Abstract
Deubiquitination is a major form of post-translational protein modification involved in the regulation of protein homeostasis and various cellular processes. Deubiquitinating enzymes (DUBs), comprising about five subfamily members, are key players in deubiquitination. USP10 is a USP-family DUB featuring the classic USP domain, which performs deubiquitination. Emerging evidence has demonstrated that USP10 is a double-edged sword in human cancers. However, the precise molecular mechanisms underlying its different effects in tumorigenesis remain elusive. A possible reason is dependence on the cell context. In this review, we summarize the downstream substrates and upstream regulators of USP10 as well as its dual role as an oncogene and tumor suppressor in various human cancers. Furthermore, we summarize multiple pharmacological USP10 inhibitors, including small-molecule inhibitors, such as spautin-1, and traditional Chinese medicines. Taken together, the development of specific and efficient USP10 inhibitors based on USP10’s oncogenic role and for different cancer types could be a promising therapeutic strategy.
1. Introduction
Dysfunctional protein expression is associated with many diseases including cancer. Proteins, especially in eukaryotic cells, maintain normal cell function under steady-state conditions, 80% of which is mediated by the ubiquitin–proteasome system (UPS) [1]. Ubiquitin is an 8.5 kDa protein containing 76 amino acids that produces a polyubiquitin chain on a target protein, mainly through its seven lysines at the N-terminal and methionine sites. Ubiquitin usually binds to a target protein through glycine at the C-terminal and leads to its degradation through the 26S proteasome system [2,3]. In this process, three kinds of ubiquitin enzymes play a significant role: E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin ligase. E3 ubiquitin ligase is target-specific and regulates the interaction of ubiquitin with target proteins. More details about the structure and function of UPS have been well summarized elsewhere [4].
Deubiquitination is a process that is the opposite of ubiquitination, and deubiquitination enzymes (DUBs) play an important role in this process. DUBs remove or cleave the isopeptides between the substrate protein and ubiquitin to achieve deubiquitination. This process of reversal had not been widely studied until now [5,6]. Although some researchers have found that DUBs affect a variety of cellular processes, their underlying mechanisms, targets, and inhibitors for specific diseases remain largely unknown [7]. The regulation of DUBs and the abundance of their downstream target proteins is considered a novel and promising cancer-treatment strategy [8].
Although hundreds of DUBs have been identified, the characteristics and functions of many in human diseases remain unclear [9]. USP10 has been widely studied and found to be involved in various cellular processes, including DNA repair, cell-cycle regulation [10], autophagy [11], and immune and inflammatory responses. Especially for inflammation, Li et al. demonstrated that USP10 can elevate proinflammatory factors in endometriosis (EM) and that Cai’s Neiyi Prescription (CNYP) can reduce EM-induced inflammation by inhibiting the mRNA and protein expression of USP10 [12]. However, it is known that USP10 maintains normal cell function by controlling the protein balance through the ubiquitin–proteasome degradation pathway [13]. In immune system diseases, such as asthma and systemic sclerosis, USP10 influences inflammatory responses by inhibiting T-box transcription factor (T-bet) ubiquitination and stabilizing its expression, and thus, it has also been found that quercetin, an inhibitor of USP10, alleviates asthma via enhancing the ubiquitination and promoting the degradation of T-bet [14]. Other studies have also demonstrated that USP10 deubiquitinates AMPKα, regulating energy metabolism [15], and stabilizes the CD36 protein, thus promoting the development of atherosclerosis [16].
In recent years, mounting evidence has also indicated that USP10 plays a crucial role in tumorigenesis [9,17,18,19,20,21,22,23]. The overexpression of USP10 can inhibit the formation of stress granules (SGs), thus restraining the development of a tumor [19,24]. In addition, two well-known tumor suppressors, P53 and SIRT6, can be regulated and considered as substrates of USP10 [25,26]. The p53 protein is a key transcription factor and plays an important role in the DNA-damage response, cell-cycle arrest, and apoptosis, which are closely associated with tumorigenesis. Under normal conditions, the E3 ubiquitin ligase MDM2 maintains a basal low level of p53 by promoting its ubiquitination and 26S proteasomal degradation. In 2020, Yuan et al. revealed that USP10 is a novel regulator of p53. The depletion of USP10 significantly reduced p53’s stabilization by increasing its ubiquitination. Moreover, the expression of p53’s downstream target genes, including p21 and Bax, was also downregulated [27]. Mechanistically, USP10 can reverse the activity of Mdm2 through deubiquitinating p53 in the cytoplasm, causing the return of p53 from the cytoplasm to the nucleus and affecting the nuclear output of the p53 protein. Moreover, USP10 plays an essential role in stabilizing the p53 protein in neurodegenerative diseases, and several proteins regulate the cell cycle by regulating USP10, affecting the stability of p53. For example, miR-191, a factor promoting the proliferation of pancreatic cancer cells, acts by negatively regulating USP10 and thereby reducing the content of p53 [28]. In thyroid cancer and gastric cancer, DZNep, an essential component of PRC2, also regulates p53 by regulating the content of USP10 [29,30]. Interestingly, a negative-feedback loop between USP10 and p53 was observed by Luo et al., who found that miR-138 could decrease the activity of USP10 through specifically binding to the 3′-UTR of USP10’s mRNA following an increase in the expression of the p53 protein, but in turn, p53 reduced the expression of miR-138, forming a feedback pathway [31]. It is worth noting that, in addition to regulating DNA damage via p53 ubiquitination, USP10 also stabilizes MSH2 (an important DNA mismatch-repair protein) and TRAF6 (an activator of NF-κB), which affects DNA repair and other physiological processes during DNA damage [23,25,27,32,33]. In line with this, some studies found that the disruption of the interaction between USP10 and p53 inhibited cancer cells’ viability or tumor growth. For example, mycoplasma DnaK particles inhibited the anticancer activity of p53 through binding to USP10 [34]. The inhibition of the proline hydroxylase PHD3 could significantly reduce the binding of USP10 to p53 [35]. Resveratrol promotes the p53-mediated apoptosis of cancer cells by acting on the USP10-binding protein G3BP1 [36]. IGF2BP3 (insulin-like growth factor 2 mRNA binding protein 3) inhibits p53 expression through disrupting the interaction between USP10 and p53 and promotes lung cancer progression [37]. Although some substrates and regulators of USP10 have been reported, the role of USP10 in tumorigenesis remains poorly understood. Herein, we focus on the involvement of USP10, along with its substrates and upstream regulators, in cancer cells. We expect that the summary of such knowledge will help us to develop new inhibitors or strategies to target USP10 in human cancers in the future.
2. USP10 Is a USP-Family Deubiquitinating Ligase
The USP family is a relatively large group (with more than 50 known members) of deubiquitinating enzymes [38,39]. As the largest and most diverse family of DUBs, the USPs have many similarities with and differences from other DUBs, such as OTUs (ovarian tumor-related proteases), JAMMs (Jad1/Pad/MPN domain-containing metalloenzymes), UCHs (ubiquitin C-terminal hydrolases), MJDs/Josephin (Machado–Joseph disease proteases), and the ZUP1/ZUFSP (zinc finger with UFM1-specific peptidase) family. All these DUBs can reverse ubiquitination by cleaving the peptide or isopeptide bond and removing ubiquitin from its substrates. However, they can be differentially classified into cysteine proteases and metalloproteases. For example, the JAMMs are known to be metalloproteases, but the OTUs, USPs, MJDs/Josephin, and UCHs are considered cysteine proteases [40]. Interestingly, among the cysteine-protease DUBs, the USPs are larger than the other DUBs [9]. More importantly, they consist of one specific USP domain (also known as the catalytic domain) and three other conserved subdomains, which form a hand-like structure including a thumb, finger, and palm [41]. Different domains or subdomains play important roles in regulating the enzyme activity.
All the USPs contain short motifs (two short conserved fragments, a lysine box, and a histidine box) and a finger-like structure containing β-lamellar structures that coordinate and support ubiquitin’s ligands, mediating the interaction with the ubiquitin substrate. The thumb is composed of the central catalytic helix, nucleophilic cysteine, and core proteasome structure. The palm contains aspartic acid, histidine residues, and the conserved β-sheet structure of the protease core. Therefore, the thumb–palm gap provides the catalytic center for the USP domain [9,13,15,42]. Recent results from NMR structural studies show that the active site undergoes rearrangements upon Ub binding to a USP. Subsequently, a conformational change in the catalytic domain promotes the catalytic hydrolysis of Ub from the ubiquitinated proteins.
The USPs are grouped into five subfamilies based on the ubiquitin domain’s architecture: Ub-associated domain (UBA domain) (5 members), Ub-interacting motif (UIM) (3 members), zinc-finger Ub-specific protease domain (ZnF-UBP domain) (10 members) (Figure 1), and the common potential ubiquitin domain (UBL domain) (17 members) (Figure 2). All these members have very similar ubiquitin-related domains (different from the USP domain). However, USP10 is a member of the family only including the USP domain (about 23 members) (Figure 3) and having no UBA, UIM, ZnF-UBP, or UBL domain (Figure 1, Figure 2 and Figure 3) [9,13].
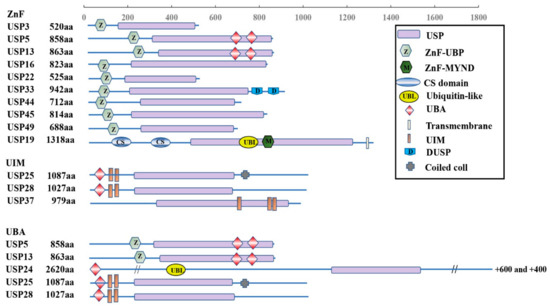
Figure 1.
Schematic illustration of ZnF, UIM, and UBA subfamily architectures.
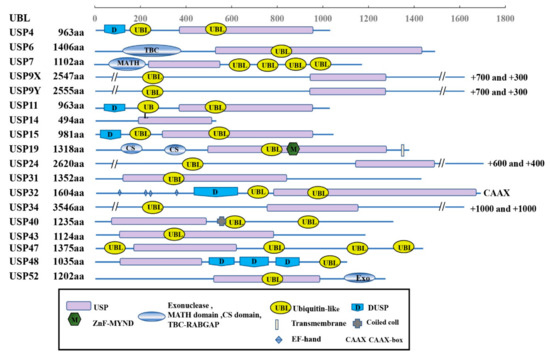
Figure 2.
