High-Throughput Sequencing Reveals Transcriptome Signature of Early Liver Development in Goat Kids
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction
2.3. Library Preparation and Sequencing
2.4. Read Mapping and Transcriptome Assembly
2.5. Differential Expression Analysis and Functional Enrichment
2.6. Construction of the Protein–Protein Interaction (PPI) Network
2.7. Quantitative Real-Time PCR (qRT-PCR) Validation
3. Results
3.1. Sequencing Data Summary
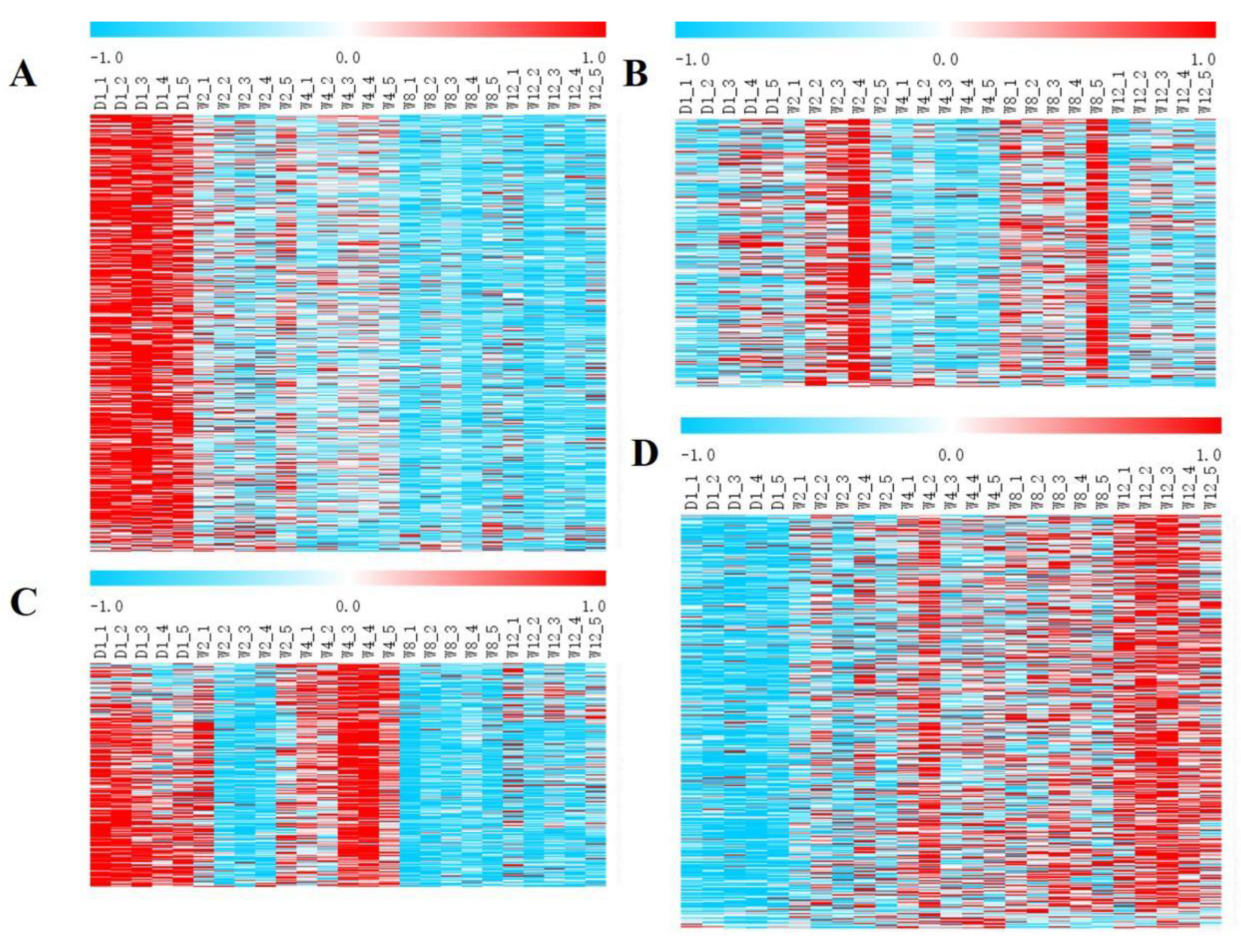
3.2. Identification and Analysis of DEGs
3.3. GO and KEGG Enrichment Analysis
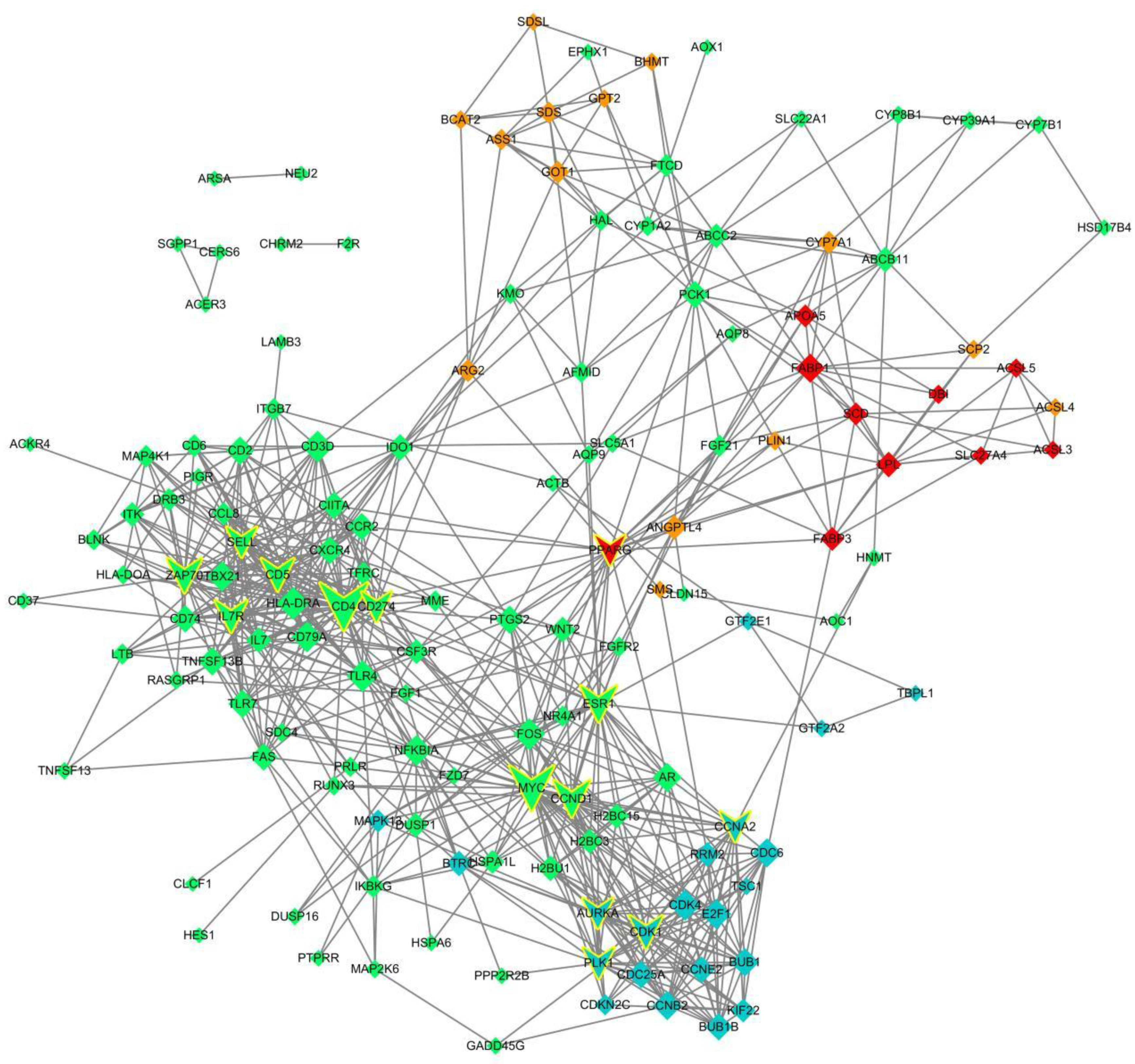
3.4. Screening of the Key Regulatory Genes
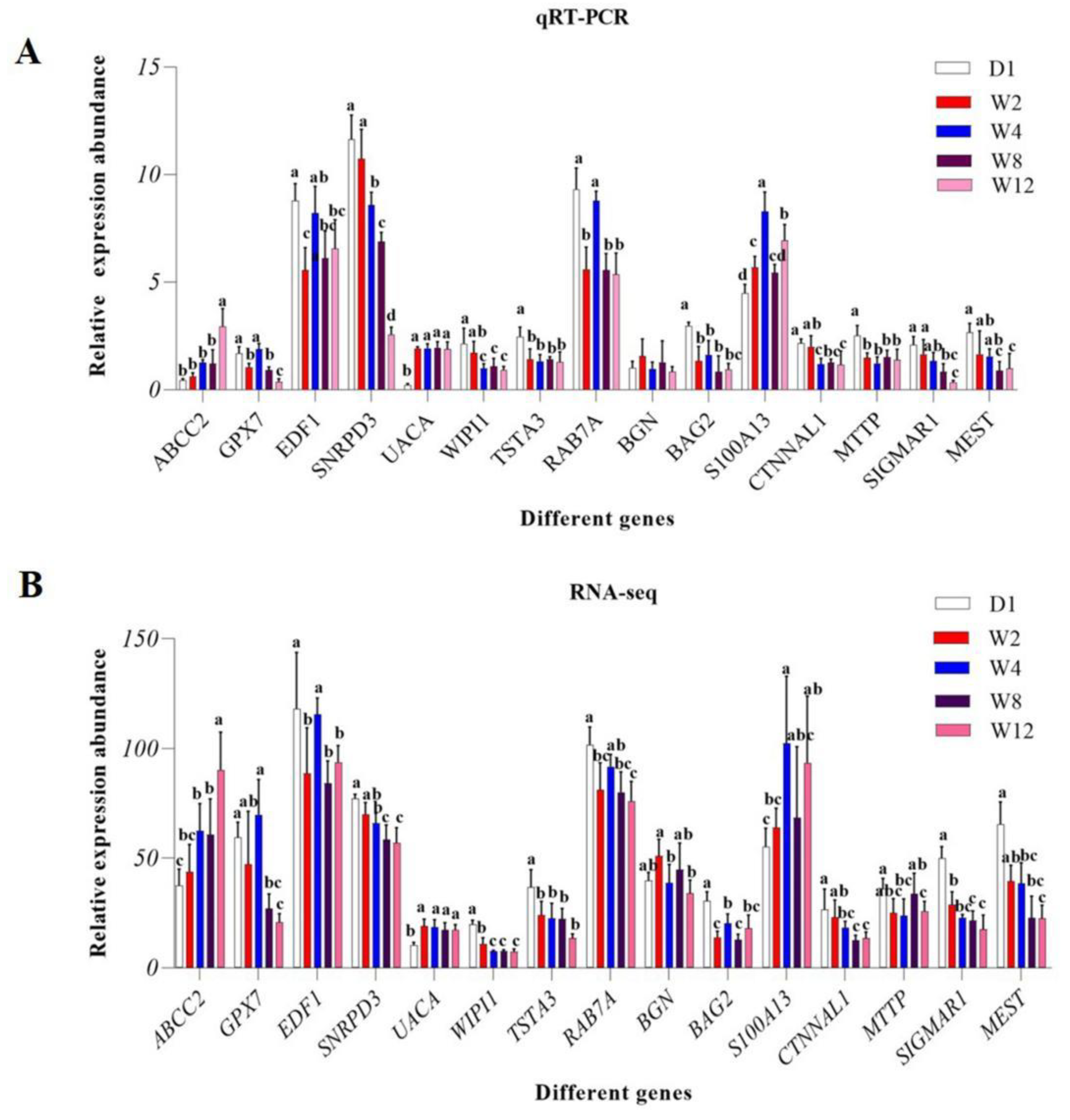
3.5. Validation of DEGs by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calne, R.Y. Immunological tolerance—The liver effect. Immunol. Rev. 2000, 174, 280–282. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Jiang, X.; Sun, D.; Han, G.; Wang, F.; Ye, M.; Wang, L.; Zou, H. Glycoproteomics Analysis of Human Liver Tissue by Combination of Multiple Enzyme Digestion and Hydrazide Chemistry. J. Proteome Res. 2009, 8, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Grune, T.; Reinheckel, T.; Joshi, M.; Davies, K.J.A. Proteolysis in cultured liver epithelial cells during oxidative stress. Role of the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1995, 270, 2344–2351. [Google Scholar] [CrossRef] [PubMed]
- Stamataki, Z.; Swadling, L. The liver as an immunological barrier redefined by single-cell analysis. Immunology 2020, 160, 157–170. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, X.; Huang, J.; Zeng, Y.; Liu, W.; Geng, C.; Li, K.W.; Yang, D.; Wu, S.; Wei, H.; et al. Relationships between Hematopoiesis and Hepatogenesis in the Midtrimester Fetal Liver Characterized by Dynamic Transcriptomic and Proteomic Profiles. PLoS ONE 2009, 4, e7641. [Google Scholar] [CrossRef]
- Lee, J.S.; Ward, W.O.; Knapp, G.; Ren, H.; Vallanat, B.; Abbott, B.; Ho, K.; Karp, S.J.; Corton, J.C. Transcriptional ontogeny of the developing liver. BMC Genom. 2012, 13, 33. [Google Scholar] [CrossRef]
