Effect of Humantenine on mRNA m6A Modification and Expression in Human Colon Cancer Cell Line HCT116
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Reagent
2.2. Cell Culture and Treatment
2.3. RNA Preparation
2.4. High-Throughput m6A MeRIP-seq and mRNA-seq
2.5. Sequencing Data Analysis
2.6. Molecular Docking
3. Results
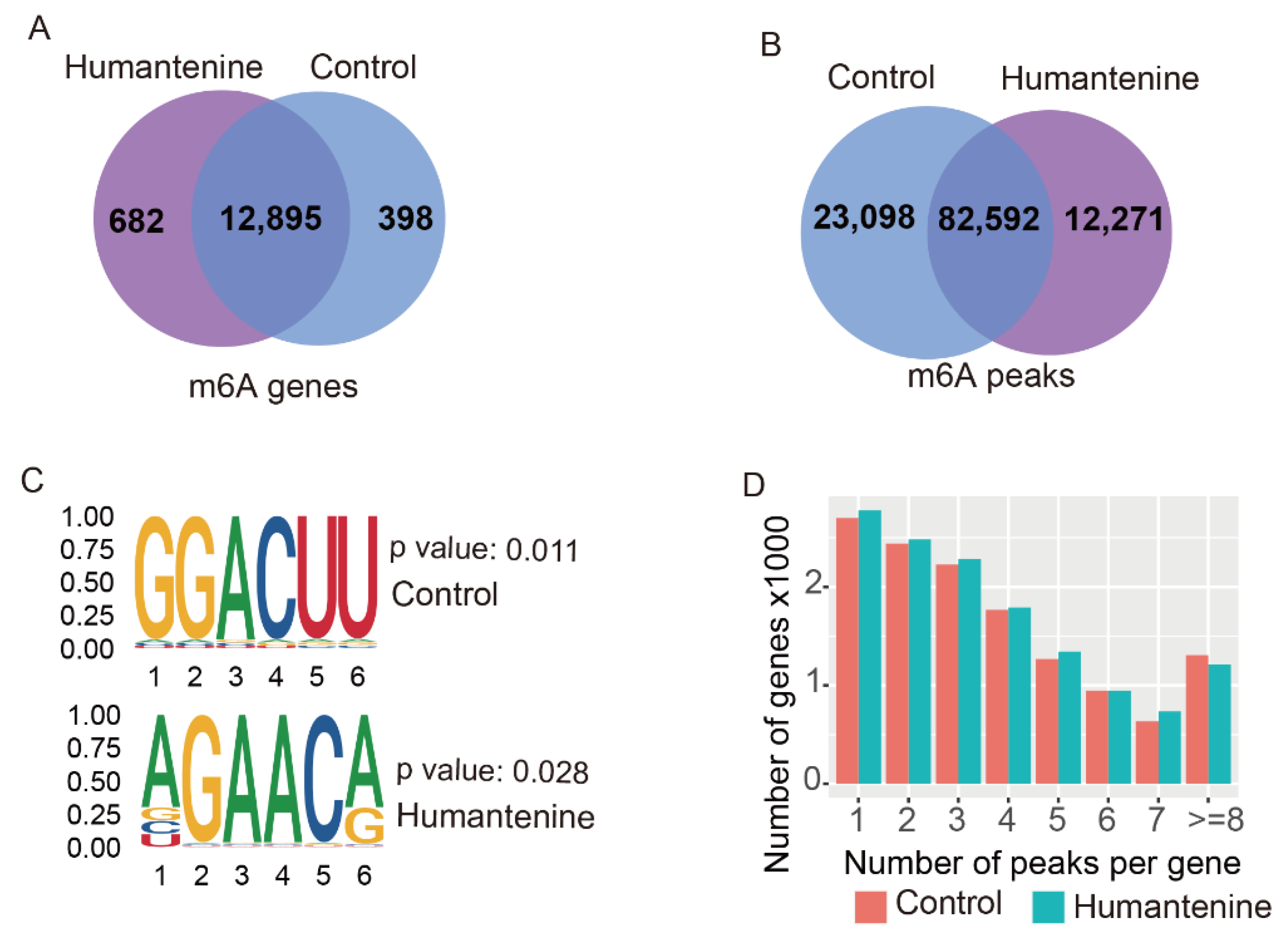