Schematic illustration of UBL-related subfamily architectures.
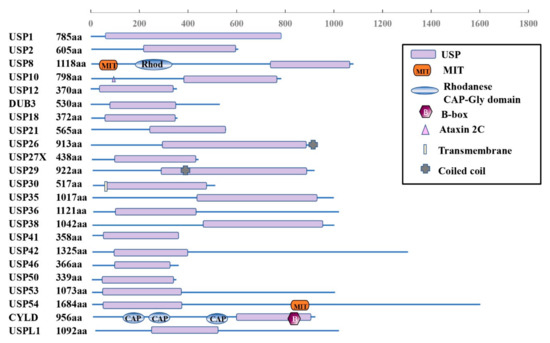
Figure 3.
Schematic illustration of USP10-related subfamily architectures.
The domain structures of the ZnF, UIM, and UBA subfamilies are shown in Figure 1. There is a zinc finger Ub-specific protease domain (ZnF-UBP domain) in ten members (upper), Ub-interacting motif (UIM) in three members (middle), and Ub-associated domain (UBA domain) in five members (lower). However, USP5 and USP13 from the ZnF family, together with USP25 and USP28 from the UIM family, also have UBA domains.
The domain structures of the UBL-related subfamily are shown in Figure 2. All the members have the domain of the ubiquitin-specific proteases (USPs) and ubiquitin-like domain (UBL domain). In addition, different members have other additional domains, such as transmembrane and coiled-coil domains.
The domain structure of the USP10-related subfamily is shown in Figure 3. All the members have the domain of the ubiquitin-specific proteases (USPs). In addition, USP8 and USP54 also contain the domain of microtubule-interacting and trafficking proteins (MIT) or rhodanese (Rhod). Both USP26 and USP19 include coiled-coil domains. However, USP30 contains a transmembrane domain and CYLD includes the CAP-Gly domain (CAP) and B-Box (B).
The human USP10 gene encodes 798 amino acids forming a typical USP domain. The USP10 consists of a larger N-terminus region, a USP catalytic structure domain (about 380 amino acids, starting at 415 amino acids from the N-terminus [43]), and a smaller C-terminus region (Figure 1) [44]. The USP10 proteins are highly conserved among humans and other mammals. For example, there is about 99% percent identity between the amino acid sequences of the human and rat or mouse forms [9].
Similar to other DUBs, USP10 can deubiquitinate and cleave Ub from the C-termini of substrates through four steps. The first step is the USP binding to the ubiquitin COOH terminus via its USP domain, causing a conformational change in the catalytic domain. Then, the conserved residues of the group (Cys, His, Asp, and Asn) form a catalytic triad in a specific manner. Next, the deprotonated thiol group conducts a nucleophilic attack on the carbonyl carbon, and the active site is transferred from its initial position. Finally, the non-anchoring Ub is removed from the target proteins [9,45]. It is known that the USPs specifically cleave ubiquitin sections or process ubiquitin chains based on the substrates. For example, both USP21 and USP7 cleave K6-linked ubiquitin chains. USP7 can also cleave K11-, K33-, K48-, and K63-linked chains [9,45]. However, whether and how USP10 can also remove K11-, K48-, and K63-linked ubiquitin chains remains largely unknown. Recently, Yuan et al. demonstrated that USP10 stabilized Smad4 via directly binding to Smad4 and removing a Lys48-linked polyubiquitin chain [46]. Another previous study from Hu et al. also showed that the depletion of USP10 increased the K48-linked polyubiquitination of HDAC6 in the non-small-cell lung cancer (NSCLC) cell line H1299. In addition, He et al. discovered that USP10 reduced the K63-linked polyubiquitination of PTEN in the NSCLC A549 cell line [47]. All these modifications affect the protein’s stability when targeted by USP10.
It has been reported that many other proteins, such as TP53 RPS2, RPS3, RPS10, and LC3B, can be deubiquitinated by USP10 and are known substrates (as summarized in Table 1). Although many of their ubiquitin sites have been identified, the mechanism of ubiquitin recognition by USP10 remains elusive. It will be intriguing to assess how and which ubiquitin linkage could be recognized and cleaved by USP10 in vitro and in vivo. More importantly, the characteristics of the specificity for chains other than K48- and K63-linked ubiquitin of USP10 is also worthy of future investigation.

Table 1.
Summary of the identified deubiquitination substrates for USP10.
3. Regulation of USP10
As for most deubiquitinating enzymes, the activity of USPs can be regulated by multiple mechanisms, including those acting at the transcriptional and post-translational levels. Previous studies showed that a subset of USP-family deubiquitinating enzymes including USP1, USP4, USP8, and USP13 could be phosphorylated by CDK1, AKT, or CLK3 kinase, preventing the USPs and substrates binding [45]. For example, the CDK1-mediated serine phosphorylation of USP1 disrupts the interaction between USP1 and UAF1. CLK3 mediated the phosphorylation of USP13 at Y708, promoting its binding to c-Myc, which is an important transcription factor and also known as an oncogene in many cancers [71]. In line with the above studies, USP10’s activity can be regulated by phosphorylation at its N-terminus domain. Deng et al. revealed that AMPKα directly mediated the phosphorylation of Ser76 at the USP10 N-terminus and increased its activity. Interestingly, USP10 could regulate the deubiquitination of AMPKα, creating a positive-feedback pathway [15] (Figure 4). Yuan et al. demonstrated that ATM could phosphorylate USP10 at Thr42 and Ser337 upon the DNA-damage response and USP10 was translocated into the nucleus, where the N-terminus of USP10 (amino acids 1–100) binds to p53 and inhibits its ubiquitination [27]. In addition, Luo et al. reported that co-stimulation by BCR and TLR1/2 initiated the AKT-dependent phosphorylation of T674 of USP10; subsequently, USP10 entered the nucleus and stabilized the AID protein (also see Figure 4 and Table 2) [72]. In addition to phosphorylation directly controlling the activation of USP10, a previous study from the Deng group revealed that, in keloid cells, TRAF4 inhibited USP10-mediated p53 deubiquitination and degradation through disrupting the access of p53 to USP10 [73]. Liu et al. found that USP10 stabilized beclin-1 by interacting with and inhibiting its ubiquitination. Very interestingly, they also pointed out that beclin-1 controlled the stability of USP10 by regulating the stability of USP13, which can deubiquitinate USP10 (also see Figure 4) [53]. Another finding regarding feedback regulation was reported by several groups [33,53,74,75]. They found that MCPIP1 directly binds to USP10 and serves as a bridge between USP10 and TANK [33]. MCPIP1 forms a complex with USP10 and TANK, which mediates the deubiquitination of the TRAF6 K63 chain [76] and inhibits NF-κB activation upon DNA damage [33]. On the other hand, genotoxic-stress-induced NF-κB activation enhanced MCPIP1 transcription [75]. In addition, Lin et al. demonstrated that USP10 enhanced the stability of SIRT6 through interacting with its N-terminal regulatory domain and deubiquitinating SIRT6, and then inhibited the transcriptional activity of the c-Myc oncogene through SIRT6 [25]. Consistent with the latest findings, Zhou et al. found that USP13 also affected the ubiquitination of c-Myc and resulted in enhancing its stability [77]. However, feedback regulation between USP10 and c-Myc was also found. c-Myc directly binds to the second E-box sequence of the USP10 gene and activates the transcription of USP10 [69]. All these findings suggest that regulatory feedback mechanisms play an important role in signaling pathway regulation and maintaining a balance in protein levels. They also encourage us to identify many more modifiers of USP10, such as kinases and DUBs, and uncover the mechanisms of USP10’s dynamic regulation. This highlights that identifying more regulators of USP10 will be necessary for clarifying its role in disease.
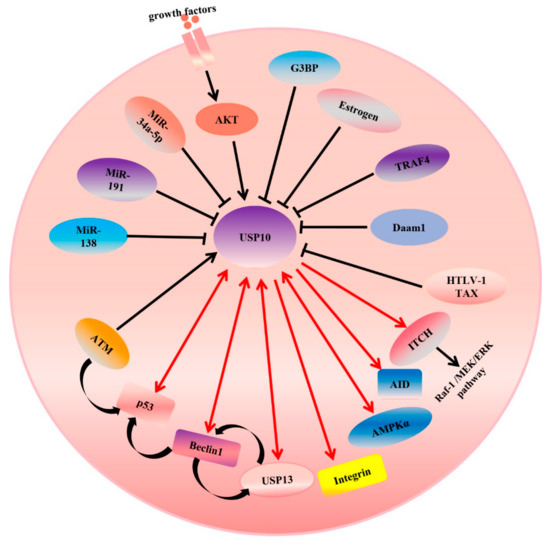
Figure 4.
USP10 targets different signaling pathways in cancer cells.

Table 2.
Summary of the identified forms of regulation for USP10.
An arrowhead from USP10 indicates positive regulation of the substrate protein. Single arrowheads with red lines indicate that USP10 only regulates the substrates, while double arrowheads with red lines indicate that they can regulate each other. For example, USP10 regulate the activity or expression of integrin, AID, ITCH, p53, USP13, and so on. Conversely, p53 and USP13 also can regulate USP10 forming feedback loop. An arrowhead toward USP10 indicates a positive regulation of USP10’s deubiquitinating ligase activity, while a blunt head toward USP10 indicates negative regulation of such. As shown, both AKT and ATM increase the activity of USP10. However, overexpression of miR-138, miR-191 and miR-34a-5p inhibits the mRNA and protein expression levels of USP10. A curved arrowhead with a black color indicates positive regulation.