- Peng, L.; Piekos, S.C.; Guo, G.L.; Zhong, X.-B. Role of Farnesoid X Receptor in the Determination of Liver Transcriptome during Postnatal Maturation in Mice. Nucl. Recept. Res. 2017, 4, 101308. [Google Scholar] [CrossRef][Green Version]
- Chen, W.; Yan, Q.; Yang, H.; Zhou, X.; Tan, Z. Effects of restrictions on maternal feed intake on the immune indexes of umbilical cord blood and liver Toll-like receptor signaling pathways in fetal goats during pregnancy. J. Anim. Sci. Biotechnol. 2019, 10, 29. [Google Scholar] [CrossRef]
- Zhou, X.; Yang, H.; Yan, Q.; Ren, A.; Kong, Z.; Tang, S.; Han, X.; Tan, Z.; Salem, A.Z.M. Evidence for liver energy metabolism programming in offspring subjected to intrauterine undernutrition during midgestation. Nutr. Metab. 2019, 16, 20. [Google Scholar] [CrossRef]
- Beath, S. Hepatic function and physiology in the newborn. Semin. Neonatol. 2003, 8, 337–346. [Google Scholar] [CrossRef]
- Cui, J.Y.; Gunewardena, S.S.; Yoo, B.; Liu, J.; Renaud, H.J.; Lu, H.; Zhong, X.-B.; Klaassen, C.D. RNA-Seq Reveals Different mRNA Abundance of Transporters and Their Alternative Transcript Isoforms During Liver Development. Toxicol. Sci. 2012, 127, 592–608. [Google Scholar] [CrossRef] [PubMed]
- Gruppuso, P.A.; Sanders, J.A. Regulation of liver development: Implications for liver biology across the lifespan. J. Mol. Endocrinol. 2016, 56, R115–R125. [Google Scholar] [CrossRef] [PubMed]
- Grijalva, J.; Vakili, K. Neonatal liver physiology. Semin. Pediatr. Surg. 2013, 22, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Heck, L.J.; Erenberg, A. Serum glucose levels in term neonates during the first 48 hours of life. J. Pediatr. 1987, 110, 119–122. [Google Scholar] [CrossRef]
- Srinivasan, G.; Pildes, R.; Cattamanchi, G.; Voora, S.; Lilien, L. Plasma glucose values in normal neonates: A new look. J. Pediatr. 1986, 109, 114–117. [Google Scholar] [CrossRef]
- Sperling, M.A.; Delamater, P.V.; Phelps, D.; Fiser, R.H.; Oh, W.; Fisher, D.A. Spontaneous and amino acid-stimulated glucagon secretion in the immediate postnatal period. Relation to glucose and insulin. J. Clin. Investig. 1974, 53, 1159–1166. [Google Scholar] [CrossRef]
