Abstract
Triple quadrupole mass spectrometry coupled to liquid chromatography (LC-TQ-MS) can detect and quantify modified nucleosides present in various types of RNA, and is being used increasingly in epitranscriptomics. However, due to the low resolution of TQ-MS and the structural complexity of the many naturally modified nucleosides identified to date (>160), the discrimination of isomers and mass-analogs can be problematic and is often overlooked. This study analyzes 17 nucleoside standards by LC-TQ-MS with separation on three different analytical columns and discusses, with examples, three major causes of analyte misidentification: structural isomers, mass-analogs, and isotopic crosstalk. It is hoped that this overview and practical examples will help to strengthen the accuracy of the identification of modified nucleosides by LC-TQ-MS.
1. Introduction
Research into post-transcriptional RNA modification is increasingly focusing on its critical impacts on RNA decay, translational efficiency, subcellular localization, quality control, RNA–protein interactions, and disease development [1,2,3]. Intensely mined information from the epitranscriptome reveals that dynamic modification of RNA is a response to physiological and environmental changes, although the biological consequences of such modifications are still being elucidated [3,4,5]. Methylation of key transcripts, for example, N6-methyladenosine (m6A), has been observed in budding yeast and was crucial to the initiation of meiosis during nitrogen starvation [6]. YTHDF1, an m6A reader protein that enhances translational efficiency by recruiting eukaryotic initiation factor 3, was found to be concentrated on stress granules and triggered stalled translation when arsenite was added to induce oxidative stress in HeLa cells [7]. YTHDF2 (an m6A reader) and FTO (an m6A eraser) competitively bind to the 5′-UTR of messenger (m)RNA and regulate m6A methylation in a mouse embryonic fibroblast (MEF) cell line, which facilitates cap-independent translation of specific transcripts under stress conditions [8]. On transfer RNAs (tRNAs), modifications at positions 34 (the wobble position), 37, and 58 can be dynamic and related to environmental factors. When taurine supply was limited, τm5U34 (5-taurinomethyluridine) and τm5s2U34 (5-taurinomethyl-2-thiouridine) on five mitochondrial transfer (mt)RNAs switched to cmnm5U34 (5-carboxymethylaminomethyluridine) and cmnm5s2U (5-carboxymethylaminomethyl-2-thiouridine) with unknown consequences [9]. Bicarbonate-free air culturing of HEK293T cells triggered the downregulation of t6A37 (N6-threonylcarbamoyladenosine) on mt-tRNAs decoding ANN codons and probably impaired the translation of mitochondrial respiratory complex I [10]. m1A58 (1-methyladenosine) in human cytoplasmic tRNAs was influenced by the glucose concentration of the culture medium via an FTO (m1A eraser)-dependent pathway that adjusted the decoding preference [11].
To determine the identity, quantity, and location of RNA modifications, RNA sequencing (NGS or NNGS approaches) [12,13,14], oligonucleotide mass spectrometry [15,16], and nucleoside mass spectrometry [17] are frequently used. Nuclear magnetic resonance spectroscopy has also recently been used to observe the dynamic incorporation of RNA modifications into nascent tRNA [18,19]. A generic protocol to qualitatively and quantitatively analyze modified nucleosides has been developed using high-performance liquid chromatography coupled to triple quadrupole mass spectrometry (LC-TQ-MS) [20]. Total RNA or a purified fraction is hydrolyzed and dephosphorylated to the free nucleosides using alkaline phosphatase, phosphodiesterase I, and nuclease P1. Two-step digestion is recommended as basic pH is suitable for alkaline phosphatase, while phosphodiesterase I and nuclease P1 are more effective in acidic conditions. However, some studies recommend an acidic environment for the complete digestion because certain RNA modifications are sensitive to pH. For instance, ct6A (cyclic N6-threonylcarbamoyladenosine) quickly epimerizes to t6A in mild alkaline buffer, causing an 18 Da mass increase [21]. The enzymes are then removed from the digestion mixture using a 10 kDa cut-off centrifugal filter unit, and the resulting nucleosides are vacuum-dried and dissolved in an appropriate solvent, usually 90% (v/v) acetonitrile or water, depending on the downstream liquid chromatography. Hydrophilic interaction liquid chromatography (HILIC) [22] and reversed-phase chromatography (RPC) [23] with semi-micro flow rates (0.1–1 mL/min) have been used to separate nucleosides, while micro-flow HPLC (5–50 μL/min) has recently been recommended for higher sensitivity without loss of reproducibility [24].
