsnes, J.; Hinck, C.S.; Krzysiak, T.C.; Henen, M.A.; Pakhomova, O.; Mendoza, V.; et al. Binding Properties of the Transforming Growth Factor-β Coreceptor Betaglycan: Proposed Mechanism for Potentiation of Receptor Complex Assembly and Signaling. Biochemistry 2016, 55, 6880–6896. [Google Scholar] [CrossRef]
- Groeneveld, M.E.; Bogunovic, N.; Musters, R.J.P.; Tangelder, G.J.; Pals, G.; Wisselink, W.; Micha, D.; Yeung, K.K. Betaglycan (TGFBR3) up-regulation correlates with increased TGF-β signaling in Marfan patient fibroblasts in vitro. Cardiovasc. Pathol. 2018, 32, 44–49. [Google Scholar] [CrossRef]
- H3BV60 (H3BV60)—Protein—InterPro. Available online: http://www.ebi.ac.uk/interpro/protein/UniProt/H3BV60/ (accessed on 23 November 2021).
- Diestel, U.; Resch, M.; Meinhardt, K.; Weiler, S.; Hellmann, T.V.; Mueller, T.D.; Nickel, J.; Eichler, J.; Muller, Y.A. Identification of a Novel TGF-β-Binding Site in the Zona Pellucida C-terminal (ZP-C) Domain of TGF-β-Receptor-3 (TGFR-3). PLoS ONE 2013, 8, e67214. [Google Scholar] [CrossRef][Green Version]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef]
- Neptune, E.R.; Frischmeyer, P.A.; Arking, D.E.; Myers, L.; Bunton, T.E.; Gayraud, B.; Ramirez, F.; Sakai, L.Y.; Dietz, H.C. Dysregulation of TGF-β activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 2003, 33, 407–411. [Google Scholar] [CrossRef]
- Judge, D.P.; Biery, N.J.; Keene, D.R.; Geubtner, J.; Myers, L.; Huso, D.L.; Sakai, L.Y.; Dietz, H.C. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J. Clin. Investig. 2004, 114, 172–181. [Google Scholar] [CrossRef]
- Chaudhry, S.S.; Cain, S.A.; Morgan, A.; Dallas, S.L.; Shuttleworth, C.A.; Kielty, C.M. Fibrillin-1 regulates the bioavailability of TGFβ1. J. Cell Biol. 2007, 176, 355–367. [Google Scholar] [CrossRef]
- Huang, F.; Chen, Y.G. Regulation of TGF-β receptor activity. Cell Biosci. 2012, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dietz, H.C. Marfan syndrome: From molecules to medicines. Am. J. Hum. Genet. 2007, 81, 662–667. [Google Scholar] [CrossRef] [PubMed]
| Samples | Mode of Inheritance | Analysis_ID | Candidate Genes (n) |
|---|---|---|---|
| Fam1_all | Dominant | 1 | 7 |
| Recessive | 2 | 8 | |
| Fam 1.1 | Dominant | 3 | 37 |
| Recessive | 4 | 170 | |
| Fam 1.2 | Dominant | 5 | 14 |
| Recessive | 6 | 31 | |
| Fam 3 | Dominant | 7 | 49 |
| Recessive | 8 | 188 | |
| Fam 5 | Dominant | 9 | 5 |
| Recessive | 10 | 6 | |
| Severe-Not_Mild | Dominant | 11 | 12 |
| Recessive | 12 | 15 | |
| Mild-Not_Severe | Dominant | 13 | 95 |
| Recessive | 14 | 97 |
| Approach | Samples | Mode of Inheritance | Candidate Genes |
|---|---|---|---|
| Familial | Fam 1.1 | Dominant | PPARD |
| Recessive | FBN1 | ||
| Fam 3 | Dominant | MAP3K1 | |
| Recessive | JAG1 | ||
| Non- familial | Severe vs. Mild | Dominant | MAP3K1, PTPRJ |
| Recessive | PTPRJ | ||
| Mild vs. Severe | Dominant | KIAA1462, TNFSF18, TGFBR3L | |
| Recessive | TNFSF18, KIAA1462 |
| Candidate Genes | Expression in Aorta | Genomic Coordinates (GRCh37/hg19) | Variants | MAF in gnomAD | Variant Type | SIFT | ACMG Classification |
|---|---|---|---|---|---|---|---|
| PPARD | Yes | Chr6:35387913 | NM_001171818.2:c.140G>A | 4.003 × 10−5 | missense | tolerated-low confidence | Likely benign |
| FBN1 | Yes | Chr15:48808467 | NM_000138.5:c.1240C>A | N.R. | missense | tolerated | Likely pathogenic |