- Platt, M.W.; Deshpande, S. Metabolic adaptation at birth. Semin. Fetal Neonatal Med. 2005, 10, 341–350. [Google Scholar] [CrossRef]
- Ferre, P.; Decaux, J.-F.; Issad, T.; Girard, J. Changes in energy metabolism during the suckling and weaning period in the newborn. Reprod. Nutr. Dev. 1986, 26, 619–631. [Google Scholar] [CrossRef]
- Hawdon, J.M.; Platt, M.W.; Aynsley-Green, A. Patterns of metabolic adaptation for preterm and term infants in the first neonatal week. Arch. Dis. Child. 1992, 67, 357–365. [Google Scholar] [CrossRef]
- Shaffer, E.A.; Zahavi, I.; Gall, D.G. Postnatal development of hepatic bile formation in the rabbit. Am. J. Dig. Dis. 1985, 30, 558–563. [Google Scholar] [CrossRef]
- Henning, S.J. Postnatal development: Coordination of feeding, digestion, and metabolism. Am. J. Physiol. Liver Physiol. 1981, 241, G199–G214. [Google Scholar] [CrossRef] [PubMed]
- Little, J.M.; Richey, J.E.; Van Thiel, D.H.; Lester, R. Taurocholate pool size and distribution in the fetal rat. J. Clin. Investig. 1979, 63, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Barnard, J.A.; Ghishan, F.K.; Wilson, F.A. Ontogenesis of taurocholate transport by rat ileal brush border membrane vesicles. J. Clin. Investig. 1985, 75, 869–873. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, Z.; Shen, Z.; Shen, H.; Aschenbach, J.R. Transcriptomic analyses suggest a dominant role of insulin in the coordinated control of energy metabolism and ureagenesis in goat liver. BMC Genom. 2019, 20, 854. [Google Scholar] [CrossRef]
- Yang, C.; Zhou, X.; Yang, H.; Gebeyew, K.; Yan, Q.; Zhou, C.; He, Z.; Tan, Z. Transcriptome analysis reveals liver metabolism programming in kids from nutritional restricted goats during mid-gestation. PeerJ 2021, 9, e10593. [Google Scholar] [CrossRef]
- Chen, Q.; Hua, C.; Niu, L.; Geng, Y.; Cai, L.; Tao, S.; Ni, Y.; Zhao, R. Exploring differentially expressed genes related to metabolism by RNA-Seq in goat liver after dexamethasone treatment. Gene 2018, 659, 175–182. [Google Scholar] [CrossRef]