In addition to USP10 being regulated by the proteins of host cells, previous studies of the Takahashi group showed that HTLV-1 Tax interacted with amino acids 727–798 at the C-terminal of USP10 and inhibited USP10’s deubiquitination activity and function. For example, inhibiting USP10 reduces the production of reactive oxygen species and suppresses apoptosis and the formation of SGs (stress granules) [78]. On the other hand, it has been found that USP10 can be regulated at the transcriptional level. Generally, a microRNA binds to the 3-untranslated region (3′-UTR) of an mRNA in a sequentially specific manner, inducing the mRNA’s degradation or inhibiting its translation. Luo et al. revealed that USP10 is a target of miR-138 and that the 3′-UTR conserved domain of USP10 directly interacts with miR-138. The overexpression of miR-138 inhibits the mRNA and protein expression levels of USP10 (also see Figure 4 and Table 2). Moreover, they also found that p53, which can be deubiquitinated by USP10, negatively regulates the expression of miR-138 through binding to the miRNA’s promoter region [31]. In other words, there is feedback regulation. Another microRNA, miR-191, is considered an upstream regulator of USP10 in PDAC (pancreatic cancer) cells [28]. All the regulators of USP10 are summarized in Table 2.
Zhang et al. indicated that miR-34a-5p acted as a negative regulator of USP10, but the underlying mechanism of the microRNA’s action remains largely unclear [79]. More importantly, whether there is feedback regulation is also unknown. This highlights that identifying more regulators of USP10 will be essential for clarifying the mechanisms and roles in different diseases, especially for cancer.
4. USP10 in Cancers
USP10 plays multiple roles in many diseases including human cancers. It was shown that the overexpression of USP10 promoted the proliferation and metastasis of multiple tumors including adult T-cell leukemia, glioblastoma multiforme, chronic myeloid leukemia, non-small-cell lung cancer, hepatocellular carcinoma (HCC), colon cancers, etc., while it has also been reported that the expression of USP10 was reduced in gastric carcinoma (GC), HCC, and colon cancer and suppressed the development of tumorigenesis through regulating different signaling pathways, such as the p53 apoptosis pathway, suggesting that USP10 is, indeed, a double-edged sword in different cancer types or different cell contexts for the same cancer. We will further discuss the oncogenic and tumor-suppressor roles of USP10 in the following section.
4.1. USP10 as an Oncogene
It was first shown that increased expression of USP10 predicted poor survival in patients with glioblastoma multiforme (GBM) [83]. Mounting evidence has indicated that USP10 plays an oncogenic role in tumorigenesis. In adult T-cell leukemia (AML), previous studies have revealed that the human T-cell leukemia virus type 1 (HTLV-1) oncoprotein Tax interacts with USP10 and promotes ROS-dependent apoptosis and the occurrence of AML. Moreover, USP10 was also found to reduce the sensitivity of AML cells to chemotherapeutic drugs including arsenic [19,78]. Recently, the Weisberg group verified that increased expression of mutant FLT3–ITD correlated with poor prognosis and low survival. Mechanically, USP10 led to the accumulation of FLT3–ITD through deubiquitinating and stabilizing the mutant FLT3–ITD, promoting AML progression [9,61]. In addition to mutant FLT3–ITD, another spleen tyrosine kinase (SYK), a critical regulator of FLT3, was deubiquitinated and stabilized by USP10; depleting or inhibiting USP10 reduced the expression of SYK, which promoted the proliferation of Ba/F3 and MOLM14 cells [68,84].
Chronic myeloid leukemia (CML) is positively associated with abnormally high expression of the tyrosine kinase BCR-ABL. Liao et al. found that both USP10 and SKP2 are highly expressed in the primary monocytes of patients with CML than in the primary monocytes of healthy humans. Increased USP10 led to the deubiquitination and stabilization of SKP2, causing BCR-ABL activation and promoting CML cells’ proliferation [85,86]. To date, in addition to blood-related cancers, USP10 also plays oncogenic roles in many solid tumors including hepatocellular carcinoma (HCC) [46], colon cancer [68,84], renal-cell carcinoma (RCC) [27], non-small-cell lung cancer (NSCLC)[27], prostate cancer (PCa) [87,88], esophageal cancer [87,88], breast cancer [75,89], and melanoma [87,88]. In tissue samples from HCC patients, increased USP10 levels are positively correlated with the abundance of YAP/TAZ [90]. Functionally, USP10 directly deubiquitinates and stabilizes YAP/YAZ and promotes HCC progression. Additionally, other substrates of USP10 in HCC have been found, such as Smad4 and TGF-β. Yuan et al. found that USP10 directly bounds to and stabilized Smad4, promoting HCC metastasis, using a functional RNA-interference screening method; thus, USP10 is considered a prognostic and therapeutic target in Smad4-positive HCC metastatic patients [46]. In prostate cancer, high expression of USP10 indicates poor prognosis [18,91]. USP10 promotes the proliferation of PCa cell lines through binding to and increasing the stability of G3BP2, which inhibits p53 activity. On the other hand, androgen receptor (AR), a key regulator, plays a crucial role in the regulation of PCa progression. AR-related proteins including H2A, Zub1, and H2Aub1 can be deubiquitinated and stabilized by USP10. In esophageal cancer and human neuroblastoma, USP10 affects cancer cell proliferation by stabilizing PCNA and NRF-1, respectively [92]. These findings suggest that USP10 could function as an oncogene to precisely control cell proliferation in tumor cells.
In addition to the differential effects of USP10 in different cancers, USP10 can be a double-edged sword in the same cancer. For example, in colon cancer, USP10 interacted with and stabilized SIRT6 through inhibiting SIRT6’s ubiquitination, suppressing proliferation [25]. However, the USP10-mediated deubiquitination of NLRP7 or MSI2 increased its stabilization and expression, promoting the occurrence of CRC [68,84]. It will be intriguing to assess when and how USP10 could deubiquitinate different proteins in vitro and in vivo.
In contrast to the tumor-suppressor role of USP10 in p53-WT cancer cells, USP10 exerts an oncogenic function in p53-mutant cancer cells. For example, in p53-mutant RCC cells, increased USP10 expression promotes cell proliferation via deubiquitinating and stabilizing the mutant p53 [27]. Additionally, in NSCLC, the USP10-mediated deubiquitination of the oncogenic protein histone deacetylase 6 (HDAC6) leads to cisplatin resistance in patients harboring mutant p53 [93]. In addition, USP10 affects NSCLC progression through regulating EIF4G1 in a p53-independent manner [94].
In addition to USP10 controlling cell proliferation, a study from the Ouchida group revealed that USP10 promotes tumor migration or invasion. Epithelial-to-mesenchymal transition (EMT) plays an essential role in the process of tumor invasion. USP10 facilitates tumor migration by stabilizing the protein abundance of the EMT transcription factor Slug [7]. Moreover, USP10 activates the Raf-1/MEK/ERK pathway, which is an important regulator of EMT [60]. Functionally, USP10 stabilizes the ITCH E3 ligase, enhancing the stability of MEK1 and activating the Raf-1/MEK/ERK pathway. Recently, our group reported that ITCH polyubiquitinated and activated BRAF in melanoma cells in response to proinflammatory cytokines, leading to the elevation of MEK/ERK signaling [95]. This further supports the role of USP10 in activating the Raf-1/MEK/ERK pathway. In addition, Spain-1, an inhibitor of USP10, inhibits melanoma growth and improves the anticancer effect of cisplatin by inhibiting USP10 activity [87,88]. However, the underlying mechanism remains largely unknown.
4.2. USP10 as a Tumor Suppressor
To understand the role of USP10 in liver cancer, Lu et al. analyzed 74 pairs of paraffin-embedded tissues of HCC patients and adjacent non-tumor specimens (61 men and 13 women) and found that compared to low levels of USP10, high levels of USP10 predicted longer disease-free survival and overall survival. In addition, USP10’s mRNA expression was downregulated in clinical HCC tissue samples compared with adjacent non-tumor samples [96]. Mechanistically, USP10 stabilizes the expression of PTEN and AMPKα through inhibiting the Lys48-linked polyubiquitylation of PTEN and regulating the K63-linked ubiquitin chain of AMPKα; all these inhibit mTOR activation and suppress the proliferation of HCC cell lines [96]. As in HCC, USP10 was found to be downregulated in lung cancer, and the knockdown of USP10 inhibited PTEN ubiquitination and promoted tumor growth and invasion [97]. Recently, Yu et al. demonstrated that the protein and mRNA levels of the tumor suppressor KLF4 were reduced in lung cancer tissues. Moreover, the depletion of KLF4 facilitates the development of lung cancer [98]. Wang et al. further found that USP10 maintains KLF4’s stability through deubiquitinating KLF4. Similarly, MSH2 (MutS Homolog 2), another USP10 substrate, was identified in lung cancers by Zhang et al. Their results show that the depletion of USP10 in A549 increased cell survival and decreased apoptosis through destabilizing MSH2 [32]. All these results indicate that in the same cancers, USP10 affects tumor development via regulating different substrates. Growing evidence has shown that USP10 plays important tumor-suppressor roles in different cancers, such as colorectal cancer, non-small-cell lung cancer, small intestinal adenocarcinoma, epithelial ovarian cancer, colorectal cancer, renal-cell carcinoma, breast cancer, and gastric cancer [99,100,101]. In 2020, Bhattacharya summarized the partial roles of USP10 in different cancers [9]. For example, in colon cancer, USP10 interacted with and stabilized SIRT6 through suppressing SIRT6 ubiquitination [25]. Thus, increasing the expression of c-Myc and p53 further inhibits cell-cycle progression, cell growth and tumorigenesis. In addition, it has also been shown that USP10 is silenced by methylation and downregulated in the early stages of colorectal cancer [99]. Taken together, all these findings suggest that USP10 also functions as a tumor suppressor. USP10 has lower expression in GC than in para-cancer tissues. More importantly, low expression of USP10 indicates poor prognosis in GC patients, suggesting that USP10 might be a promising prognostic marker in GC [102]. Interestingly, S100A12 (also named calgranulin C) has been found to be downregulated in GC clinical samples and positively correlated with USP10 expression. Previous studies using RCC (renal-cell carcinoma) tissue microarrays indicated that USP10 expression was reduced in RCC samples compared with normal renal tissues. The reconstitution of USP10 inhibited the colony formation and cell proliferation of the CAKI-1 and CAKI-2 RCC cell lines. However, it is still unknown whether USP10 can deubiquitinate and stabilize these related proteins. Further studies need to be conducted to uncover the underlying mechanisms [102,103]. More recently, Kim et al. demonstrated that USP10 effectively suppressed curcumin-induced paraptosis in malignant breast cancer cells through a mechanism not involving the regulation of beclin-1, p53, or AMPK. All these results indicate that USP10 may also function in a deubiquitinase-independent manner [104]. Given that USP10 may be a tumor suppressor, several upstream regulators of USP10 including miR-191 and DZNep were identified in pancreatic cancer and thyroid cancer cells, separately. Mechanistically, both of them promote cell proliferation via inhibiting the expression of USP10, which further decreases the stability of p53 [29,30].