3.1. Transcriptome-Wide Detection of m6A Modification after Humantenine Treatment of HCT116 Cells
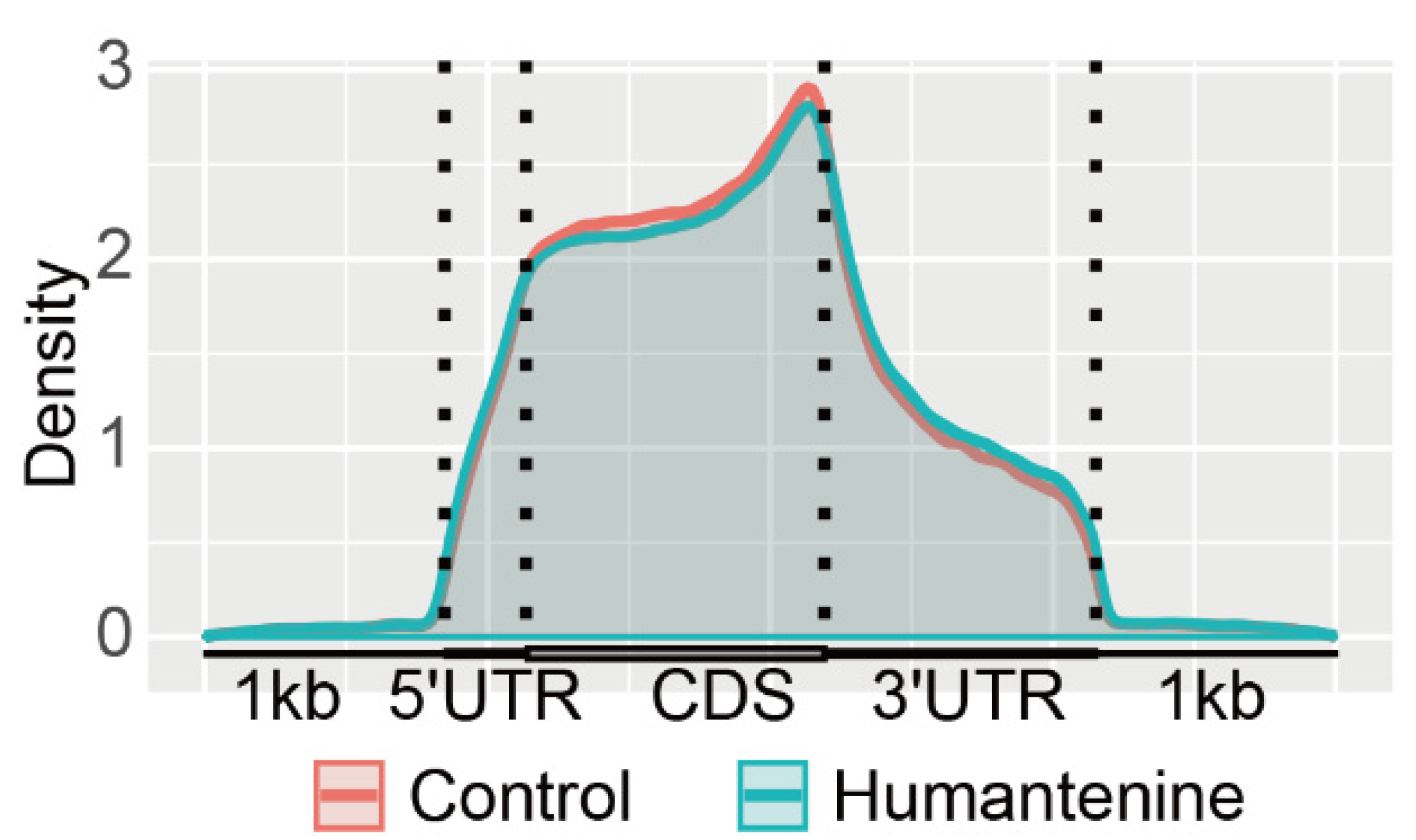
3.2. Distribution of m6A Modification in the Transcriptome