- National Committee on Livestock Genetic Resources. Animal Genetic Resources in China: Sheep and Goats, 1st ed.; China Agricultural Press: Beijing, China, 2011; pp. 287–289. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 1087. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020, 49, D605–D612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Kikuguchi, C.; Nishijima, S.; Nagashima, T.; Takahashi, A.; Okada, M.; Yamamoto, T. Postnatal liver functional maturation requires Cnot complex-mediated decay of mRNAs encoding cell cycle and immature liver genes. Development 2019, 146, dev168146. [Google Scholar] [CrossRef]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Baeyens, D.A.; McGraw, D.W.; Jacobi, S.E.; Cornett, L.E. Transcription of the beta2-adrenergic receptor gene in rat liver is regulated during early postnatal development by an upstream repressor element. J. Cell Physiol. 1998, 175, 333–340. [Google Scholar] [CrossRef]
- Tatewaki, H.; Tsuda, H.; Kanaji, T.; Yokoyama, K.; Hamasaki, N. Characterization of the human protein S gene promoter: A possible role of transcription factors Sp1 and HNF3 in liver. Thromb. Haemost. 2003, 90, 1029–1039. [Google Scholar] [CrossRef]
- Yao, D.; Luo, J.; He, Q.; Xu, H.; Li, J.; Shi, H.; Wang, H.; Chen, Z.; Loor, J. Liver X receptor α promotes the synthesis of monounsaturated fatty acids in goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase 1 in an SREBP-1-dependent manner. J. Dairy Sci. 2016, 99, 6391–6402. [Google Scholar] [CrossRef]
- Xu, E.; Zhang, L.; Yang, H.; Shen, L.; Feng, Y.; Ren, M.; Xiao, Y. Transcriptome profiling of the liver among the prenatal and postnatal stages in chickens. Poult. Sci. 2019, 98, 7030–7040. [Google Scholar] [CrossRef]
- Cui, K.; Wang, B.; Ma, T.; Si, B.W.; Zhang, N.F.; Tu, Y.; Diao, Q.Y. Effects of dietary protein restriction followed by realimentation on growth performance and liver transcriptome alterations of lamb. Sci. Rep. 2018, 8, 15185. [Google Scholar] [CrossRef] [PubMed]
- Ha, N.-T.; Drögemüller, C.; Reimer, C.; Schmitz-Hsu, F.; Bruckmaier, R.; Simianer, H.; Gross, J. Liver transcriptome analysis reveals important factors involved in the metabolic adaptation of the transition cow. J. Dairy Sci. 2017, 100, 9311–9323. [Google Scholar] [CrossRef] [PubMed]