Multiple reaction monitoring (MRM) mode is used with TQ-MS when determining modified nucleosides. Collision-induced dissociation of the N-β-glycosidic bond under the appropriate collision energy (CE) yields the nucleobase production, BH2+. Ideally, synthetic standards of the modified nucleosides should be used to confirm optimum CE, ion mass–to–charge ratios (m/z), specific product ions and retention times, but their preparation is expensive, time consuming and labor intensive. The m/z values for precursor (MH+) and product (BH2+) ions can be readily calculated from their chemical structures (Figure 1). CEs for detecting modified nucleosides can be estimated using native (unmodified) adenosine (A), uridine (U), cytidine (C), and guanosine (G) standards. This may not yield exact values for the modified nucleosides but is a practical compromise. Some published LC-TQ-MS applications for RNA modifications use fewer than 20 modified nucleoside standards and determine other nucleosides using calculated MRM transitions and estimated CE values [20,25].
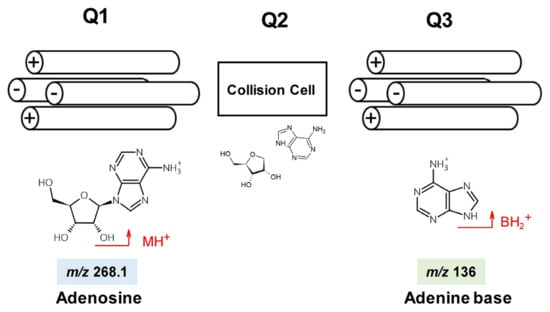
Figure 1.
Precursor (MH+) and product (BH2+) ions of adenosine selected by quadrupoles Q1 and Q3.
Over 160 natural RNA modifications have been identified to date [26]. However, this complexity and the low resolution of TQ-MS (approximately 0.5 Da) can give rise to three types of misidentification when using MH+ and BH2+ MRM transitions to determine nucleosides. Type I mistakes result from regioisomers. For instance, five natural monomethylated adenosines have been identified. These are m1A, m2A (2-methyladenosine), m6A, m8A (8-methyladenosine), and Am (2′-O-methyladenosine) (Figure 2), and have been found variously in mRNA, tRNA, and ribosomal (r)RNA [26]. Am can be discriminated by monitoring the transition m/z 282.1→136 because of its unmodified nucleobase, while m1A, m2A, m6A, and m8A all give rise to the transition m/z 282.1→150 (Figure 2). High-resolution mass spectrometry can discriminate between these four monomethylated derivatives via in-source, collision-induced dissociation (CID) and negative mode ionization that produces unique patterns of fragments [27]. However, this approach requires a library of MS2 and MS3 spectra for each isomer and sensitivity can be reduced for quantitative analysis.
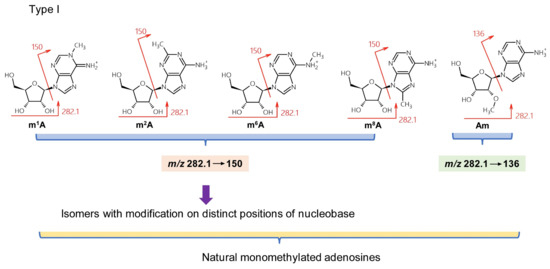
Figure 2.
Type I misidentification: natural isomers.
Although synthetic standards can strengthen the identification of nucleoside isomers, some cannot be separated (or are closely eluted) by liquid chromatography because of their structural similarity. Type II mistakes with TQ-MS are nucleoside misidentifications due to similar masses (<0.5 Da mass differences). m6,6A (N6,N6-dimethyladenosine) and f6A (N6-formyladenosine) exhibit MH+ masses of 296.1359 and 296.0995, respectively. Both are modified on the nucleobase (Figure 3). Low-resolution TQ-MS of m/z 296.1→164 cannot distinguish between these two compounds. Furthermore, m6,6A has an isomer—m2,8A (2,8-dimethyladenosine)—which can lead to the complicated situation of f6A/m6,6A being present in a eukaryotic total RNA sample, or m6,6A/m2,8A being present in a eubacterial rRNA sample [26].
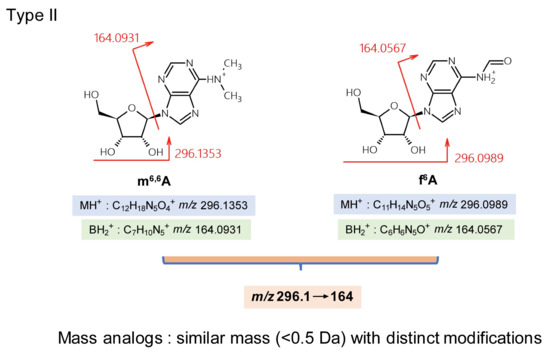
Figure 3.
Type II misidentification: mass-analogs (<0.5 Da mass difference).