| JAG1 | Yes | Chr20:10620449 | NM_000214.3:c.3355G>C | N.R. | missense | tolerated | Likely pathogenic |
| Chr20:10620450 | NM_000214.3:c.3353T>A | N.R. | missense | tolerated | Uncertain significance | ||
| MAP3K1 | Yes | Chr5:56111414 | NM_005921.2:c.14C>G | 4.225 × 10−4 | missense | deleterious-low confidence | Uncertain significance |
| Chr5:56177614 | NM_005921.2:c.2587G>T | 3.621 × 10−5 | missense | tolerated-low confidence | Benign | ||
| Chr5:.56111762 | NM_005921.2:c.362G>A | N.R. | missense | tolerated | Uncertain significance | ||
| Chr5:56168548-56168649 | NM_005921.2:c.1504_1505+101del | N.R. | frameshift deletion | neutral | Pathogenic | ||
| PTPRJ | Yes | Chr11:48166437-48166511 | NM_002843.4:c.2786_2786+73del | N.R. | frameshift deletion | unknown/ unassessed | Likely pathogenic |
| Chr11:48002530-48002532 | NM_002843.4:c.66_68del | N.R. | in-frame deletion | damaging | Uncertain significance | ||
| Chr11:48161067G | NM_002843.4:c.2182G>A | 1 × 10−3 | missense | deleterious | Uncertain significance | ||
| KIAA1462 | Yes | Chr10:30316501-30316503 | NM_020848.4:c.2574_2576del | N.R. | frameshift deletion | unknown/ unassessed | Uncertain significance |
| Chr10:30318653 | NM_020848.4:c.424C>T | 1 × 10−3 | missense | tolerated | Likely benign | ||
| Chr10:30316499-30316500 | NM_020848.4:c.2577_2578insACTGCTGCT | in-frame insertion | unknown/ unassessed | Uncertain significance | |||
| TNFSF18 | No | Chr17:173010746 | NM_005092.4:c.361G>A | 4.788 × 10−5 | missense | tolerated | Uncertain significance |
| Chr17:173010656 | NM_005092.4:c.451A>G | 3.989 × 10−6 | missense | tolerated | Uncertain significance | ||
| Chr17:173010651-173010652 | NM_005092.4:c.455_456insTTG | N.R. | frameshift insertion | unknown/ unassessed | Uncertain significance | ||
| TGFBR3L | Yes | Chr19:7981648-7981650 | NM_001195259.2:c.418_420del | N.R. | in-frame deletion | unknown/ unassessed | Uncertain significance |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jimenez, Y.; Paulsen, C.; Turner, E.; Iturra, S.; Cuevas, O.; Lay-son, G.; Repetto, G.M.; Rojas, M.; Calderon, J.F. Exome Sequencing Identifies Genetic Variants Associated with Extreme Manifestations of the Cardiovascular Phenotype in Marfan Syndrome. Genes 2022, 13, 1027. https://doi.org/10.3390/genes13061027
Jimenez Y, Paulsen C, Turner E, Iturra S, Cuevas O, Lay-son G, Repetto GM, Rojas M, Calderon JF. Exome Sequencing Identifies Genetic Variants Associated with Extreme Manifestations of the Cardiovascular Phenotype in Marfan Syndrome. Genes. 2022; 13(6):1027. https://doi.org/10.3390/genes13061027
Chicago/Turabian StyleJimenez, Yanireth, Cesar Paulsen, Eduardo Turner, Sebastian Iturra, Oscar Cuevas, Guillermo Lay-son, Gabriela M. Repetto, Marcelo Rojas, and Juan F. Calderon. 2022. "Exome Sequencing Identifies Genetic Variants Associated with Extreme Manifestations of the Cardiovascular Phenotype in Marfan Syndrome" Genes 13, no. 6: 1027. https://doi.org/10.3390/genes13061027
APA StyleJimenez, Y., Paulsen, C., Turner, E., Iturra, S., Cuevas, O., Lay-son, G., Repetto, G. M., Rojas, M., & Calderon, J. F. (2022). Exome Sequencing Identifies Genetic Variants Associated with Extreme Manifestations of the Cardiovascular Phenotype in Marfan Syndrome. Genes, 13(6), 1027. https://doi.org/10.3390/genes13061027