- Mukiibi, R.; Vinsky, M.; Keogh, K.; Fitzsimmons, C.; Stothard, P.; Waters, S.M.; Li, C. Liver transcriptome profiling of beef steers with divergent growth rate, feed intake, or metabolic body weight phenotypes1. J. Anim. Sci. 2019, 97, 4386–4404. [Google Scholar] [CrossRef] [PubMed]
- Heimes, A.; Brodhagen, J.; Weikard, R.; Becker, D.; Meyerholz, M.; Petzl, W.; Zerbe, H.; Schuberth, H.-J.; Hoedemaker, M.; Schmicke, M.; et al. Cows selected for divergent mastitis susceptibility display a differential liver transcriptome profile after experimental Staphylococcus aureus mammary gland inoculation. J. Dairy Sci. 2020, 103, 6364–6373. [Google Scholar] [CrossRef]
- Xiong, Q.; Tao, H.; Zhang, N.; Zhang, L.; Wang, G.; Li, X.; Suo, X.; Zhang, F.; Liu, Y.; Chen, M. Skin transcriptome profiles associated with black- and white-coated regions in Boer and Macheng black crossbred goats. Genomics 2019, 112, 1853–1860. [Google Scholar] [CrossRef]
- Mirza, A.Z.; AlThagafi, I.I.; Shamshad, H. Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur. J. Med. Chem. 2019, 166, 502–513. [Google Scholar] [CrossRef]
- Janani, C.; Kumari, B.R. PPAR gamma gene—A review. Diabetes Metab. Syndr. 2015, 9, 46–50. [Google Scholar] [CrossRef]
- Canbay, A.; Bechmann, L.; Gerken, G. Lipid Metabolism in the Liver. Z. Gastroenterol. 2007, 45, 35–41. [Google Scholar] [CrossRef]
- Luckheeram, R.V.; Zhou, R.; Verma, A.D.; Xia, B. CD4+T Cells: Differentiation and Functions. Clin. Dev. Immunol. 2012, 2012, 925135. [Google Scholar] [CrossRef]
- Yu, M.; Xu, W.; Jie, Y.; Pang, J.; Huang, S.; Cao, J.; Gong, J.; Li, X.; Chong, Y. Identification and validation of three core genes in p53 signaling pathway in hepatitis B virus-related hepatocellular carcinoma. World J. Surg. Oncol. 2021, 19, 66. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Santamaría, D.; Barrière, C.; Cerqueira, A.; Hunt, S.; Tardy, C.; Newton, K.; Cáceres, J.F.; Dubus, P.; Malumbres, M.; Barbacid, M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 2007, 448, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Haneke, K.; Schott, J.; Lindner, D.; Hollensen, A.K.; Damgaard, C.K.; Mongis, C.; Knop, M.; Palm, W.; Ruggieri, A.; Stoecklin, G. CDK1 couples proliferation with protein synthesis. J. Cell Biol. 2020, 219, e201906147. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Liu, S.; Sun, H.; Qin, X.; Zhou, N.; Zheng, W.; Zhang, M.; Zhou, H.; Tuersunayi, A.; Duan, C.; et al. All-trans-retinoic acid shifts Th1 towards Th2 cell differentiation by targeting NFAT1 signalling to ameliorate immune-mediated aplastic anaemia. Br. J. Haematol. 2020, 191, 906–919. [Google Scholar] [CrossRef] [PubMed]
- Jenne, C.N.; Kubes, P. Immune surveillance by the liver. Nat. Immunol. 2013, 14, 996–1006. [Google Scholar] [CrossRef]
- Li, H.; Ding, P.; Peng, B.; Ming, Y.-Z. Cross-talk between hepatic stellate cells and T lymphocytes in liver fibrosis. Hepatobiliary Pancreat. Dis. Int. 2021, 20, 207–214. [Google Scholar] [CrossRef]
- Iritani, B.M.; Eisenman, R.N. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. USA 1999, 96, 13180–13185. [Google Scholar] [CrossRef]
- Farrell, A.S.; Sears, R.C. MYC Degradation. Cold Spring Harb. Perspect. Med. 2014, 4, a014365. [Google Scholar] [CrossRef]
- Dang, C.V. A Time for MYC: Metabolism and Therapy. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 79–83. [Google Scholar] [CrossRef]
- Beaulieu, M.-E.; Castillo, F.; Soucek, L. Structural and Biophysical Insights into the Function of the Intrinsically Disordered Myc Oncoprotein. Cells 2020, 9, 1038. [Google Scholar] [CrossRef] [PubMed]
- Carroll, P.A.; Freie, B.W.; Mathsyaraja, H.; Eisenman, R.N. The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 2018, 12, 412–425. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Up | Down |
|---|---|---|
| ACTB | CACCACACCTTCTACAAC | TCTGGGTCATCTTCTCAC |
| ABCC2 | AATACAACGGCACCAACTATCCATCC | CATAGACACTCCAGATGTTCGCTACG |
| GPX7 | GGAGCAGGACTTCTACGACTTCAAG | CCACATTCACCACCAGGGACAC |
| EDF1 | GTGGTGGATGAAAATGCAGTAG | TTGAAAATCCACAGCCAATGAG |
| SNRPD3 | GTATGACAGGCTATGACCCGTTGC | CGTTCTGGAAGATCAGCGACACTC |
| UACA | GACACCGCTTGTTCTGGCTACTC | GCAACCGTACTCGCATCCTAGC |
