Abstract
N6-methyladenosine (m6A) RNA modification is a conserved mechanism to regulate gene expression that plays vital roles in the development of plants. However, the m6A RNA modification in forest trees remains limited. Here, we performed a complete analysis of m6A writers, erasers and readers in Poplar 84K, including gene location, gene structures, conserved motifs, phylogenetic relationships, promoter analysis, expression profiles and the homology modeling. We have identified 61 m6A pathway genes in Poplar 84K (Populus alba × Populus glandulosa), including 14 m6A writers, 14 m6A erasers and 33 m6A readers. Phylogenetic analysis indicated that the m6A writers and erasers were clustered into four groups and m6A readers were clustered into two groups. Promoter analysis showed that m6A pathway genes were mainly responsive to low oxygen followed by ABA and ethylene. The expression of the identified m6A pathway genes showed tissue-specific expression patterns in leaves, xylem, phloem and roots. Moreover, 17 genes were significantly up-regulated and 13 genes were significantly down-regulated in poplar overexpressing the transcription factor LBD15. Homology modeling and molecular docking results suggested that PagFIP37b was most likely to be regulated by LBD15, and the qPCRshowed that PagFIP37b were up-regulated in the LBD15-oe plants. The results provide insights that aid in the future elucidation of the functions of these m6A pathway genes and the epigenetic regulation mechanism of these genes in Poplar 84K.
1. Introduction
The RNA epigenetic modification plays a significant role in the regulation of gene expression [1,2]. Up to now, more than 100 modifications have been reported in RNA [3], and N6-methyladenosine (m6A) RNA methylation is the most abundant intermediate chemical modification of post-transcriptional gene regulation in eukaryotes. The m6A modification occurs at the sixth N atom of adenine, and the m6A accounts for up to 80% of RNA methylation modifications in eukaryotic cells and 50% of methylated modifications in mRNA [4]. m6A is widely studied in eukaryotes species, such as yeast, plants, flies, mammals and viral RNAs with a nuclear phase [5].
m6A modification affects almost every stage of mRNA metabolism, as it provides a binding site for effector proteins that regulate the stability, splicing and translation of mRNA [6,7]. Liu and Pan confirmed that the m6A could recognize RNAs all the time through identifying the m6A responsive RNA-binding protein [8]. The core proteins that participate in the m6A pathway are divided into three groups named writers (methyltransferases), erasers (demethylases) and readers [6,9]. In mammals, METTL3 (methyltransferase-like 3), METTL14 (methyltransferase-like 14) and WTAP serve as writers, FTO belongs to erasers, the readers included ALKBH5 and YTH (YTH domain family 2) [6,10,11]. In plants, the MTA (homologue of human METTL3), MTB (homologue of human METTL14), VIR, HAKA, and FIP (ortholog of human WTAP) were identified as plant m6A writers [5,7,12,13,14]. ALKBH9B and ALKBH10B proteins were considered as erasers to remove the methylation modification in the nucleus [5,9,10]. The reader proteins mainly included ECT2/3/4 and CPSF30 [6,9]. m6A has been examined in many species, including Arabidopsis [11], maize [12], wheat [13], oat [14], rice [15], sea buckthorn and apple [16,17]. Recently, the function of some m6A pathway genes in plants has been studied. The inactivation of MTA could reduce m6A modification and lead to a failure of embryonic development and reduced apical dominance [11,18]. Shoot meristems require FIP37 to be maintained and are continuously produced in Arabidopsis [19]. ECT2/3 are necessary to regulate the formation and timing of leaf morphogenesis [6]. These studies indicated that the m6A modifications in mRNA play a crucial role in plant development.
m6A RNA methylation is a conserved mechanism to enrich and control gene expression and plays an important role in organisms. The distribution pattern of methylation sites and the consensus sequence of m6A seem to be conserved in human and yeast; both the modification sites are enriched at the 3′untranslated regions (3′UTRs). In addition, m6A in human is also enriched around stop codons and within internal long exons [5]. However, plants may have evolved with unique mechanisms in m6A methylation machinery. In plants, it has been shown that m6A is usually enriched around the stop codon, 3′UTRs, long exons, TSS (the transcription start site) and TES (transcription end site) [16,20,21,22]. The writers in Arabidopsis could recognize the consensus motif RRACH [23]. However, not all RRACH motifs in plants are associated with m6A modification, and the molecular mechanism of recognition is also undefined [8].