Overall, all the above studies suggest that USP10 can act as a tumor suppressor via different molecular and cellular mechanisms.
5. Targeting USP10 in Human Cancers
Although USP10 is a double-edged sword and plays a dual role in tumorigenesis, it is widely considered a well-known oncogene in specific contexts as described above. In light of the important oncogenic role of USP10 in many cancers, inhibiting the expression or activity of USP10 is expected to be a promising therapeutic strategy for curing a range of human tumors. In line with the notion, many researchers have developed multiple screening methods to identify small-molecule inhibitors for cancer treatment, such as activity-based probes, Ub-7-amino-4-methylcoumarin(AMC), Ub-phospholipase A2 (PLA2), time-resolved fluorescence resonance energy transfer (TR-FRET), SDS-PAGE–Coomassie, and many others [105]. Different methods have different advantages and disadvantages. For example, UB-PLA2 cannot be used to screen inhibitors of the UCH family, but TR-FRET is very sensitive for screening UCH-family inhibitors. However, compared with UB-PLA2, a major problem is the lack of commercial kits available for TR-FRET methods. More details about these methods have been reviewed by Chen et al. [105]. In recent years, several inhibitors have been developed based on the structure or enzyme activity of USPs, such as GW7647 for USP1 [106], Q29 for USP2 [107], PR619 for USP2/4/5/7/8/15/20/28/47 [105], H9X19818 for USP7/10 [108], and VLX1570 for USP14 [109]. Among them, VLX1570 was approved for clinical trials as the first DUB inhibitor [109].
Given VLX1570’s potential to promote lung-tissue damage, caused by the accumulation of the drug’s metabolites, it has been demonstrated to lead to many adverse effects, especially severe respiratory insufficiency [105,110]. Therefore, screening and developing small-molecule inhibitors specifically targeting the USPs could be a potential and promising strategy for human cancer therapy. Below, we focus on the progress regarding USP10 inhibitors.
5.1. Spautin-1
In 2011, it was reported for the first time that spautin-1 was a highly potent autophagy inhibitor, as demonstrated using imaging-based screening. Spautin-1 inhibits autophagy by reducing the activity of USP10 and USP13, which promotes a major aspect of autophagy: degradation by Vps34 complexes. In addition, spautin-1 also downregulated the expression of USP10 and USP13 at the protein level [53]. Given the role of spautin-1 in autophagy, spautin-1 can improve the therapeutic efficacy of IM for CML patients. Mechanically, spautin-1 significantly inhibits imatinib mesylate (IM)-induced autophagy in CML cells by downregulating beclin-1 and enhances IM-induced apoptosis by inactivating PI3K/AKT and GSK-3β [111]. CFTR plays an important role in cystic fibrosis (CF) [112]. Although it has been shown that USP10 deubiquitinates and degrades CFTR [64], Pesce et al. found that spautin-1 could not affect the expression of CFTR, indicating that spautin-1 affected CF progression in a USP10-independent manner. However, the underlying mechanism is largely unknown. Interestingly, spautin-1 was recently shown to suppress cell growth or kill cancer cell lines in an autophagy-independent manner. A report from the Yang group revealed that spatutin-1 promoted immunogenic cell death (ICD) through the activation of the JUN transcription factor in response to mitochondrial oxidative injury, ultimately resulting in the upregulation of many cytokines including CXCL10 (C-X-C motif chemokine ligand 10) and IL-6 (interleukin-6) [113]. In addition, the inhibition of USP10 by spautin-1 significantly suppressed the migration and metastasis of HCC. Further evidence revealed that USP10 directly binds to Smad4 and stabilizes Smad4 through deubiquitination [46].
Moreover, spautin-1 suppresses the survival of ovarian cancer, prostate cancer, melanoma, and NSCLC cells in a USP10-independent manner [114]. More recently, spautin-A41, an analog of spautin-1, was developed by Elsocht et al. They found that both the autophagy-inhibiting effect and induction of microsomal stability were much better than those of spautin-1 [115]. However, there is still no evidence showing that spautin-1 or spautin-A41 directly binds to USP10. Further investigation is obviously warranted to determine whether spautin-1 and spautin-A41 could be used in clinical trials for cancer therapy in vivo.
5.2. P22077 and HBX19818 (or Analogs)
Similar to spautin-1, another two DUB inhibitors were found in 2001. Altun et al. found that PR-619 and P22077 inhibited DUB activity using an activity-based chemical proteomics screening method. They further validated that PR-619 inhibited a broad range of DUBs, but P22077 only targeted USP7. Almost simultaneously, another USP7 inhibitor, HBX19818, and its analogs were developed using biochemical assays and an activity-based protein-profiling strategy. It has been demonstrated that P22077 inhibits the proliferation of neuroblastoma, colon cancer, ovarian cancer, and lung cancer cells via different mechanisms including the induction of p53-mediated apoptosis [116,117,118]. Recently, P22077 was reported to inhibit the growth and metastasis of melanoma by activating the ATM/ATR signaling pathway [119]. Notably, P22077, HBX19818, and HBX19818 analogs including Compounds 3, 7, and 9 also inhibit USP10 in mutant-FLT3-expressing cells. Among the HBX19818 analogs, Compounds 3 and 9 are more specific for USP10 but not USP7. Furthermore, Compound 3 also has a stronger inhibitory activity at low concentrations than the others. In addition, the results indicate that the IC50 of HBX19818 for USP10 is 14 μM and that of P22077 is 6 μM in these cells. Additionally, the inhibitory activity is lower than that for USP7. Importantly, both of them can directly bind to and inhibit USP10. As expected, P22077, HBX19818, and its analogs inhibited the growth of acute myeloid leukemia harboring an FLT3 mutation [61]. On the other hand, it has been reported that USP10 is closely related to the DNA-damage response. It would be intriguing to explore whether a synergistic therapy combining USP10 and an effector of DNA damage would be a more efficient strategy for cancer treatment.
5.3. Wu-5
Wu-5, a novel USP10 inhibitor, was found by screening an in-house compound library, and it was shown to overcome FLT3-inhibitor resistance and enhance the anti-AML effect of crenolanib. Mechanistically, Wu-5 inhibits USP10 activity through interacting with USP10 in cells. Subsequently, it reduces the expression of the downstream effector AMPKα, suppressing the growth of MV4-11 cells [120]. However, further in-depth research is required to determine whether Wu-5 has broad-range effects and specificity in human cancer cells and clinical efficacy.
5.4. Quercetin
Quercetin (C15H10O7) is a pentahydroxyflavone, widely present in many fruits and vegetables. It exerts many effects combating different diseases, such as cancer, immunity diseases, and cardiovascular diseases. Dysregulation of the T-box transcriptional factor T-bet can cause many immune-mediated diseases [121,122]. A previous study by Pan et al. revealed that quercetin reduced the expression of USP10, which interacts with and maintains the level of T-bet, resulting in T-bet downregulation [14]. Therefore, quercetin was considered a T-bet inhibitor. Many studies have demonstrated that quercetin inhibits the growth of many cancer cells including those of breast cancer, colon cancer, lung cancer, and other cancers [122]. Although several clinical trials have been published, there is still no direct clinical evidence showing that it has any therapeutic effects in human cancers. Therefore, it is necessary to further clarify the role of quercetin and explore whether the inhibition of USP10 is the major mechanism for cancer treatment. It will also be interesting to screen many more flavonoids and develop quercetin derivatives targeting USP10 for anticancer therapy.
5.5. Traditional Chinese Medicine
Traditional Chinese Medicine plays a crucial role in the treatment of many diseases including cancer. In 2009, a medicine called Cai’s Neiyi Prescription (CNYP) was invented by Cai; it inhibits inflammation via inducing the apoptosis of endometrial stromal cells. Furthermore, they found that CNYP reduces USP10’s mRNA and protein expression. Altogether, it has been demonstrated that CNYP is an inhibitor of USP10, indicating that CNYP is a promising anticancer drug [12]. Given that Traditional Chinese Medicine has relatively few adverse effects, it will also be intriguing to test whether CNYP inhibits cancer cells’ proliferation or metastasis in vitro and in vivo, with the potential for its earlier use in patients.
5.6. UbV.10.1
Different from other USP10 inhibitors, UbV.10.1 is a mixture of proteins or peptides that have high affinity for USP10 and inhibit its activity. It was identified by screening a phage-displayed ubiquitin variant (UbV) library and considered an inhibitor of endogenous USP10 in cells. The overexpression of UbV.10.1 facilitates p53’s export from the nucleus to the cytoplasm and degradation through the inhibition of USP10 [123]. However, it is still unknown whether it can inhibit cancer cells’ proliferation or metastasis, etc. Additional in-depth investigation is necessary before using this inhibitor for clinical trials.
5.7. DZNep
3-deazaneplanocin A (DZNep) was developed based on the activity of S-adenosylhomocysteine hydrolase. It exerts anticancer effects in many cancers including blood and solid cancers through inhibiting EZH2 or other methyltransferase activity [124]. A previous report revealed that DZNEP suppressed the proliferation of TP53-wild-type cells but not TP53-mutant-type cells. One reason is that it can activate the p53 pathway by increasing USP10 expression, but it is more toxic to the majority of cancer cell lines; it has not been approved by the Food and Drug Administration (FDA). In 2020, another EZH2 inhibitor, tazemetostat, was approved by the FDA for metastatic epithelioid sarcoma treatment [125]. It is encouraging us to explore whether tazemetostat or other EZH2 inhibitors cure cancers in an USP10-independent manner. It also very interesting to test whether tazemetostat will be more efficient together with other pathway drugs such as P53, etc.