| WIPI1 | CAGTTCAGTCGCTCAGTCGTGTC | GGAAACTGTTGGTTGTACAGAC |
| TSTA3 | GCTACTCCGTGTATGTGTACAA | TAACGAAGGAATTCATGATGCC |
| RAB7A | CAGGAGGCAACACGGTGAAGAC | CACAGACAGACGAGATGGCTTGATG |
| BGN | GTAAACCACCTGCACCTAAAAG | CCTCCACAGTACACAGTACAAT |
| BAG2 | CTTTGAGAGAAGCAGCAACTG | TGACACTTCAACGGTGAGAG |
| S100A13 | TGTGCTTAGTTGCTCAGTCGTGTC | AGGTTGGATCTCTGGGTTGGGAAG |
| CTNNAL1 | TGGGAGATGGTGAAGGACAGAGAAG | TTGTGGTTGTTCAGTCGCTCAGTC |
| MTTP | CAGTTCAGTCGCTCAGTCGTGTC | CGAAAGAGGATGAGATGGCTGGATG |
| SIGMAR1 | ATGCCCTCCTTTCTGCCGTTTG | GAAGGTGCCTGAGATGATGGTATCG |
| MEST | ATACCCGACCTTCTGAGAGTGAGC | CCCACCCAGCGTCTTCTAAACTTC |
| Sample Name | Known mRNA Num | Novel mRNA Num | All mRNA Num |
|---|---|---|---|
| D1 | 23,861 (73.37%) | 7324 | 31,185 |
| W2 | 24,022 (76.58%) | 7347 | 31,369 |
| W4 | 23,901 (76.57%) | 7315 | 31,216 |
| W8 | 23,725 (76.37%) | 7339 | 31,064 |
| W12 | 23,799 (76.42%) | 7344 | 31,143 |
| Total | 29,976 (79.94%) | 7521 | 37,497 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Xuan, R.; Wang, A.; Li, Q.; Zhao, Y.; Du, S.; Duan, Q.; Wang, Y.; Ji, Z.; Guo, Y.; et al. High-Throughput Sequencing Reveals Transcriptome Signature of Early Liver Development in Goat Kids. Genes 2022, 13, 833. https://doi.org/10.3390/genes13050833
Zhao X, Xuan R, Wang A, Li Q, Zhao Y, Du S, Duan Q, Wang Y, Ji Z, Guo Y, et al. High-Throughput Sequencing Reveals Transcriptome Signature of Early Liver Development in Goat Kids. Genes. 2022; 13(5):833. https://doi.org/10.3390/genes13050833
Chicago/Turabian StyleZhao, Xiaodong, Rong Xuan, Aili Wang, Qing Li, Yilin Zhao, Shanfeng Du, Qingling Duan, Yanyan Wang, Zhibin Ji, Yanfei Guo, and et al. 2022. "High-Throughput Sequencing Reveals Transcriptome Signature of Early Liver Development in Goat Kids" Genes 13, no. 5: 833. https://doi.org/10.3390/genes13050833
APA StyleZhao, X., Xuan, R., Wang, A., Li, Q., Zhao, Y., Du, S., Duan, Q., Wang, Y., Ji, Z., Guo, Y., Wang, J., & Chao, T. (2022). High-Throughput Sequencing Reveals Transcriptome Signature of Early Liver Development in Goat Kids. Genes, 13(5), 833. https://doi.org/10.3390/genes13050833