The m6A level was affected by the expression of m6A pathway genes. It has been shown that the m6A levels were reduced through down-regulation of the m6A pathway genes [23]. However, knowledge of RNA modification in forest trees remains limited. Poplar 84K has become a model plant for forest tree studies, since it is a fast-growing poplar with an available whole genome sequence, high transformability and economic and ecological value. In our previous studies, we have reported that an ortholog gene of AtLBD15 from Eucalyptus grandis takes part in the leaf development in Poplar 84K [24] and analyzed the differential m6A modification sites between Poplar 84K and the LBD15 overexpression plants [22]. In order to further elucidation of the molecular mechanism of m6A modification associated with LBD15, here, we systematically identified m6A writers, erasers and readers genome-wide in Poplar 84K. The gene structures, gene location, conserved motifs and phylogenetic relationships were identified, and the homology modeling of these proteins and LBD15 were analyzed. In addition, the tissue-specific expression profiles and the expression of these genes in Poplar 84K and the LBD15 overexpression plants were also investigated. The results provide insight to further elucidate the functions of the genes of m6A pathway and the epigenetic regulation mechanism of m6A in Poplar 84K.
2. Materials and Methods
2.1. Plant Materials
Poplar 84K was planted in the greenhouse located at the Chinese Academy Forestry, Beijing, China. Leaves, xylem, phloem and roots were sampled from 6-month-old plants and stored in liquid nitrogen for RNA isolation. The shoots cultivated for 6 weeks were collected from Poplar 84K and LBD15 overexpression plants and stored in liquid nitrogen until use. Three independent biological replicates were performed from three whole plants for each sample.
2.2. Identification of m6A Pathway Genes in Poplar 84K
All the m6A pathway genes from Arabidopsis and Oryza sativa protein sequences were downloaded from GenBank (Table S3) and used as a query to search for homologous genes in Poplar 84K genome through tBLASTn with an e-value of 1 × 10−5 [25]. The conserved domains of the candidate genes were analyzed in NCBI-CDD [26]. The molecular weight (Mw) and isoelectric point (pI) of these proteins were investigated using the ExPASy online software [27]. Their subcellular localization was predicted based on the Busca online software [28].
2.3. Phylogenetic Tree, Gene Structure, Conserved Motifs, Promoter Prediction and Protein Interaction Analysis
The chromosomal location and gene structure of these m6A pathway genes were obtained from the genome annotation files, which were downloaded from (https://www.ncbi.nlm.nih.gov/genome/87686, accessed on 1 June 2022). The chromosome physical location of those genes was displayed by MapGene2Chromosome v2.0 [29], and the gene structure was shown using a Gene Structure Display Server [30]. The MEME tool was used to predict the conserved motif of the candidate protein sequences [31]. The maximum motif number was set to 5, and other parameters were left on the default settings. The phylogenetic trees and dN/dS analyzed were constructed by MEGA7.0 software [32] using the muscle and neighbor-joining (NJ) method with the bootstrap value of 1000 [33]. The upstream 2000 base pair (bp) genomic DNA sequences from the transcription start sites of m6A pathway genes were predicted in PlantCare database [34] to identify the putative cis-regulatory elements.
2.4. Quantitative Real-Time Reverse Transcription-PCR (qRT-PCR)
Total RNA of leaves, roots, xylem and phloem from Poplar 84K and LBD15-oe plants were isolated by RNA Easy Fast Plant Tissue Kit (TIANGEN, Beijing, China). The cDNA was generated by reverse transcription using FastKing RT Kit (TIANGEN, Beijing, China). The qRT-PCR analysis of m6A pathway genes were performed with SYBR® rapid quantitative PCR Kit (KAPA KK4601, Pleasanton, CA, USA) using the methods described previously [35]. Pagactin was used as a reference [24]. The primers of all the genes were listed in Table S5, and the results were analyzed using the 2−ΔΔCt method [24].