Type III misidentification arises from isotopic crosstalk, which is often not considered. For instance, the low-resolution isotopic distributions of positively ionized adenosine and inosine are shown in Figure 4. The transition 269.1→137 used to monitor inosine is subject to interference from the same transition arising from an isotopologue of adenosine. When adenosine and inosine are eluted simultaneously by a short HPLC method, the signal for inosine would be amplified significantly by isotopic mass of adenosine. Given the abundance of isotopic masses of small molecules composed of the elements C, H, O, N, and S, nucleosides differing in mass by one or two units are likely to interfere with each other. Based on the natural abundance of isotopes of these elements (13C 1.11%, 2H 0.0115%, 18O 0.205%, 15N 0.364%, and 34S 4.21%), such mass differences mainly arise from 13C and 34S. Such mass-analogs are not rare among naturally modified nucleosides: crosstalk between mcm5U (5-methoxycarbonylmethyluridine, 317.1→185) [28], nchm5U (5-carbamoylmethyl-2-thiouridine, 318.1→186) [29], and cm5s2U (5-carboxymethyl-2-thiouridine, 319.1→187) [30] is a good example and is illustrated in Figure 5.
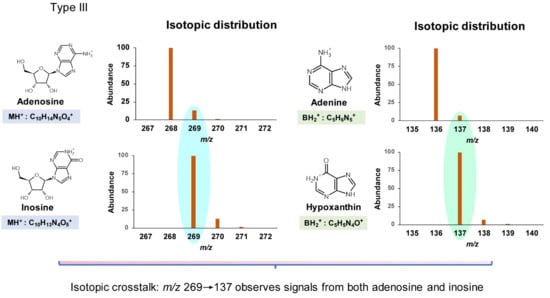
Figure 4.
Type III misidentification: isotopic crosstalk. Isotopic distribution was calculated using the online tool https://www.sisweb.com/mstools/isotope.htm (accessed on 1 March 2022).
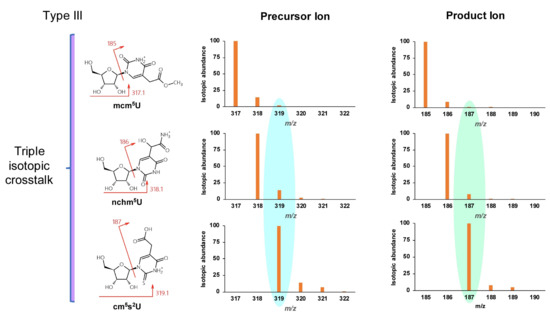
Figure 5.
Example of triple isotopic crosstalk type III misidentification between mcm5U, mchm5U, and cm5s2U.
These three types of misidentifications can occur in the same RNA sample; thus, correctly identifying a nucleoside by TQ-MS is not straightforward. This study illustrates this complexity by describing the analysis of 17 nucleosides and modified nucleosides using three commercially available liquid chromatography columns. Ongoing discussion of the separation of commonly modified nucleosides using reversed-phase and HILIC chromatography, and of the signals derived from TQ-MS, will help to minimize the misidentifications described above.
2. Materials and Methods
LC-MS grade formic acid and acetonitrile were purchased from Thermo Fisher (Shanghai, China) and LC-MS grade water from Sigma (Shanghai, China) for the preparation of mobile phases and standard solutions. Nucleoside standards were purchased from respected commercial manufacturers and agencies as follows: uridine (U), cytidine (C), 4-thiouridine (s4U), and N6-methyladenosine (m6A) from Aladdin (Shanghai, China); 1-methyladenosine (m1A), 1-methylinosine (m1I), N6,N6-dimethyladenosine (m6,6A), N6-formyladenosine (f6A), pseudouridine (Y), 5-hydroxyuridine (ho5U), 2-thiouridine (s2U), 5-methyldihydrouridine (m5D), 2-thiocytidine (s2C), and N1-methylguanosine (m1G) from TRC (Toronto, Canada); 2-methyladenosine (m2A) from Howei Pharm (Guangzhou, China); N2-methylguanosine (m2G) from TopScience (Rizhao, China); and 7-methylguanosine (m7G) from Sigma (Shanghai, China).
Stock solutions of nucleosides (10–100 mM) were prepared in dimethyl sulfoxide and stored at −20 °C. Mixed standard solutions were prepared by diluting stock solutions with 0.1% formic acid in acetonitrile/water (90/10, v/v) for HILIC chromatography, or water containing 0.1% formic acid for reversed-phase chromatography.