6. Conclusions and Perspectives
Since its discovery in 2001, USP10’s roles in regulating diverse cellular processes and different diseases, especially cancers, have been widely investigated [9,51]. In brief, USP10 is a double-edged sword in affecting human tumorigenesis due to the complex cellular contexts. Therefore, given the oncogenic role of USP10, the development of more specific USP10 inhibitors could be a potential and promising strategy for cancer treatment. To date, only Compounds 3 and 9 of the HBX19818 analogs are more specific for USP10, while other inhibitors, including spautin-1 (the first to be developed for USP10), are not specific for USP10 and have at least two targets [61,105]. Bearing this notion in mind, it is necessary and urgent to screen or develop much more specific USP10 inhibitors. In addition to directly targeting USP10, alternative approaches also need to be considered, such as the inhibition of USP10 by regulating its upstream regulators, including USP13, estrogen, and microRNAs (as summarized in Table 2). On the other hand, due to their nature and low side effects, using natural agents such as quercetin and Traditional Chinese Medicine as USP10 inhibitors could be a safe and effective strategy for cancer therapy. Although some upstream regulators and substrates of USP10 have been identified, in-depth investigations of both the underlying mechanisms of USP10-mediated tumorigenesis and the development of corresponding drugs are urgently needed. Mounting evidence indicates that enzymes also exert multiple functions in an enzymatic-activity-independent fashion, such as EZH2. Therefore, notably, we also need to pay more attention to exploring the additional role of USP10 outside its deubiquitinase activity. In light of the double-sided role of USP10 in tumorigenesis, it would be fascinating to explore the physiological role of USP10 in the progression of different tumors by using conditional knockout (KO) or knock-in mouse models. Looking forward, cancer-type-specific USP10 animal models will accelerate and improve the development of specific USP10 inhibitors. We believe that inhibitors of USP10 with the best specificity and efficacy will be developed and used in clinical trials for the therapy of different cancers in the near future.
Author Contributions
Conceptualization, T.H. and S.J.; writing—original draft preparation, L.T., T.H. and X.L. (Xiao Liu); writing—review and editing, L.T., T.H., X.L. (Xiao Liu), and X.J.; visualization: X.J., K.Z., Y.W. and X.L. (Xiumin Li); supervision: T.H. and S.L-J.; funding acquisition: T.H. and S.J. All authors have read and agreed to the published version of the manuscript.
Funding
T.H. was supported in part by the Henan National Science Fund for Excellent Young Scholars (No. 212300410067), the National Natural Science Foundation of China (Nos. 82002731 and 82172891), and the Doctoral Foundation of Xinxiang Medical University (No. XYBSKYZZ202001). S.J. was supported in part by the National Natural Science Foundation of China (No. 82074360) and Young Tai shan Scholars Program of Shandong Province (No. tsqn201909200). The APC was funded by the Henan National Science Fund for Excellent Young Scholars (No. 212300410067).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank all the members from the Han lab for the critical comments and helpful suggestions. We apologize for being unable to cite many important papers in this field due to space limitations.
Conflicts of Interest
The authors declare no conflict of interests.
References
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Kleiger, G.; Mayor, T. Perilous journey: A tour of the ubiquitin–proteasome system. Trends Cell Biol. 2014, 24, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C. Reversible Monoubiquitination of PCNA: A Novel Slant on Regulating Translesion DNA Synthesis. Mol. Cell 2006, 22, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Çetin, G.; Klafack, S.; Studencka-Turski, M.; Krüger, E.; Ebstein, F. The Ubiquitin–Proteasome System in Immune Cells. Biomolecules 2021, 11, 60. [Google Scholar] [CrossRef]
- Baek, K.-H. Conjugation and deconjugation of ubiquitin regulating the destiny of proteins. Exp. Mol. Med. 2003, 35, 1–7. [Google Scholar] [CrossRef]
- Zhang, L.; Gong, F. Involvement of USP24 in the DNA damage response. Mol. Cell. Oncol. 2016, 3, e1011888. [Google Scholar] [CrossRef][Green Version]
- Ouchida, A.T.; Kacal, M.; Zheng, A.; Ambroise, G.; Zhang, B.; Norberg, E.; Vakifahmetoglu-Norberg, H. USP10 regulates the stability of the EMT-transcription factor Slug/SNAI2. Biochem. Biophys. Res. Commun. 2018, 502, 429–434. [Google Scholar] [CrossRef]
- Lei, H.; Shan, H.; Wu, Y. Targeting deubiquitinating enzymes in cancer stem cells. Cancer Cell Int. 2017, 17, 101. [Google Scholar] [CrossRef]
- Bhattacharya, U.; Neizer-Ashun, F.; Mukherjee, P.; Bhattacharya, R. When the chains do not break: The role of USP10 in physiology and pathology. Cell Death Dis. 2020, 11, 1033. [Google Scholar] [CrossRef]
- Lawson, A.P.; Bak, D.W.; Shannon, D.A.; Long, M.J.C.; Vijaykumar, T.; Yu, R.; El Oualid, F.; Weerapana, E.; Hedstrom, L. Identification of deubiquitinase targets of isothiocyanates using SILAC-assisted quantitative mass spec-trometry. Oncotarget 2017, 8, 51296–51316. [Google Scholar] [CrossRef]
- Jung, Y.; Kim, H.D.; Yang, H.W.; Kim, H.J.; Jang, C.-Y.; Kim, J. Modulating cellular balance of Rps3 mono-ubiquitination by both Hel2 E3 ligase and Ubp3 deubiquitinase regulates protein quality control. Exp. Mol. Med. 2017, 49, e390. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhu, Y.; Zhang, T.; Hang, Y.; Chen, Q.; Jin, Y. Cai’s Neiyi Prescription promotes apoptosis and inhibits inflammation in endometrial stromal cells with endome-triosis through inhibiting USP10. Biotechnol. Appl. Biochem. 2019, 66, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Chen, Z.; Wang, L.; Chen, C.; Li, D.; Wan, H.; Li, B.; Shi, G. Deubiquitination and stabilization of T-bet by USP10. Biochem. Biophys. Res. Commun. 2014, 449, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Yang, X.; Qin, B.; Liu, T.; Zhang, H.; Guo, W.; Lee, S.B.; Kim, J.J.; Yuan, J.; Pei, H.; et al. Deubiquitination and Activation of AMPK by USP10. Mol. Cell 2016, 61, 614–624. [Google Scholar] [CrossRef]
- Xia, X.; Hu, T.; He, J.; Xu, Q.; Yu, C.; Liu, X.; Shao, Z.; Liao, Y.; Huang, H.; Liu, N. USP10 deletion inhibits macrophage-derived foam cell formation and cellular-oxidized low density lipoprotein uptake by promoting the degradation of CD36. Aging 2020, 12, 22892–22905. [Google Scholar] [CrossRef]
- Shen, W.; Zhang, Z.; Ma, J.; Lu, D.; Lyu, L. The Ubiquitin Proteasome System and Skin Fibrosis. Mol. Diagn. Ther. 2021, 25, 29–40. [Google Scholar] [CrossRef]
- Ballar Kirmizibayrak, P.; Erbaykent-Tepedelen, B.; Gozen, O.; Erzurumlu, Y. Divergent Modulation of Proteostasis in Prostate Cancer. Adv. Exp. Med. Biol. 2020, 1233, 117–151. [Google Scholar] [CrossRef]
- Aulas, A.; Finetti, P.; Lyons, S.M.; Bertucci, F.; Birnbaum, D.; Acquaviva, C.; Mamessier, E. Revisiting the Concept of Stress in the Prognosis of Solid Tumors: A Role for Stress Granules Proteins? Cancers 2020, 12, 2470. [Google Scholar] [CrossRef]
- Anisimov, S.; Takahashi, M.; Kakihana, T.; Katsuragi, Y.; Kitaura, H.; Zhang, L.; Kakita, A.; Fujii, M. G3BP1 inhibits ubiquitinated protein aggregations induced by p62 and USP10. Sci. Rep. 2019, 9, 12896. [Google Scholar] [CrossRef]
- Kwon, S.-K.; Saindane, M.; Baek, K.-H. p53 stability is regulated by diverse deubiquitinating enzymes. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Satija, Y.K.; Bhardwaj, A.; Das, S. A portrayal of E3 ubiquitin ligases and deubiquitylases in cancer. Int. J. Cancer 2013, 133, 2759–2768. [Google Scholar] [CrossRef] [PubMed]
- Emami, S. Interplay between p53-family, their regulators, and PARPs in DNA repair. Clin. Res. Hepatol. Gastroenterol. 2011, 35, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Panas, M.D.; Schulte, T.; Thaa, B.; Sandalova, T.; Kedersha, N.; Achour, A.; McInerney, G.M. Viral and Cellular Proteins Containing FGDF Motifs Bind G3BP to Block Stress Granule Formation. PLoS Pathog. 2015, 11, e1004659. [Google Scholar] [CrossRef]