2.5. Homology Modeling of 3D Structures, Molecular Docking and Protein Docking
The 3D structures of m6A pathway proteins are important for investigating their gene function. The 3D structure of m6A pathway proteins and LBD15 were predicted using the homology modeling method. The PDB database [36] was used to find the best template, and the model was built by Swiss-Model [37]. The predicted cis-elements of m6A pathway proteins that can interact with LBD15 were performed by JASPAR [38]. The highest scoring cis-element was used to dock with LBD15 through Autodock vina [39]. The protein docking of PagMT families was performed using ZDOCK [40], and PDBePISA [41] was used to analyze the docking results. Pymol software was used for evaluation of quality, and the equation linking affinity (Ka) and ligand free energy of binding ΔGwere calculated by formula:ΔG = −RTln (KA/C) (T is temperature in kelvin, C is 1 M concentration and R = 8.314 J/mol/K) [42].
2.6. Statistical Methods
In this study, one-way ANOVA was used to perform the statistical analysis for the differential tissues expression of m6A pathway genes through IBM SPSS 19 software. We ranked all averages from highest to lowest and marked the letter a after the highest average; then, the mean was compared with the following means, and any mean that was not significantly different was marked with the letter a, while any mean that was significantly different was marked with the letter b, and so on until the smallest average has a marked letter and stops. Moreover, the statistical analysis of the expression between Poplar 84K and LBD15-oe plants were calculated by T-test was using IBM SPSS 19 software. p < 0.05 (*) was considered statistically significant, while p < 0.01 (**) was considered extremely significant.
3. Results
3.1. Genome-Wide Identification of m6A Pathway Genes in Poplar 84K
The protein sequences involved in the m6A pathway in plants including Arabidopsis and O.sativa were downloaded from GenBank. tBLASTn analysis of these protein sequences against the genome of Poplar 84K were performed to identify putative m6A pathway genes. As a result, a total of 61 m6A pathway-like genes were identified in Poplar 84K, including 14 m6A erasers (14 PagALKBHs), 14 m6A writers (8 PagMTs, 2 PagFIP37s, 2 PagVIRILIZERs and 2 PagHAKAIs), 33 m6A readers (28 PagECTs and 5 PagCPSF30s). Analysis of gene feature showed that the length of the ORF (open-reading frame) varied from 1783 to 7182 bp of m6A erasers, 1285 to 17,347 bp of m6A writers, and 3379 to 16,254 bp of m6A readers, respectively. The amino acid length varied from 240 to 770 aa for m6A erasers, 288 to 2179 aa for m6A writers, and 398 to 975 aa for m6A readers, respectively. The average molecular weight (Mw) is 44.41, 82.22 and 69.89 kDa, respectively, and the theoretical pI of m6A pathway genes ranged from 4.62 to 9.22. The prediction of subcellular localization displayed that 59 genes were located in the nucleus, and another 2 genes were located in cytoplasm (Table 1).

Table 1.
Sequences feature of m6A pathway genes in Poplar 84K.
3.2. Phylogenetic Analysis of m6A Pathway Genes in Poplar 84K
To investigate the phylogenetic relationship of m6A pathway genes in Poplar 84K, the phylogenetic tree was constructed using the protein sequence of the N6-methyladenosine writers, erasers and readers in Poplar 84K with the corresponding genes in Arabidopsis and O. sativa, respectively. The results showed that the writers in Poplar 84K were clustered into four groups, including PagMT, PagFIP37, PagVIRILIZER and PagHAKAI families (Figure 1A). The PagMT group was divided into three subgroups: PagMTA, PagMTB and PagMTC. The erasers in Poplar 84K only contained the PagALKBH family, which was clustered into four groups (Figure 1B). The erasers in Poplar 84K had a distribution in each group. The phylogenetic analysis showed that the readers in Poplar 84K were classified into two groups, PagECT and PagCPSF30 (Figure 1C). PagECT family contained a conserved YTHDF domain, whereas the PagCPSF30 family contained a conserved YTHDC domain. This is consistent with previous studies from other plants, suggesting that the readers have a conserved role in the plants.

Figure 1.
Phylogenetic trees of m6A pathway genes from Poplar 84K, A. thaliana and O. sativa (A) Writers; (B) Erasers; (C) Readers. The phylogenetic trees were constructed using MEGA 7.0 by the neighbor-joining (NJ) method with 1000 bootstrap replicates. The groups of m6A pathway genes from Poplar 84K are shown in different colors.