Mass spectrometry was conducted on a QTRAP 6500 LC-MS system (AB Sciex, Redwood, CA, USA). Mobile phase A consisted of 0.1% (v/v) formic acid in water, and mobile phase B was acetonitrile containing 0.1 % (v/v) formic acid. Reversed-phase chromatographic separations were run on a Waters Atlantis T3 column (2.1 mm2 × 150 mm2, 3 µm) and a Supelco Discovery HS F5 column (2.1 mm2 × 150 mm2, 3 µm). The gradient program (0.1 mL/min flow rate) was as follows: 0–6 min, 0% B; 6–35 min, 0–90% B; 35–40 min, 90% B; 40–40.1 min, 90–0% B; and 40.1–50 min, 0% B. HILIC separations were conducted on a Waters Acquity UPLC BEH amide column (2.1 mm2 × 150 mm2, 1.7 µm) at 0.1 mL/min flow rate using the elution gradient: 0–5 min, 90% B; 5–35 min, 90–40% B; 35–40 min, 40% B; 40–40.1 min, 40–90% B; and 40.1–50 min, 90% B. Column temperatures were maintained at 36 ºC, the autosampler at room temperature, and the injection volume was 1 μL. MS and MS/MS detection used an electrospray ionization (ESI) source in positive ion mode and the following optimized parameters: ion-spray voltage 5.5 kV, source temperature 350 °C, curtain gas 30 psi, collision gas 8 psi, ion source gas 1 at 30 psi, ion source gas 2 at 1 psi, entrance potential 10 V, collision cell exit potential 13 V, and dwell time 10 ms. MultiQuant 3.0.3 software (AB Sciex, Redwood, CA, USA) was used for data analysis.
3. Results and Discussion
3.1. Analytical Chromatography Columns
Three analytical columns were evaluated for their ability to resolve modified nucleosides over a 50 min chromatographic runtime: Acquity BEH amide (1.7 μm, 2.1 mm × 150 mm; Waters), Discovery HS F5 (3 μm, 2.1 mm × 150 mm; Supelco), and Atlantis T3 (3 μm, 2.1 mm × 150 mm; Waters) (Figure 6). The Acquity BEH amide (HILIC) column was packed with ethylene-bridged hybrid particles covalently attached by trifunctionally-bonded amide groups, while the linker structure of BEH amide is not published by the Waters Corporation. The Discovery HS F5 column was filled with spherical silica gel and a propyl spacer-linked pentafluorophenyl (PFP) stationary phase. The Atlantis T3 column was an octadecyl silica-based (ODS), reversed-phase C18 column with optimized pore diameter, C18-ligand density, and end-capping. Excellent performance in the separation of highly polar chemicals, including carbohydrates and nucleoside triphosphates, has been reported using these columns [31,32,33].
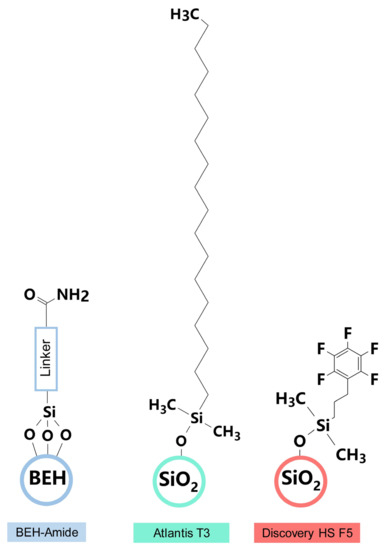
Figure 6.
Packing materials and stationary phases of Acquity BEH amide, Discovery HS F5, and Atlantis T3 analytical chromatography columns.
3.2. Adenosine Derivatives
One pmol each of m1A, m2A, m6A, m1I (1-methylinosine), m6,6A, and f6A were mixed and resolved on the three columns (Figure 7). Positive mode mass pair 282.1→150 was monitored to detect m1A, m2A, and m6A, 283.1→151 for m1I, and 296.1→164 for m6,6A and f6A. The methylated adenosines were eluted in the order m6A/m2A/m1A under HILIC conditions, but m1A/m2A/m6A under PFP and ODS conditions (Figure 7A). With the PFP column, m2A and m6A did not achieve baseline separation, which may lead to type I misidentification.
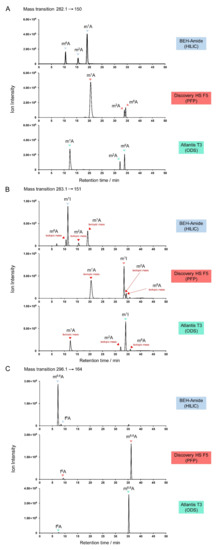
Figure 7.
Potential misidentification of adenosine derivatives. (A) Separation of natural monomethylated adenosine isomers m1A, m2A, and m6A on various columns. (B) Monitoring of 1-methylinosine (m1I; m/z 283.1→151) showing crosstalk from isotopes of m1A, m2A, and m6A. (C) Choice of chromatographic column is influenced by the similarity of m6,6A and f6A retention times in their shared MRM channel (m/z 296.1→164).