- Lin, Z.; Yang, H.; Tan, C.; Li, J.; Liu, Z.; Quan, Q.; Kong, S.; Ye, J.; Gao, B.; Fang, D. USP10 Antagonizes c-Myc Transcriptional Activation through SIRT6 Stabilization to Suppress Tumor Formation. Cell Rep. 2013, 5, 1639–1649. [Google Scholar] [CrossRef] [PubMed]
- Simeoni, F.; Tasselli, L.; Tanaka, S.; Villanova, L.; Hayashi, M.; Kubota, K.; Isono, F.; Garcia, B.A.; Michishita-Kioi, E.; Chua, K.F. Proteomic analysis of the SIRT6 interactome: Novel links to genome maintenance and cellular stress signaling. Sci. Rep. 2013, 3, 3085. [Google Scholar] [CrossRef]
- Yuan, J.; Luo, K.; Zhang, L.; Cheville, J.C.; Lou, Z. USP10 Regulates p53 Localization and Stability by Deubiquitinating p53. Cell 2010, 140, 384–396. [Google Scholar] [CrossRef]
- Liu, H.; Xu, X.-F.; Zhao, Y.; Tang, M.-C.; Zhou, Y.-Q.; Lu, J.; Gao, F.-H. MicroRNA-191 promotes pancreatic cancer progression by targeting USP10. Tumor Biol. 2014, 35, 12157–12163. [Google Scholar] [CrossRef]
- Cui, B.; Yang, Q.; Guan, H.; Shi, B.; Hou, P.; Ji, M. PRIMA-1, a Mutant p53 Reactivator, Restores the Sensitivity of TP53 Mutant-Type Thyroid Cancer Cells to the Histone Methylation Inhibitor 3-Deazaneplanocin A. J. Clin. Endocrinol. Metab. 2014, 99, E962–E970. [Google Scholar] [CrossRef]
- Cheng, L.L.; Itahana, Y.; Lei, Z.D.; Chia, N.-Y.; Wu, Y.; Yu, Y.; Zhang, S.L.; Thike, A.A.; Pandey, A.; Rozen, S.; et al. TP53 Genomic Status Regulates Sensitivity of Gastric Cancer Cells to the Histone Methylation Inhibitor 3-Deazaneplanocin A (DZNep). Clin. Cancer Res. 2012, 18, 4201–4212. [Google Scholar] [CrossRef]
- Luo, Z.; Zhang, M.; Cui, R.; Tili, E.; Kim, T.; Lee, T.J.; Peng, Y.; Croce, C. A negative feedback regulatory loop between miR-138 and TP53 is mediated by USP10. Oncotarget 2019, 10, 6288–6296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, C.; Tong, D.; Xiang, S.; Williams, K.; Bai, W.; Li, G.-M.; Bepler, G.; Zhang, X. Ubiquitin-specific Peptidase 10 (USP10) Deubiquitinates and Stabilizes MutS Homolog 2 (MSH2) to Regulate Cellular Sensitivity to DNA Damage. J. Biol. Chem. 2016, 291, 10783–10791. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Huang, X.; Xin, H.-B.; Fu, M.; Xue, A.; Wu, Z.-H. TRAF Family Member-associated NF-κB Activator (TANK) Inhibits Genotoxic Nuclear Factor κB Activation by Facilitating Deubiquitinase USP10-dependent Deubiquitination of TRAF6 Ligase. J. Biol. Chem. 2015, 290, 13372–13385. [Google Scholar] [CrossRef] [PubMed]
- Zella, D.; Curreli, S.; Benedetti, F.; Krishnan, S.; Cocchi, F.; Latinovic, O.S.; Denaro, F.; Romerio, F.; Djavani, M.; Charurat, M.E.; et al. Mycoplasma promotes malignant transformation in vivo, and its DnaK, a bacterial chaperone protein, has broad oncogenic properties. Proc. Natl. Acad. Sci. USA 2018, 115, E12005–E12014. [Google Scholar] [CrossRef]
- Rodriguez, J.; Herrero, A.; Li, S.; Rauch, N.; Quintanilla, A.; Wynne, K.; Krstic, A.; Acosta, J.C.; Taylor, C.; Schlisio, S.; et al. PHD3 Regulates p53 Protein Stability by Hydroxylating Proline 359. Cell Rep. 2018, 24, 1316–1329. [Google Scholar] [CrossRef]
- Oi, N.; Yuan, J.; Malakhova, M.; Luo, K.; Li, Y.; Ryu, J.; Zhang, L.; Bode, A.M.; Xu, Z.; Lou, Z.; et al. Resveratrol induces apoptosis by directly targeting Ras-GTPase-activating protein SH3 domain-binding protein 1. Oncogene 2015, 34, 2660–2671. [Google Scholar] [CrossRef]
- Zhao, W.; Lu, D.; Liu, L.; Cai, J.; Zhou, Y.; Yang, Y.; Zhang, Y.; Zhang, J. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) promotes lung tumorigenesis via attenuating p53 stability. Oncotarget 2017, 8, 93672–93687. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef]
- Bomberger, J.M.; Barnaby, R.L.; Stanton, B.A. The deubiquitinating enzyme USP10 regulates the endocytic recycling of CFTR in airway epithelial cells. Channels 2010, 4, 150–154. [Google Scholar] [CrossRef]
- Adsi, H.; Levkovich, S.A.; Haimov, E.; Kreiser, T.; Meli, M.; Engel, H.; Simhaev, L.; Karidi-Heller, S.; Colombo, G.; Gazit, E.; et al. Chemical Chaperones Modulate the Formation of Metabolite Assemblies. Int. J. Mol. Sci. 2021, 22, 9172. [Google Scholar] [CrossRef]
- Chen, R.; Pang, X.; Li, L.; Zeng, Z.; Chen, M.; Zhang, S. Ubiquitin-specific proteases in inflammatory bowel disease-related signalling pathway regulation. Cell Death Dis. 2022, 13, 139. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Li, P.; Song, L.; Jeffrey, P.D.; Chernova, T.A.; Wilkinson, K.D.; Cohen, R.E.; Shi, Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005, 24, 3747–3756. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.D. Ubiquitination and deubiquitination: Targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 2000, 11, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Reed, B.J.; Locke, M.; Gardner, R.G. A Conserved Deubiquitinating Enzyme Uses Intrinsically Disordered Regions to Scaffold Multiple Protein Interaction Sites. J. Biol. Chem. 2015, 290, 20601–20612. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbe, S. Deubiquitylases from Genes to Organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef]
- Yuan, T.; Chen, Z.; Yan, F.; Qian, M.; Luo, H.; Ye, S.; Cao, J.; Ying, M.; Dai, X.; Gai, R.; et al. Deubiquitinating enzyme USP10 promotes hepatocellular carcinoma metastasis through deubiquitinating and stabilizing Smad4 protein. Mol. Oncol. 2020, 14, 197–210. [Google Scholar] [CrossRef]
- He, Y.; Jiang, S.; Mao, C.; Zheng, H.; Cao, B.; Zhang, Z.; Zhao, J.; Zeng, Y.; Mao, X. The deubiquitinase USP10 restores PTEN activity and inhibits non–small cell lung cancer cell proliferation. J. Biol. Chem. 2021, 297, 101088. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Xiao, L.; Zhai, C.; Yi, T.; Wang, G.; Wang, E.; Ji, X.; Hu, L.; Shen, G.; et al. Usp10 Modulates the Hippo Pathway by Deubiquitinating and Stabilizing the Transcriptional Coactivator Yorkie. Int. J. Mol. Sci. 2019, 20, 6013. [Google Scholar] [CrossRef]
- Faus, H.; Meyer, H.-A.; Huber, M.; Bahr, I.; Haendler, B. The ubiquitin-specific protease USP10 modulates androgen receptor function. Mol. Cell. Endocrinol. 2005, 245, 138–146. [Google Scholar] [CrossRef]
- Kashiwaba, S.-I.; Kanao, R.; Masuda, Y.; Kusumoto-Matsuo, R.; Hanaoka, F.; Masutani, C. USP7 Is a Suppressor of PCNA Ubiquitination and Oxidative-Stress-Induced Mutagenesis in Human Cells. Cell Rep. 2015, 13, 2072–2080. [Google Scholar] [CrossRef]
- Jochemsen, A.G.; Shiloh, Y. USP10: Friend and Foe. Cell 2010, 140, 308–310. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Kitaura, H.; Kakita, A.; Kakihana, T.; Katsuragi, Y.; Nameta, M.; Zhang, L.; Iwakura, Y.; Nawa, H.; Higuchi, M.; et al. USP10 Is a Driver of Ubiquitinated Protein Aggregation and Aggresome Formation to Inhibit Apoptosis. iScience 2018, 9, 433–450. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, H.; Kim, M.; Xu, L.; Li, Y.; Zhang, L.; Cai, Y.; Norberg, H.V.; Zhang, T.; Furuya, T.; et al. Beclin1 Controls the Levels of p53 by Regulating the Deubiquitination Activity of USP10 and USP13. Cell 2011, 147, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.; Sugino, T.; Nolte, H.; Andrade, J.; Zimmermann, B.; Shi, C.; Doddaballapur, A.; Ong, Y.T.; Wilhelm, K.; Fasse, J.W.D.; et al. Deubiquitinase USP10 regulates Notch signaling in the endothelium. Science 2019, 364, 188–193. [Google Scholar] [CrossRef]
- Simpson-Holley, M.; Kedersha, N.; Dower, K.; Rubins, K.H.; Anderson, P.; Hensley, L.E.; Connor, J.H. Formation of Antiviral Cytoplasmic Granules during Orthopoxvirus Infection. J. Virol. 2011, 85, 1581–1593. [Google Scholar] [CrossRef]
- Soncini, C.; Berdo, I.; Draetta, G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 2001, 20, 3869–3879. [Google Scholar] [CrossRef]
- Meyer, C.; Garzia, A.; Morozov, P.; Molina, H.; Tuschl, T. The G3BP1-Family-USP10 Deubiquitinase Complex Rescues Ubiquitinated 40S Subunits of Ribosomes Stalled in Translation from Lysosomal Degradation. Mol. Cell 2020, 77, 1193–1205.e5. [Google Scholar] [CrossRef]
- Hu, C.; Zhang, M.; Moses, N.; Hu, C.-L.; Polin, L.; Chen, W.; Jang, H.; Heyza, J.; Malysa, A.; Caruso, J.A.; et al. The USP10-HDAC6 axis confers cisplatin resistance in non-small cell lung cancer lacking wild-type p53. Cell Death Dis. 2020, 11, 328. [Google Scholar] [CrossRef]
- Guturi, K.K.N.; Bohgaki, M.; Bohgaki, T.; Srikumar, T.; Ng, D.; Kumareswaran, R.; El Ghamrasni, S.; Jeon, J.; Patel, P.; Eldin, M.S.; et al. RNF168 and USP10 regulate topoisomerase IIα function via opposing effects on its ubiquitylation. Nat. Commun. 2016, 7, 12638–12713. [Google Scholar] [CrossRef]