The dN/dS value of the m6A pathway genes were calculated by the MEGA7.0 software to detect molecular selection effects (Table S1). As a result, the m6A pathway genes except for PagCPSF30s had a dN/dS < 1, indicating that they had undergone strong purifying selection. There were both dN/dS > 1 and dN/dS < 1 in the PagCPSF30 family, suggesting it had undergone purifying selection and position section. The results suggested that the m6A pathway genes were highly conserved during the evolutionary process with the exception of PagCPSF30s.
3.3. Analysis of Chromosomal Location
To further understand the evolutionary relationship of the 61 m6A pathway genes, the chromosomal location of these genes was determined. The result showed that these genes were distributed on 31 chromosomes (Figure 2). Among them, chromA01 and chromG01 contain the largest number of m6A pathway genes; PagCPSF30b, PagCPSF30c, PagMTA1, PagECT1 and PagECT19 were located on chromA01. Additionally, PagCPSF30d, PagCPSF30e, PagMTA2, PagECT5 and PagECT24 were also located on chromG01. ChromA14 and chromG14 contained four m6A pathway genes, respectively. PagALKBH6Bb, PagALKBH7Ba, PagMTA3 and PagECT21 were located on chromA14, and PagALKBH7Bb, PagALKBH6Ba, PagMTA4 and PagECT18 were located on chromG14. The result suggested that these genes were generated by segmental duplication, which was the single driving force in the evolutionary process of m6A pathway genes in Poplar 84K.

Figure 2.
Genes location on chromosomes. The chromosomes in genome A are shown in red, and the chromosomes in genome G are shown in blue.
3.4. Gene Structure and Conserved Motif Analysis
The exon–intron pattern is an important feature for a gene and can provide important evidence for gene functional diversification, so the exon–intron patterns of m6A pathway genes were determined (Figure 3). The intron numbers in writers varied from 2 to 27. For erasers, the intron number varied from 3 to 6. The intron number of the readers clustered in the PagECT group varied from 5 to 11, and those clustered in PagCPSF30 group varied from 5 to 7. The PagCPSF30 gene family has the largest intron size. The results showed that the genes clustered into the same clade in the phylogenetic tree had similar exon–intron patterns, indicating the conservation of these genes during evolution.

Figure 3.
The gene structure and conserved motifs of m6A pathway genes. (A) Exons, UTRs, introns and intron phases are shown. (B) Motifs are represented by boxes.
The conserved motif analysis results showed that a total of 15 motifs were identified in the m6A pathway genes in Poplar 84K and the genes clustered with the same group had similar motifs, which is consistent with the results of the gene structure and the phylogenetic tree. We found that some motifs were highly conserved in the m6A pathway genes in Poplar 84K; for example, the motif 1 was found in all the erasers, motif 7, motif 9 and motif 10 were present in all the readers and motif 15 was present in all writers.
3.5. Promoter Analysis of m6A Pathway Genes
The cis-elements of these genes are shown in Figure 4 and Table S2. CAAT-box and TATA-box were found in all the m6A pathway genes. The main cis-acting elements were predicted to respond to abiotic stress, hormones and inducers such as methyl jasmonate (MeJA), light, anaerobic, ethylene, salicylic acid (SA), drought, low temperature, abscisic acid (ABA), gibberellin and auxin. The largest number of cis-elements were light-response elements, and they were found in all the m6A pathway genes. For erasers, 13 genes had ABA response elements, while 12 genes had anaerobic response and ethylene response elements. In readers, 33 genes have anaerobic response elements. In writers, anaerobic response elements were found in all the members. The results showed that m6A pathway genes were mainly responsive to low oxygen followed by ABA and ethylene.
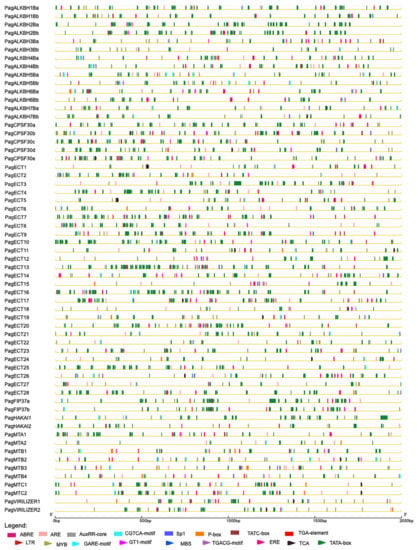
Figure 4.