When m/z 283.1→150 was used to detect m1I, type III misidentification (isotopic crosstalk) could occur. Interfering signals from isotopes of m1A, m2A, and m6A were evident, adjacent to the primary m1I peak (Figure 7B). In the case of HILIC separation, a small m6A peak eluted close to m1I, while a m2A peak co-eluted with m1I under PFP conditions. Isotopic crosstalk should be carefully ruled out, especially when the target analyte is in low abundance and may be masked by an isotopologue of another nucleoside.
m6,6A and f6A exhibited similar retention times in the HILIC method (Figure 7C), although baseline separation was achieved. TQ-MS is not sufficiently sensitive to distinguish the mass difference between these two nucleosides (0.0364 Da); thus, the transition 296.1→164 acquired signals from both compounds. It is noteworthy that, although one pmol of each standard was injected, the signal for f6A was much lower than for m6,6A. This can be explained by the N6-formyl group of the f6A nucleobase tending to be deprotonated rather than protonated. This detection weakness under positive mode ionization increases the chance of f6A being misidentified as m6,6A in the absence of synthetic standards. Therefore, it is recommended to use a PFP or ODS column to detect m6,6A and f6A because of the large difference in retention times under these conditions (Figure 7C).
3.3. Uridine and Cytidine Derivatives
The m/z differences between protonated uridine and cytidine (m/z 245.1 and 244.1), and between their nucleobases (m/z 113 and 112) are approximately one. Thus, monitoring of uridine derivatives can be subject to inferring signals from isotopes of cytidine derivatives. For example, the transition m/z 259.1→127 can detect m5U (5-methyluridine), m3U (3-methyluridine), and isotopes of m5C (5-methylcytidine), m4C (N4-methylcytidine), and m3C (3-methylcytidine). Type I, II, and III misidentifications are possible, in some cases, in a single chromatogram monitoring uridine derivatives. Two standard mixtures were prepared to illustrate this complexity: firstly, 1 pmol each of U (uridine), C (cytidine), and Y (pseudouridine); and secondly, 1 pmol each of s2U (2-thiouridine), s4U (4-thiouridine), ho5U (5-hydroxyuridine), m5D (5-methyldihydrouridine), and s2C (2-thiocytidine). Figure 8A illustrates the monitoring of uridine (m/z 245.1→113) with isotopic crosstalk from cytidine. However, although the mass of pseudouridine is identical to uridine, the protonated nucleobase of pseudouridine is not seen. Instead, pseudouridine is uniquely identified by the transitions m/z 245.1→209/179/155. The CID fragmentation of pseudouridine is shown in Figure 8B [34]. The product ions of pseudouridine derivatives, such as m1Y (1-methylpseudouridine) and Ym (2’-O-methylpseudouridine), can be predicted using this fragmentation pattern to avoid interference with uridine derivatives.
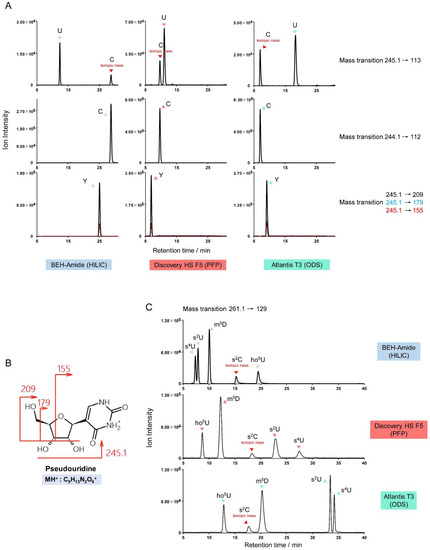
Figure 8.
Potential misidentification of cytidine and uridine derivatives. (A) Separation of cytidine (labelled C), uridine (labelled U), and pseudouridine (labelled Y) on various columns, showing cytidine crosstalk. (B) The unique fragmentation pattern of pseudouridine can be used to predict the product ions of pseudouridine derivatives. (C) The MRM channel m/z 261.1→129 detects the signal from s2U, s4U, ho5U, m5D, and isotopic s2C, and illustrates three types of potential misidentification.
Type I, II, and III potential misidentifications can be observed simultaneously in the mixture of s2U, s4U, ho5U, m5D, and s2C standards. Figure 8C shows how peaks for all five modified nucleosides appear in one MRM channel (m/z 261.1→129): s2U and s4U are structural isomers (type I); s2U, s4U, ho5U, and m5D have similar precursor and product ion masses (<0.5 Da difference) (type II); and a cross-talking isotopologue of s2C can be observed in the primary s2U/s4U/ho5U/m5D channel (type III).