- Chen, Q.; Hang, Y.; Zhang, T.; Tan, L.; Li, S.; Jin, Y. USP10 promotes proliferation and migration and inhibits apoptosis of endometrial stromal cells in endome-triosis through activating the Raf-1/MEK/ERK pathway. Am. J. Physiol. Cell Physiol. 2018, 315, C863–C872. [Google Scholar] [CrossRef]
- Weisberg, E.L.; Schauer, N.; Yang, J.; Lamberto, I.; Doherty, L.; Bhatt, S.; Nonami, A.; Meng, C.; Letai, A.; Wright, R.; et al. Inhibition of USP10 induces degradation of oncogenic FLT3. Nat. Chem. Biol. 2017, 13, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Meng, C.; Weisberg, E.; Case, A.; Lamberto, I.; Magin, R.S.; Adamia, S.; Wang, J.; Gray, N.; Liu, S.; et al. Inhibition of the deubiquitinase USP10 induces degradation of SYK. Br. J. Cancer 2020, 122, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Piatnitskaia, S.; Takahashi, M.; Kitaura, H.; Katsuragi, Y.; Kakihana, T.; Zhang, L.; Kakita, A.; Iwakura, Y.; Nawa, H.; Miura, T.; et al. USP10 is a critical factor for Tau-positive stress granule formation in neuronal cells. Sci. Rep. 2019, 9, 10591. [Google Scholar] [CrossRef]
- Bomberger, J.M.; Barnaby, R.L.; Stanton, B.A.; Yang, H.S.; Kim, E.; Lee, S.; Park, H.J.; Cooper, D.S.; Rajbhandari, I.; Choi, I. The Deubiquitinating Enzyme USP10 Regulates the Post-endocytic Sorting of Cystic Fibrosis Transmembrane Conductance Regulator in Airway Epithelial Cells. J. Biol. Chem. 2009, 284, 18778–18789. [Google Scholar] [CrossRef]
- Boulkroun, S.; Ruffieux-Daidié, D.; Vitagliano, J.-J.; Poirot, O.; Charles, R.-P.; Lagnaz, D.; Firsov, V.; Kellenberger, S.; Staub, O. Vasopressin-inducible ubiquitin-specific protease 10 increases ENaC cell surface expression by deubiq-uitylating and stabilizing sorting nexin 3. Am. J. Physiol. Renal Physiol. 2008, 295, F889–F900. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, S.R.; Tedeso, L.J.; Wang, L.; Bernstain, A.J. The deubiquitylase USP10 regulates integrin β1 and β5 and fibrotic wound healing. J. Cell Sci. 2017, 130, 3481–3495. [Google Scholar] [PubMed]
- Jia, R.; Bonifacino, J.S. The ubiquitin isopeptidase USP10 deubiquitinates LC3B to increase LC3B levels and autophagic activity. J. Biol. Chem. 2021, 296, 100405. [Google Scholar] [CrossRef]
- Li, B.; Qi, Z.-P.; He, D.-L.; Chen, Z.-H.; Liu, J.-Y.; Wong, M.-W.; Zhang, J.-W.; Xu, E.-P.; Shi, Q.; Cai, S.-L.; et al. NLRP7 deubiquitination by USP10 promotes tumor progression and tumor-associated macrophage polarization in colorectal cancer. J. Exp. Clin. Cancer Res. 2021, 40, 126. [Google Scholar] [CrossRef]
- Ko, A.; Han, S.Y.; Choi, C.H.; Cho, H.; Lee, M.-S.; Kim, S.-Y.; Song, J.S.; Hong, K.-M.; Lee, H.-W.; Hewitt, S.M.; et al. Oncogene-induced senescence mediated by c-Myc requires USP10 dependent deubiquitination and stabilization of p14ARF. Cell Death Differ. 2018, 25, 1050–1062. [Google Scholar] [CrossRef]
- Wang, X.; Xia, S.; Li, H.; Wang, X.; Li, C.; Chao, Y.; Zhang, L.; Han, Z. The deubiquitinase USP10 regulates KLF4 stability and suppresses lung tumorigenesis. Cell Death Differ. 2020, 27, 1747–1764. [Google Scholar] [CrossRef]
- Villamil, M.A.; Liang, Q.; Chen, J.; Choi, Y.S.; Hou, S.; Lee, K.H.; Zhuang, Z. Serine Phosphorylation Is Critical for the Activation of Ubiquitin-Specific Protease 1 and Its Interaction with WD40-Repeat Protein UAF1. Biochemistry 2012, 51, 9112–9123. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhang, X.; Chen, R.; Li, R.; Liu, Y.; Zhang, J.; Liu, Q.; Si, M.; Liu, J.; Wu, B.; et al. USP10 regulates B cell response to SARS-CoV-2 or HIV-1 nanoparticle vaccines through deubiquitinating AID. Signal Transduct. Target Ther. 2022, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.C.; Zhu, D.H.; Chen, Y.J.; Huang, T.Y.; Peng, Y.; Liu, S.Y.; Lu, P.; Xue, Y.H.; Xu, Y.P.; Yang, B.; et al. TRAF4 Promotes Fibroblast Proliferation in Keloids by Destabilizing p53 via Interacting with the Deubiquitinase USP10. J. Invest Dermatol. 2019, 139, 1925–1935.e5. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Shi, Y.; Iwai, K.; Wu, Z.H. LUBAC regulates NF-kappaB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 2011, 30, 3741–3753. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Shi, Y.; Xue, J.; Miao, R.; Huang, S.; Wang, T.; Wu, J.; Fu, M.; Wu, Z.H. USP10 inhibits genotoxic NF-kappaB activation by MCPIP1-facilitated deubiquitination of NEMO. EMBO J. 2013, 32, 3206–3219. [Google Scholar] [CrossRef]
- Hinz, M.; Stillman, M.; Çöl Arslan, S.; Khanna, K.K.; Dittmar, G.; Scheidereit, C. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol. Cell. 2010, 40, 63–74. [Google Scholar] [CrossRef]
- Esposito, M.; Akman, H.B.; Giron, P.; Ceregido, M.A.; Schepers, R.; Paez, L.C.R.; La Monaca, E.; De Greve, J.; Coux, O.; De Trez, C.; et al. USP13 controls the stability of Aurora B impacting progression through the cell cycle. Oncogene 2020, 39, 6009–6023. [Google Scholar] [CrossRef]
- Takahashi, M.; Higuchi, M.; Makokha, G.N.; Matsuki, H.; Yoshita, M.; Tanaka, Y.; Fujii, M. HTLV-1 Tax oncoprotein stimulates ROS production and apoptosis in T cells by interacting with USP10. Blood 2013, 122, 715–725. [Google Scholar] [CrossRef]
- Zhang, B.; Li, H.; Li, D.; Sun, H.; Li, M.; Hu, H. Long noncoding RNA Mirt2 upregulates USP10 expression to suppress hepatic steatosis by sponging miR-34a-5p. Gene 2019, 700, 139–148. [Google Scholar] [CrossRef]
- Phillips, A.T.; Boumil, E.F.; Venkatesan, A.; Castro, N.; Bernstein, A.M. Daam1 negatively regulates USP10 activity. bioRxiv 2022. [Google Scholar] [CrossRef]
- Bolaender, A.; Zatorska, D.; He, H.; Joshi, S.; Sharma, S.; Digwal, C.S.; Patel, H.J.; Sun, W.; Imber, B.S.; Ochiana, S.O.; et al. Chemical tools for epichaperome-mediated interactome dysfunctions of the central nervous system. Nat. Commun. 2021, 12, 4669. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Tao, H.; Wu, Q.; Huang, S.-Y.; Xiao, Y. Modeling of the Long-Term Epidemic Dynamics of COVID-19 in the United States. Int. J. Environ. Res. Public Health 2021, 18, 7594. [Google Scholar] [CrossRef] [PubMed]
- Grunda, J.M.; Nabors, L.; Palmer, C.; Chhieng, D.C.; Steg, A.; Mikkelsen, T.; Diasio, R.B.; Zhang, K.; Allison, D.; Grizzle, W.E.; et al. Increased Expression of Thymidylate Synthetase (TS), Ubiquitin Specific Protease 10 (USP10) and Survivin is Associated with Poor Survival in Glioblastoma Multiforme (GBM). J. Neuro-Oncol. 2006, 80, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Liu, T.; Liu, X.; Zhu, F.; Zhu, F.; Liu, X.; Peng, Z. USP10 regulates Musashi-2 stability via deubiquitination and promotes tumour proliferation in colon cancer. FEBS Lett. 2019, 593, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Liu, N.; Xia, X.; Guo, Z.; Li, Y.; Jiang, L.; Zhou, R.; Tang, D.; Huang, H.; Liu, J. USP10 modulates the SKP2/Bcr-Abl axis via stabilizing SKP2 in chronic myeloid leukemia. Cell Discov. 2019, 5, 24. [Google Scholar] [CrossRef]
- Ren, R. Mechanisms of BCR–ABL in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer 2005, 5, 172–183. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, J.L.; Liang, L.; Liu, N.; Qi, M.; Zhao, S.; Su, J.; Liu, J.; Peng, C.; Chen, X.; et al. Potent USP10/13 antagonist spautin-1 suppresses melanoma growth via ROS-mediated DNA damage and ex-hibits synergy with cisplatin. J. Cell. Mol. Med. 2020, 24, 4324–4340. [Google Scholar] [CrossRef]
- Liao, Y.; Guo, Z.; Xia, X.; Liu, Y.; Huang, C.; Jiang, L.; Wang, X.; Liu, J.; Huang, H. Inhibition of EGFR signaling with Spautin-1 represents a novel therapeutics for prostate cancer. J. Exp. Clin. Cancer Res. 2019, 38, 157. [Google Scholar] [CrossRef]
- Kumari, N.; Saei, A.; Lalith, C.; Serra, V.; Jha, S.; Eichhorn, P. Identifying the oncogenic role of USP10 as the regulator of PTEN function in breast cancer. Ann. Oncol. 2018, 29, iii10–iii11. [Google Scholar] [CrossRef]
- Zhu, H.; Yan, F.; Yuan, T.; Qian, M.; Zhou, T.; Dai, X.; Cao, J.; Ying, M.; Dong, X.; He, Q.; et al. USP10 Promotes Proliferation of Hepatocellular Carcinoma by Deubiquitinating and Stabilizing YAP/TAZ. Cancer Res. 2020, 80, 2204–2216. [Google Scholar] [CrossRef]