The cis-regulatory elements in the promoter of 61 m6A pathway genes. The cis-regulatory elements were represented bytriangle and rectangle in different colors.
3.6. Expression Profiles of m6A Pathway Genes
The expression of m6A pathway genes in differential tissues was investigated using qPCR (Figure 5). The results showed that the expression of these genes displayed differential expression patterns. All the writer genes were highly expressed in leaves, especially, the PagFIP37 genes which had the highest expression in leaves. The readers that clustered into the same subgroup displayed similar expression patterns; for instance, PagECT1, PagECT3, PagECT4 and PagECT5 were mainly expressed in roots, while PagECT20, PagECT22, PagECT25 and PagECT27 showed the highest expression levels in leaves. PagECT19 had the highest expression level in the phloem. As for erasers, the different erasers showed divergent expression patterns, such as PagALKBH4s and PagALKBH5s, which were mainly expressed in phloem, and PagALKBH3s showed significantly high expression in leaves.

Figure 5.
qPCR validation of 61 m6A pathway genes in tissues. Figure (A)–(BI) showed the expression level of 61 genes in differential tissues. One-way ANOVA was calculated using IBM SPSS 19 software. The a, b, c and d indicated whether the difference was significant. The same letter marked in the same gene among different tissues indicated no significant difference, and different letters indicated significant difference.
The expression of the m6A pathway genes in Poplar 84K and LBD15-oe plants was also investigated by qPCR (Figure 6). The results showed that the expression of the m6A pathway genes was regulated in the LBD15-oe plants. There were 17 genes that were significantly up-regulated; among them, the expressions of PagALKBH4Ba, PagECT7 and PagECT13 were up-regulated more than 2-fold, while PagHAKAI2, PagFIP37b, PagALKBH2Ba, PagECT2, PagECT9, PagECT15, PagECT16, PagECT23, PagECT28 and PagCPSF30a were up-regulated more than 1.5-fold in the LBD15-oe plants. Conversely, 13 genes were significantly down-regulated; for example, PagECT12 and PagALKBH3Bb were down-regulated more than 2-fold in the LBD15-oe plants. The results implied that the regulatory role of m6A modification may be associated with the expression of LBD15.

Figure 6.
qPCR validation of 61 m6A pathway genes in Poplar 84K and LBD15-oe plants. Figure (A)–(BI) showed the expression level of 61 genes in Poplar 84K and LBD15-oe plants. A t-test was calculated using IBM SPSS 19 software. * represent p < 0.05 and ** represent p < 0.01.
3.7. Homology Modeling, Molecular Docking and Protein Docking
The protein model of the m6A pathway genes and LBD15 were built by homology modeling, as shown in Figure 7 and Figure 8A. The template and RMSD are shown in Table S3. The RMSD (root-mean-square deviation) is less than 1Å with respect to the templates, suggesting that the homology modeling was reliable. As a result, we obtained the 3D structure of a total of 56 proteins of the m6A pathway genes; five other proteins were not modeled, as there was no homologous template to build in the database. In the 3D structures, the α-helix, β-fold and random coil were signed in different colors. All the structures of readers contained five α-helixes and four adjacent β-folds and formed highly similar spatial structures. The 3D structures of erasers varied among groups based on the metal ions; for example, PagALKBH1Ba contained one Mn ion, while PagALKBH1Bb contained one Mn ion and one Zn ion in the structure, and PagALKBH7Bs and PagALKBH3Bs contained one Fe ion. For the writers, PagMTCs displayed as dimers and contained two centrosymmetric monomeric proteins. The proteins that clustered into the same group had highly similar 3D structures, indicating that these genes were highly conserved in the plants.

Figure 7.
The homology modeling of m6A pathway genes in Poplar 84K. The α-helix, β-fold and random coil are shown, and the ball representsmetal ions.

Figure 8.
(A) The homology modeling of LBD15. (B) The structure of chain A in LBD15.
The LBD15 protein contains a typical LOB domain displayed as dimers, which contained two centrosymmetric monomeric chains (chain A and chain B) (Figure 8A), and the monomeric chain consisted of five α-helixes (Figure 8B). The most reliable cis-elementin promoter of the m6A pathway genes that up-regulated more than 1.5-fold in the LBD15-oe plants were predicted, and the molecular docking of these cis-elements with LBD15 was performed. The results of cis-elements docking with LBD15 are shown in Table 2 and Figure S1. Among those 13 genes, PagFIP37b could be the most reliable to interact with LBD15 according to the Affinity.