3.4. Guanosine Derivatives
Obstacles to the identification of guanosine derivatives center on the possible isomers, particularly among the monomethylated (m1G, m2G, m7G), dimethylated (m2,2G, m2,7G), and trimethylated (m2,2Gm, m2,7Gm) guanosines. Figure 9 shows the elution sequence of m1G, m2G, and m7G on various columns, from which the elution sequence of the dimethylated and trimethylated derivatives can be extrapolated. Figure 9 also illustrates the importance of analytical column choice, with only HILIC being capable of resolving the monomethylated guanosines.
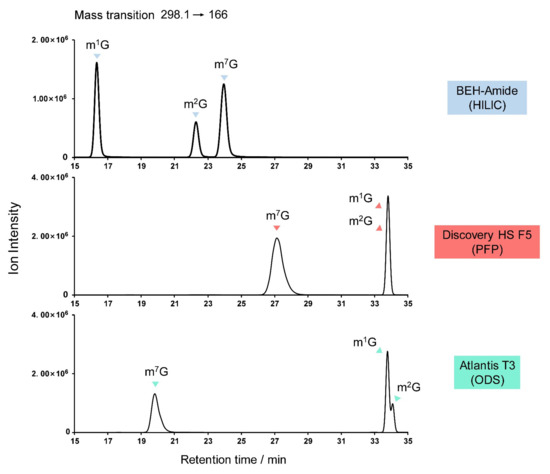
Figure 9.
Potential misidentification of guanosine derivatives, showing the separation of the natural monomethylated isomers m1G, m2G, and m7G on various columns.
4. Conclusions
Monitoring the fragmentation of protonated nucleosides into their respective nucleobases using the MRM mode of TQ-MS is useful for detecting RNA modifications. However, it is not practical to synthesize standards for many of these modified nucleosides and, consequently, three types of misidentification can result. Firstly, structural isomers can appear in the same MRM channel and may be closely eluted under HPLC (type I misidentification). Secondly, mass-analogs differing by less than 0.5 Da might also appear in the same channel and may be confused with other analytes (type II). Finally, analytes with mass differences of one or two Da can cause isotopic crosstalk (type III). This study applied a long HPLC runtime of 50 min but some nucleosides could still not be clearly resolved on-column. Therefore, the authors suggest that reports of rapid LC-MS methods for the detection of nucleosides (typically with 5–10 min runtimes) should be examined to rule out the possibility of analyte misidentifications arising from overlapping peaks. Stable isotope labeling of nucleosides can help to prevent type II and III misidentifications, as the mass shifts due to 13C or 15N provide additional molecular composition information [35,36,37]. Potential misidentifications of frequently modified nucleosides are listed in Table 1, Table 2 and Table 3.

Table 1.
Nucleosides with potential type I misidentification (natural isomers).

Table 2.
Nucleosides with potential type II misidentification (mass-analogs).

Table 3.
Nucleosides with potential type III misidentification (isotopic crosstalk).
Modified nucleosides can contain hydrophobic nucleobases (e.g., those with isopentenyl moieties) and hydrophilic nucleobases (e.g., those with hydroxy moieties), making reversed-phase and HILIC methods suitable for the separation of different groups of nucleosides. The PFP column (Discovery HS F5) separated uridine and cytidine derivatives effectively, and the HILIC column (Acquity BEH amide) was helpful for adenosine and guanosine derivatives. The chromatographic column should be carefully chosen and the HPLC method optimized depending on the target analyte(s). The analytical sensitivity of the nucleosides under positive mode LC-TQ-MS conditions is usually A>G>C>U (uridine being difficult to protonate under positive mode ionization due to its low pKa value). Negative mode ionization, derivatization [38], and the careful selection of productions should be considered to improve sensitivity. The degradation, oxidation [39], and spontaneous chemical derivatization [40] of nucleosides during pre-treatment procedures should also be taken into account. We recommend the establishment of strict standards for nucleoside analysis.
Author Contributions
X.L.: Conceptualization, Methodology, Data curation, Investigation, Validation; Q.Z. (Qianhui Zhang): Methodology, Visualization, Investigation, Software; Y.Q.: Methodology, Validation; Q.Z. (Qisheng Zhong): Methodology, Validation; D.L.: Methodology; X.W.: Methodology; P.F.: Conceptualization, Supervision; H.L.: Conceptualization, Methodology, Writing—original draft, Writing—review & editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Hainan Provincial Natural Science Foundation of China (Grant No. 2019RC003, 220MS004), the Program of Hainan Association for Science and Technology Plans to Youth R & D Innovation (Grant No. QCXM202013), and the National Natural Science Foundation of China (Grant No. 31900436).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data supporting this study’s findings are available on http://www.doi.org/10.11922/sciencedb.01462 (accessed on 1 March 2022) with appropriate protection periods.