- Draker, R.; Sarcinella, E.; Cheung, P. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 2011, 39, 3529–3542. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Ou, C.; Liu, M.; Xiao, W.; Wen, C.; Sun, F. NRAGE promotes cell proliferation by stabilizing PCNA in a ubiquitin–proteasome pathway in esophageal carcinomas. Carcinogenesis 2014, 35, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Li, D.; Yu, T.; Huang, Y.; Yan, H.; Gu, L.; Yuan, J. Association and clinical implication of the USP10 and MSH2 proteins in non-small cell lung cancer. Oncol. Lett. 2019, 17, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wei, M.; Li, B.; Liu, Y.; Lu, Y.; Tang, Z.; Lu, T.; Yin, Y.; Qin, Z.; Xu, Z. Functional role of eukaryotic translation initiation factor 4 gamma 1 (EIF4G1) in NSCLC. Oncotarget 2016, 7, 24242–24251. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Han, T.; Fang, B.; Zhang, G.; Zhang, C.; Roberts, E.R.; Izumi, V.; Zheng, M.; Jiang, S.; Yin, X.; et al. K27-linked ubiquitination of BRAF by ITCH engages cytokine response to maintain MEK-ERK signaling. Nat. Commun. 2019, 10, 1870. [Google Scholar] [CrossRef]
- Lu, C.; Ning, Z.; Wang, A.; Chen, D.; Liu, X.; Xia, T.; Tekcham, D.S.; Wang, W.; Li, T.; Liu, X.; et al. USP10 suppresses tumor progression by inhibiting mTOR activation in hepatocellular carcinoma. Cancer Lett. 2018, 436, 139–148. [Google Scholar] [CrossRef]
- Sun, J.; Li, T.; Zhao, Y.; Huang, L.; Sun, H.; Wu, H.; Jiang, X. USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Mol. Cell. Biochem. 2018, 441, 1–7. [Google Scholar] [CrossRef]
- Yu, T.; Chen, X.; Zhang, W.; Liu, J.; Avdiushko, R.; Napier, D.L.; Liu, A.X.; Neltner, J.M.; Wang, C.; Cohen, D.; et al. KLF4 regulates adult lung tumor-initiating cells and represses K-Ras-mediated lung cancer. Cell Death Differ. 2016, 23, 207–215. [Google Scholar] [CrossRef]
- Kim, K.; Huh, T.; Park, Y.; Koo, D.-H.; Kim, H.; Hwang, I.; Choi, C.H.; Yi, J.M.; Chung, J.-Y. Prognostic significance of USP10 and p14ARF expression in patients with colorectal cancer. Pathol. Res. Pract. 2020, 216, 152988. [Google Scholar] [CrossRef]
- Han, G.H.; Chay, D.B.; Yi, J.M.; Cho, H.; Chung, J.-Y.; Kim, J.-H. Loss of Both USP10 and p14ARF Protein Expression Is an Independent Prognostic Biomarker for Poor Prognosis in Patients with Epithelial Ovarian Cancer. Cancer Genom. Proteom. 2019, 16, 553–562. [Google Scholar] [CrossRef]
- Song, J.S.; Yi, J.M.; Cho, H.; Choi, C.H.; Park, Y.; Chung, E.J.; Song, J.; Chung, J.-Y.; Hong, S.M. Dual loss of USP10 and p14ARF protein expression is associated with poor prognosis in patients with small intestinal adenocarcinoma. Tumour Biol. 2018, 40, 1010428318808678. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Wu, H.-X.; Zhan, N.; Huang, Y.-B.; Wang, Z.-S.; Yang, G.-F.; Wang, P.; Fu, G.-H. Prognostic significance of USP10 as a tumor-associated marker in gastric carcinoma. Tumor Biol. 2013, 35, 3845–3853. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zeng, Z.; Yu, T.; Qin, J.; Wu, J.; Song, J.-C.; Zhou, Z.-Y.; Yuan, J.-P. Expression and clinical implication of S100A12 in gastric carcinoma. Tumor Biol. 2014, 37, 6551–6559. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, D.M.; Woo, H.G.; Kim, K.D.; Lee, H.J.; Kwon, Y.-J.; Choi, K.S. RNAi Screening-based Identification of USP10 as a Novel Regulator of Paraptosis. Sci. Rep. 2019, 9, 4909. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, Y.; Zhou, H. Advances in the Development Ubiquitin-Specific Peptidase (USP) Inhibitors. Int. J. Mol. Sci. 2021, 22, 4546. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Dexheimer, T.S.; Ai, Y.; Liang, Q.; Villamil, M.A.; Inglese, J.; Maloney, D.J.; Jadhaw, A.; Simeonov, A.; Zhang, Z. Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem. Biol. 2011, 18, 390–400. [Google Scholar] [CrossRef]
- Ohayon, S.; Refua, M.; Hendler, A.; Aharoni, A.; Brik, A. Harnessing the Oxidation Susceptibility of Deubiquitinases for Inhibition with Small Molecules. Angew. Chem. Int. Ed. 2014, 54, 599–603. [Google Scholar] [CrossRef]
- Reverdy, C.; Conrath, S.; Lopez, R.; Planquette, C.; Atmanene, C.; Harpon, J.; Battaglia, V.; Vivat, V.; Sippl, W.; Colland, F. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem. Biol. 2012, 19, 467–477. [Google Scholar] [CrossRef]
- Wang, X.; D’Arcy, P.; Caulfield, T.R.; Paulus, A.; Chitta, K.; Mohanty, C.; Gullbo, J.; Chanan-Khan, A.; Linder, S. Synthesis and Evaluation of Derivatives of the Proteasome Deubiquitinase Inhibitor b-AP15. Chem. Biol. Drug Des. 2015, 86, 1036–1048. [Google Scholar] [CrossRef]
- Kharel, P.; Uprety, D.; Chandra, A.B.; Hu, Y.; Belur, A.A.; Dhakal, A. Bortezomib-Induced Pulmonary Toxicity: A Case Report and Review of Literature. Case Rep. Med. 2018, 2018, 2913124. [Google Scholar] [CrossRef]
- Shao, S.; Li, S.; Qin, Y.; Wang, X.; Yang, Y.; Bai, H.; Zhou, L.; Zhao, C.; Wang, C. Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia. Int. J. Oncol. 2014, 44, 1661–1668. [Google Scholar] [CrossRef] [PubMed]
- Pesce, E.; Sondo, E.; Ferrera, L.; Tomati, V.; Caci, E.; Scudieri, P.; Musante, I.; Renda, M.; Baatallah, N.; Servel, N.; et al. The Autophagy Inhibitor Spautin-1 Antagonizes Rescue of Mutant CFTR Through an Autophagy-Independent and USP13-Mediated Mechanism. Front. Pharmacol. 2018, 9, 1464. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, C.; Zhu, S.; Cao, L.; Kroemer, G.; Zeh, H.; Tang, D.; Kang, R. TFAM is a novel mediator of immunogenic cancer cell death. OncoImmunology 2018, 7, e1431086. [Google Scholar] [CrossRef] [PubMed]
- Correa, R.J.; Valdes, Y.R.; Peart, T.M.; Fazio, E.N.; Bertrand, M.; McGee, J.; Préfontaine, M.; Sugimoto, A.; DiMattia, G.E.; Shepherd, T.G. Combination of AKT inhibition with autophagy blockade effectively reduces ascites-derived ovarian cancer cell viability. Carcinogenesis 2014, 35, 1951–1961. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Chen, X.; Xie, L.; Yu, L.; Shen, M.; Wang, Y.; Wu, S.; Wang, J.; Lu, J.; Wei, G.; et al. Spautin-A41 Attenuates Cerulein-Induced Acute Pancreatitis through Inhibition of Dysregulated Autophagy. Biol. Pharm. Bull. 2019, 42, 1789–1798. [Google Scholar] [CrossRef]
- Li, J.; Han, Y.; Zhang, H.; Qian, Z.; Jia, W.; Gao, Y.; Zheng, H.; Li, B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem. Biophys. Res. Commun. 2019, 512, 479–485. [Google Scholar] [CrossRef]
- Fan, Y.-H.; Cheng, J.; Vasudevan, S.A.; Dou, J.; Zhang, H.; Patel, R.H.; Ma, I.T.; Rojas, Y.; Zhao, Y.; Yu, Y.; et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 2013, 4, e867. [Google Scholar] [CrossRef]
- Yuan, T.; Yan, F.; Ying, M.; Cao, J.; He, Q.; Zhu, H.; Yang, B. Inhibition of Ubiquitin-Specific Proteases as a Novel Anticancer Therapeutic Strategy. Front. Pharmacol. 2018, 9, 1080. [Google Scholar] [CrossRef]
- Xiang, M.; Liang, L.; Kuang, X.; Xie, Z.; Liu, J.; Zhao, S.; Su, J.; Chen, X.; Liu, H. Pharmacological inhibition of USP7 suppresses growth and metastasis of melanoma cells in vitro and in vivo. J. Cell. Mol. Med. 2021, 25, 9228–9240. [Google Scholar] [CrossRef]
- Yu, M.; Fang, Z.-X.; Wang, W.-W.; Zhang, Y.; Bu, Z.-L.; Liu, M.; Xiao, X.-H.; Zhang, Z.-L.; Zhang, X.-M.; Cao, Y.; et al. Wu-5, a novel USP10 inhibitor, enhances crenolanib-induced FLT3-ITD-positive AML cell death via inhibiting FLT3 and AMPK pathways. Acta Pharmacol. Sin. 2021, 42, 604–612. [Google Scholar] [CrossRef]
- Li, Y.; Yao, J.; Han, C.; Yang, J.; Chaudhry, M.T.; Wang, S.; Liu, H.; Yin, Y. Quercetin, Inflammation and Immunity. Nutrients 2016, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Farias, M.; Carrasco-Pozo, C. The Anti-Cancer Effect of Quercetin: Molecular Implications in Cancer Metabolism. Int. J. Mol. Sci. 2019, 20, 3177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sartori, M.A.; Makhnevych, T.; Federowicz, K.E.; Dong, X.; Liu, L.; Nim, S.; Dong, A.; Yang, J.; Li, Y.; et al. Generation and Validation of Intracellular Ubiquitin Variant Inhibitors for USP7 and USP10. J. Mol. Biol. 2017, 429, 3546–3560. [Google Scholar] [CrossRef]
- Wan, L.; Xu, K.; Wei, Y.; Zhang, J.; Han, T.; Fry, C.; Zhang, Z.; Wang, Y.V.; Huang, L.; Yuan, M.; et al. Phosphorylation of EZH2 by AMPK Suppresses PRC2 Methyltransferase Activity and Oncogenic Function. Mol. Cell 2018, 69, 279–291.e5. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Gong, Y.; Zhang, T.; Huang, J.; Tan, Z.; Xue, L. Finding an easy way to harmonize: A review of advances in clinical research and combination strategies of EZH2 inhibitors. Clin. Epigenet. 2021, 13, 62. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).