Table 2.
Amino acid residues participate in protein–ligand docking between LBD15 and predicted cis-elements in the promoters of m6A pathway genes.
The protein docking of PagMTs was performed. The results of protein docking are shown in Table 3; the docking of PagMTA1-PagMTC1 and PagMTA2-PagMTB1 are the most credible based on the score of ΔiG (kcal/M) and p-value. The results revealed that PagMTA-PagMTB, PagMTA-PagMTC, and PagMTC-PagMTB could bind and formed a dimer, which could help us understand how these writers work.

Table 3.
Protein–protein docking of PagMTAs, PagMTBs and PagMTCs. NHB: number of potential hydrogen bonds. NSB: number of potential salt bridges.
4. Discussion
m6A RNA methylation is the most abundant intermediate chemical modification of post-transcriptional gene regulation in eukaryotes. It is indispensable for plant growth and development through participating in the mRNA splicing, stability and translation [6,7]. In our previous study, the regulation of the m6A modification between Poplar 84K and the LBD15 overexpression plants was analyzed [22]. In order to increase our understanding of the mechanism of m6A modification and the epigenetic regulation of LBD15, here, we systematically at a genome level identified 61 m6A pathway genes in poplar 84K, including 14 m6A erasers, 14 m6A writers and 33 m6A readers. The gene structures, gene location, conserved motifs and phylogenetic tree were performed; the tissue-specific expression profiles and the expression of these genes in Poplar 84K and the LBD15 overexpression plants were investigated. In addition, the 3D structures and protein docking of the identified proteins were also analyzed. Our results provide insight into understanding the roles of these m6A pathway genes and the epigenetic regulation mechanism of these genes in Poplar 84K.
In this study, in total, we identified 61 m6A writers, erasers and readers in Poplar 84K; genome-wide, the number of the m6A pathway genes was significantly greater than the number found in O.sativa and Arabidopsis, which both contain 28 genes. This may be partially explained by the fact that Poplar 84K is a hybrid of P.alba and P.glandulosa. It has two subgenomes, which have different gene numbers based on the chromosomal localization analysis. Based on the phylogenetic tree, the 14 m6A writers in Poplar 84K were clustered into four groups, implying that the four types of writers have different functions. Among them, the PagMT group was the largest one, which contained eight members. This is distinct from its counterparts in Arabidopsis, suggesting that the PagMT group proteins potentially have more functions in Poplar 84K. The eraser is a demethylase which could remove the m6A modification in the nucleus. In Arabidopsis, the functions of ALKBH9B and ALKBH10B are well studied [5,9,10]. Six ALKBH proteins in Poplar 84K were clustered into the same clades with AtALKBH9B and AtALKBH10B, including PagALKBH2Ba, PagALKBH2Bb, PagALKBH4Ba, PagALKBH4Bb, PagALKBH6Ba and PagALKBH2Bb, indicating they may have a role similar to m6A demethylation. The 33 readers were classified into two groups according to the YTHDC domain and YTHDF domain; the result is consistent with other plants [25], suggesting that the readers are highly conserved in plants.
The m6A modification controls gene expression through its writers, readers and erasers, so the expression of the writers, readers and erasers is very important for investigating their potential functions. In this work, the expression of m6A pathway genes in four tissues including leaves, roots, xylem, and phloem was analyzed. The results revealed that all the identified genes were detected in the leaves, roots, xylem, and phloem of Poplar 84K, but they displayed differential expression profiles. The genes clustered into the same clade showed a similar expression pattern, suggesting they may play similar roles in the plant growth and development. Our previous study has reported that an ortholog of AtLBD15 is involved in the development of leaves [24]. We found that the level of m6A modification in LBD15 overexpression plants was altered. In order to further understand the mechanism of m6A modification in LBD15 overexpression plants, we detected the specific expression of m6A pathway genes in CK and LBD15-oe plants. As a result, we found that the expressions of the m6A pathway genes were regulated in the LBD15-oe plants. For example, some readers including PagECT7, PagECT13, PagECT15, PagECT16, PagECT2, PagECT9, PagECT23, PagECT28 and PagCPSF30a were up-regulated more than 1.5-fold in LBD15-oe plants. Moreover, these genes were mainly expressed in leaves, suggesting these genes are probably involved in the leaf development. It has been shown that proteins ECT2/3/4 are essential for leaf formation in Arabidopsis [8], which is consistent with our results. On the contrary, PagHAKAI2 was significantly down-regulated in the LBD15-oe plants; this is consistent with our previous work, which showed that 4260 down-regulated m6A peaks were detected in LBD15-oe plants [22]. Taken together, the level of m6A in the LBD15-oe plants was affected by LBD15 through regulating the expression of m6A pathway genes.