Acknowledgments
We thank Jia Zhou of the Chinese Academy of Tropical Agricultural Sciences for her kind technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nachtergaele, S.; He, C. The emerging biology of RNA post-transcriptional modifications. RNA Biol. 2017, 14, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dannfald, A.; Favory, J.-J.; Deragon, J.-M. Variations in transfer and ribosomal RNA epitranscriptomic status can adapt eukaryote translation to changing physiological and environmental conditions. RNA Biol. 2021, 18, 4–18. [Google Scholar] [CrossRef]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.R.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [Green Version]
- Asano, K.; Suzuki, T.; Saito, A.; Wei, F.-Y.; Ikeuchi, Y.; Numata, T.; Tanaka, R.; Yamane, Y.; Yamamoto, T.; Goto, T.; et al. Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res. 2018, 46, 1565–1583. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Miyauchi, K.; Harada, T.; Okita, R.; Takeshita, E.; Komaki, H.; Fujioka, K.; Yagasaki, H.; Goto, Y.-i.; Yanaka, K.; et al. CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat. Commun. 2018, 9, 1875. [Google Scholar] [CrossRef]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell 2016, 167, 816–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helm, M.; Motorin, Y. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Hartstock, K.; Rentmeister, A. Mapping m6A in RNA: Established methods, remaining challenges and emerging approaches. Chemistry 2019, 25, 3455–3464. [Google Scholar] [CrossRef]
- Zhao, L.-Y.; Song, J.; Liu, Y.; Song, C.-X.; Yi, C. Mapping the epigenetic modifications of DNA and RNA. Protein Cell 2020, 11, 792–808. [Google Scholar] [CrossRef]
- Schneeberger, E.M.; Breuker, K. Native top-down mass spectrometry of TAR RNA in complexes with a wild-type tat peptide for binding site mapping. Angew. Chem. Int. Ed. 2017, 129, 1274–1278. [Google Scholar] [CrossRef]
- Schürch, S. Characterization of nucleic acids by tandem mass spectrometry-The second decade (2004–2013): From DNA to RNA and modified sequences. Mass Spectrom. Rev. 2016, 35, 483–523. [Google Scholar] [CrossRef] [PubMed]
- Yoluç, Y.; Ammann, G.; Barraud, P.; Jora, M.; Limbach, P.A.; Motorin, Y.; Marchand, V.; Tisné, C.; Borland, K.; Kellner, S. Instrumental analysis of RNA modifications. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 178–204. [Google Scholar] [CrossRef]
- Catala, M.; Gato, A.; Tisné, C.; Barraud, P. 1 H, 15 N chemical shift assignments of the imino groups of yeast tRNA Phe: Influence of the post-transcriptional modifications. Biomol. NMR Assign. 2020, 14, 169–174. [Google Scholar] [CrossRef]
- Barraud, P.; Gato, A.; Heiss, M.; Catala, M.; Kellner, S.; Tisné, C. Time-resolved NMR monitoring of tRNA maturation. Nat. Commun. 2019, 10, 3373. [Google Scholar] [CrossRef] [Green Version]
- Su, D.; Chan, C.T.; Gu, C.; Lim, K.S.; Chionh, Y.H.; McBee, M.E.; Russell, B.S.; Babu, I.R.; Begley, T.J.; Dedon, P.C. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 2014, 9, 828–841. [Google Scholar] [CrossRef]
- Miyauchi, K.; Kimura, S.; Suzuki, T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013, 9, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, Y.; Miyauchi, K.; Kang, B.-i.; Suzuki, T. Nucleoside analysis by hydrophilic interaction liquid chromatography coupled with mass spectrometry. Methods Enzymol. 2015, 560, 19–28. [Google Scholar] [PubMed]
- Basanta-Sanchez, M.; Temple, S.; Ansari, S.A.; D’Amico, A.; Agris, P.F. Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells. Nucleic Acids Res. 2016, 44, e26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Y.; Zhong, Q.; Zhang, Y.; Lin, X.; Fu, P.; Lin, H. Micro-flow hydrophilic interaction liquid chromatography coupled with triple quadrupole mass spectrometry detects modified nucleosides in the transfer RNA pool of cyanobacteria. J. Sep. Sci. 2021, 44, 3208–3218. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piątkowski, P.; Bagiński, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A. MODOMICS: A database of RNA modification pathways—2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Nakayama, H.; Yamauchi, Y.; Nobe, Y.; Sato, K.; Takahashi, N.; Shalev-Benami, M.; Isobe, T.; Taoka, M. Method for direct mass-spectrometry-based identification of monomethylated RNA nucleoside positional isomers and its application to the analysis of Leishmania rRNA. Anal. Chem. 2019, 91, 15634–15643. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.; Brophy, J.A.; Chan, C.T.; Atmore, K.A.; Begley, U.; Paules, R.S.; Dedon, P.C.; Begley, T.J.; Samson, L.D. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010, 30, 2449–2459. [Google Scholar] [CrossRef] [Green Version]
- Pastore, C.; Topalidou, I.; Forouhar, F.; Yan, A.C.; Levy, M.; Hunt, J.F. Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification. J. Biol. Chem. 2012, 287, 2130–2143. [Google Scholar] [CrossRef] [Green Version]
- Patil, A.; Chan, C.; Dyavaiah, M.; Rooney, J.P.; Dedon, P. Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol. 2012, 9, 990–1001. [Google Scholar] [CrossRef] [Green Version]
- Ghfar, A.A.; Wabaidur, S.M.; Ahmed, A.Y.B.H.; Alothman, Z.A.; Khan, M.R.; Al-Shaalan, N.H. Simultaneous determination of monosaccharides and oligosaccharides in dates using liquid chromatography–electrospray ionization mass spectrometry. Food Chem. 2015, 176, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Portillo-Castillo, O.J.; Castro-Ríos, R.; Chávez-Montes, A.; González-Horta, A.; Cavazos-Rocha, N.; Waksman De Torres, N.; Garza-Tapia, M. A new RP-HPLC method as an auxiliary tool for optimization of sample preparation procedures for tracing of PPCPs of different hydrophilicities. Acta Pharm. 2021, 71, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.; Herold, N.; Keppler, O.T.; Geisslinger, G.; Ferreirós, N. Quantitation of endogenous nucleoside triphosphates and nucleosides in human cells by liquid chromatography tandem mass spectrometry. Anal. Bioanal. Chem. 2015, 407, 3693–3704. [Google Scholar] [CrossRef] [PubMed]
- Dudley, E.; Tuytten, R.; Bond, A.; Lemière, F.; Brenton, A.G.; Esmans, E.L.; Newton, R.P. Study of the mass spectrometric fragmentation of pseudouridine: Comparison of fragmentation data obtained by matrix-assisted laser desorption/ionisation post-source decay, electrospray ion trap multistage mass spectrometry, and by a method utilising electrospray quadrupole time-of-flight tandem mass spectrometry and in-source fragmentation. Rapid Commun. Mass Spectrom. 2005, 19, 3075–3085. [Google Scholar]
- Kellner, S.; Neumann, J.; Rosenkranz, D.; Lebedeva, S.; Ketting, R.F.; Zischler, H.; Schneider, D.; Helm, M. Profiling of RNA modifications by multiplexed stable isotope labelling. Chem. Comm. 2014, 50, 3516–3518. [Google Scholar] [CrossRef]
- Kellner, S.; Ochel, A.; Thüring, K.; Spenkuch, F.; Neumann, J.; Sharma, S.; Entian, K.-D.; Schneider, D.; Helm, M. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res. 2014, 42, e142. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, S.; Byrne, S.R.; Ammann, G.; Asadi Atoi, P.; Borland, K.; Brecheisen, R.; DeMott, M.S.; Gehrke, T.; Hagelskamp, F.; Heiss, M.; et al. Strategies to Avoid Artifacts in Mass Spectrometry-Based Epitranscriptome Analyses. Angew. Chem. Int. Ed. 2021, 60, 23885–23893. [Google Scholar] [CrossRef]
- Lopez Torres, A.; Yanez Barrientos, E.; Wrobel, K.; Wrobel, K. Selective derivatization of cytosine and methylcytosine moieties with 2-bromoacetophenone for submicrogram DNA methylation analysis by reversed phase HPLC with spectrofluorimetric detection. Anal. Chem. 2011, 83, 7999–8005. [Google Scholar] [CrossRef]
- Estevez, M.; Valesyan, S.; Jora, M.; Limbach, P.A.; Addepalli, B. Oxidative damage to RNA is altered by the presence of interacting proteins or modified nucleosides. Front. Mol. Biosci. 2021, 8, 631. [Google Scholar] [CrossRef]
- Jora, M.; Borland, K.; Abernathy, S.; Zhao, R.; Kelley, M.; Kellner, S.; Addepalli, B.; Limbach, P.A. Chemical amination/imination of carbonothiolated nucleosides during RNA hydrolysis. Angew. Chem. Int. Ed. 2021, 60, 3961–3966. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).