The m6A in the plants is a complex process; the functions of the m6A writers, erasers and readers in plants are still unclear [8]. The homology model and molecular docking could help us better understand the 3D structure of proteins and drive our future research. The results of molecular docking between the cis-element of m6A pathway genes and LBD15 showed that PagFIP37b could be regulated by LBD15, which will provide a direction for studying the epigenetic regulation mechanism associated with LBD15 in Poplar 84K. In Arabidopsis, it has been shown that MTA and MTB could interact with each other and form homodimers [43]. In this study, the protein–protein docking of PagMTs revealed that PagMTA-PagMTB, PagMTA-PagMTC, and PagMTC-PagMTB could bind and form a dimer. The ΔiG < 0 indicated that the docking was reliable [41]. The p-value < 0.5, implying the interface surface can be interaction-specific. The results showed that two PagMTs could form a dimer, which is consistent with Arabidopsis, suggesting this prediction was reliable. These results will provide valuable information for further study of the functions of the m6A pathway genes in the Poplar 84K.
5. Conclusions
In this study, a thorough and systematic approach, combining phylogenetic analysis, chromosomal localization, gene structure, conserved motif and promoter analysis as well as expression and 3D structures, was performed to help characterize the 61 m6A pathway genes identified in the genome of Poplar 84K. The results revealed that the m6A pathway genes in Poplar 84K were highly evolutionary conserved. The expression of the identified m6A pathway genes showed tissue-specific expression patterns in leaves, xylem, phloem and roots. These genes were regulated in the LBD15-oe plants. The results of 3D structures docking showed that PagFIP37b could be the most reliable to regulate by LBD15 and two PagMTs could form a dimer, which could help us understand how writers work. Our results provide some insight into the functions of these m6A pathway genes and the epigenetic regulation mechanism of these genes in Poplar 84K.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13061018/s1. Table S1: dn/ds analysis of writers, readers and erasers; Table S2: The cis-regulatory elements in the promoter of m6A pathway genes; Table S3: Accession numbers and amino acid sequences of m6A pathway genes from Arabidopsis thaliana and O.sativa; Table S4: The homology templates and docking score (RMSD) in protein homology modeling; Table S5: The primers used in qPCR; Figure S1: The results of molecular docking between cis-elements of m6A pathway genes and LBD15. The amino acid residues involved in docking are displayed in sticks and signed a label.
Author Contributions
Conceived and designed the experiments: F.S. and D.Q. Performed the experiments: X.S. and W.W. Analyzed the data: X.S., W.W., Y.Y. and I.W. Wrote the paper: F.S., Y.Y., I.W. and D.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Natural Science Foundation of China (grant numbers 32171809 to D.Q.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data in this study are available in the Supplementary Materials.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Czerwoniec, A.; Dunin-Horkawicz, S.; Purta, E.; Kaminska, K.H.; Kasprzak, J.M.; Bujnicki, J.M.; Grosjean, H.; Rother, K. MO-DOMICS: A database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009, 37, 118–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kierzek, E.; Kierzek, R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003, 31, 4472–4480. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Bressendorff, S.; Hansen, M.H.; Poulsen, C.; Erdmann, S.; Brodersen, P. An m6A-YTH Module Controls Developmental Timing and Morphogenesis in Arabidopsis. Plant Cell 2018, 30, 952–967. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.D. m6A-mediated translation regulation. Bbagrm 2018, 1862, 301–309. [Google Scholar] [CrossRef]
- Liu, N.; Pan, T. N6-methyladenosine–encoded epitranscriptomics. Nat. Struct. Mol. Biol. 2016, 23, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Pérez, M.; Aparicio, F.; López-Gresa, M.P.; Bellés, J.M.; Sánchez-Navarro, J.A.; Pallás, V. Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.-C.; Wei, L.-H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B is An RNA N6-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, S.; Li, H.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef] [Green Version]
- Nichols, J.L. ‘Cap’ structures in maize poly(A)-containing RNA. Biochim. Biophys. Acta 1979, 563, 490–495. [Google Scholar] [CrossRef]
- Kennedy, T.D.; Lane, B.G. Wheat embryo ribonucleates. XIII. Methyl-substituted nucleoside constituents and 5ʹ-terminal dinucleotide sequences in bulk poly (AR)-rich RNA from imbibing wheat embryos. Can. J. Biochem. Cell Biol. 1979, 57, 927–931. [Google Scholar]
- Haugland, R.A.; Cline, M.G. Post-transcriptional modifications of oat coleoptile ribonucleic acids: 5ʹ-terminal capping and methylation of internal nucleosides in poly(A)-rich RNA. Eur. J. Biochem. 1980, 104, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Li, C.; Hu, S.; Yu, J.; Song, S. Transcriptome-wide N6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol. 2014, 11, 1180–1188. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Lv, Z.; Diao, S.; Liu, H.; Duan, A.; He, C.; Zhang, J. Unique features of the m6A methylome and its response to drought stress in sea buckthorn (Hippophae rhamnoides Linn.). RNA Biol. 2021, 18, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Hou, N.; Liu, Z.; He, J. Profiling of N6-Methyladenosine (m6A) Modification Landscape in Response to Drought Stress in Apple (Malus prunifolia (Willd.) Borkh). Plants 2021, 11, 103. [Google Scholar] [CrossRef]
- Bodi, Z.; Zhong, S.; Mehra, S.; Song, J.; Graham, N.; Li, H.; May, S.; Fray, R.G. Adenosine methylation in Arabidopsis mRNA is associated with the 3’ end and reduced levels cause developmental defects. Front. Plant Sci. 2012, 3, 48. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.N.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Tang, K.; Zhang, D.; Xie, S.; Zhu, X.; Wang, Z.; Lang, Z. Transcriptome-wide high-through put deep m6A-seq reveals unique differential m6A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 2015, 16, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Shao, F.J.; Sun, X.C.; Wu, W.L.; Lu, Q.; Wilson, I.W.; Qiu, D.Y. A comparative analysis of differential N6-methyladenosine (m6A) modification between non-transgenic and LBD15 overexpressing Poplar 84K plants. Tree Genet. Genomes 2021, 17, 39. [Google Scholar] [CrossRef]
- Růžička, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S.; et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017, 215, 157–172. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Shao, F.J.; Macmillan, C.; Wilson, I.W.; Merwe, K.; Hussey, S.G.; Myburg, A.A.; Dong, X.; Qiu, D. Genome-wide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J. 2018, 16, 124–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein detabase search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, 265–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Jiangtao, C.; Yingzhen, K.; Qian, W.; Yuhe, S.; Daping, G.; Jing, L.; Guanshan, L. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan 2015, 37, 91–97. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene’ feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Li, Y.Y.; Yang, Y.F.; Wang, S.; Liu, H.; Qiu, D.Y. Cloning and Expression of Lateral Organ Boundaries Domain Genes (TcLBDs) in Taxus Chinensis. Sci. Silvae Sin. 2015, 51, 126–133. [Google Scholar]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef] [Green Version]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Pérez, N.M.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, 165–173. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK Server: Interactive Docking Prediction of Protein-Protein Complexes and Symmetric Multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. ‘Protein interfaces, surfaces and assemblies’ service PISA at the European Bioinformatics Institute. (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html) ‘Inference of macromolecular assemblies from crystalline state’. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Schiffrin, B.; Radford, S.E.; Brockwell, D.J.; Calabrese, A.N. PyXlinkViewer: A flexible tool for visualization of protein chemical crosslinking data within the PyMOL molecular graphics system. Protein Sci. 2020, 29, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.S.; Bielewicz, D.; Jarmolowski, A.; Szweykowska-Kulinska, Z. N6-methyladenosine (m6A): Revisiting the Old with Focus on New, an Arabidopsis thaliana Centered Review. Genes 2018, 30, 596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).