3.3. Differentially Methylated Genes and Differentially Expressed Genes
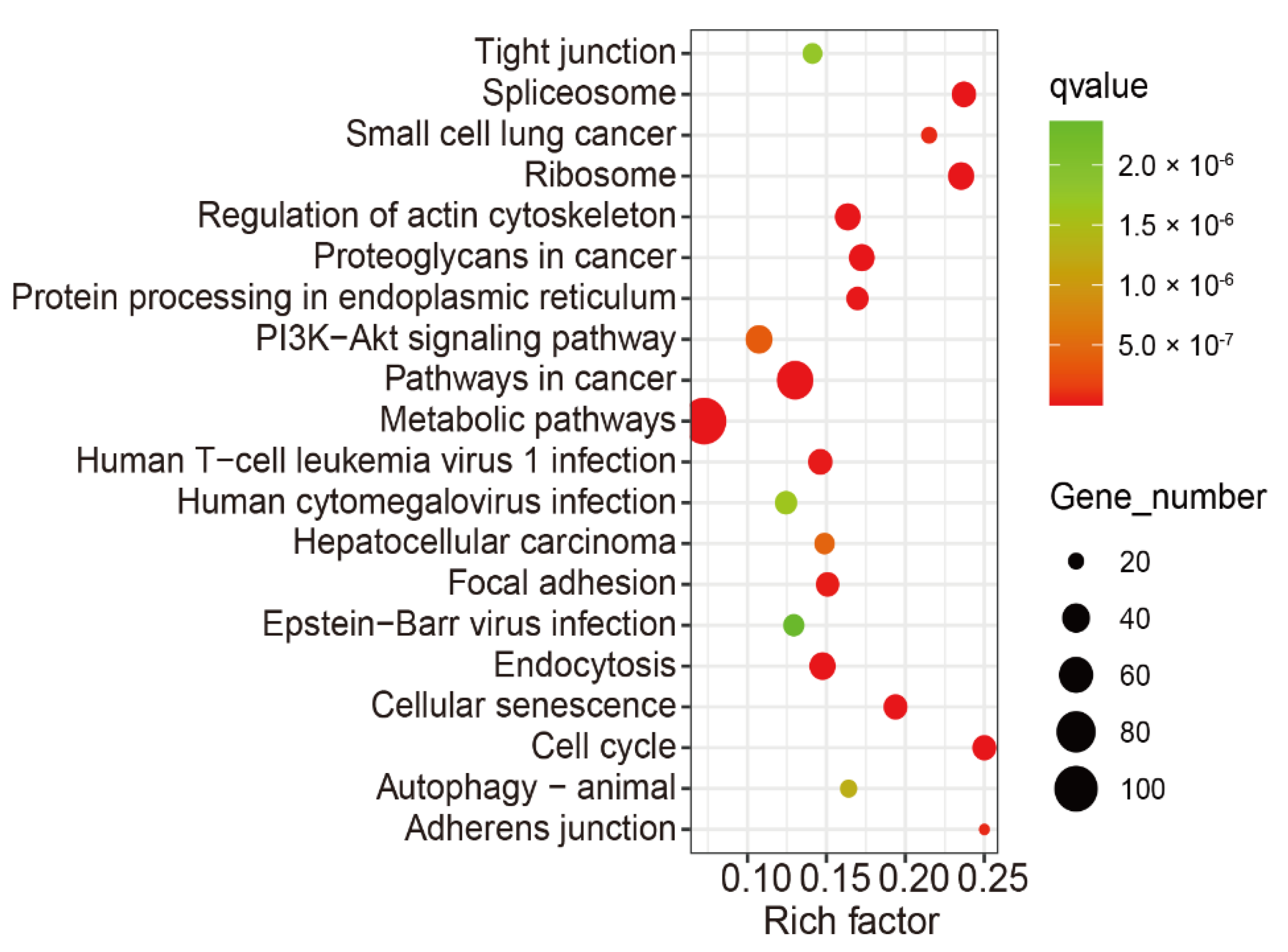
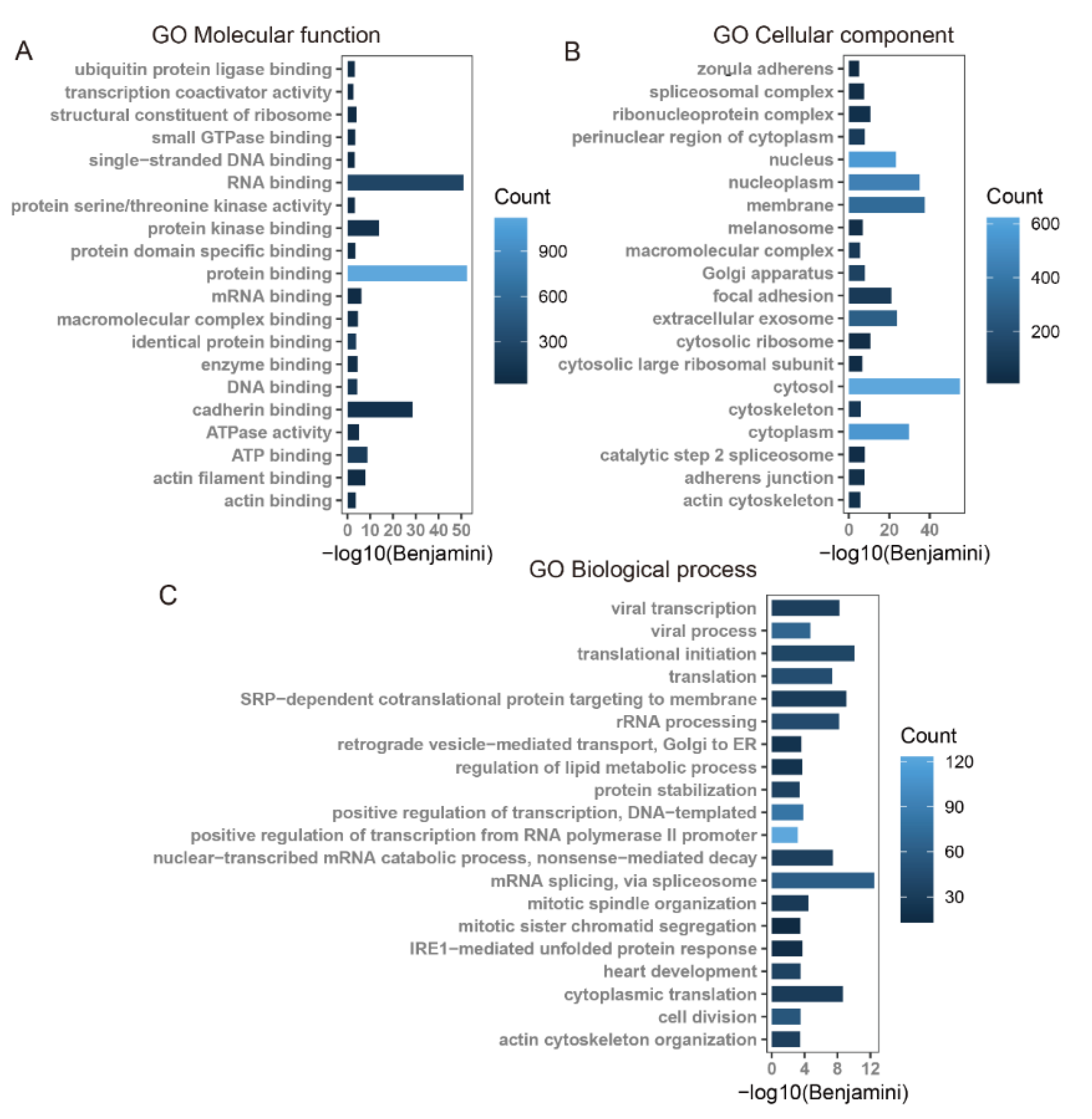
3.4. KEGG and GO Annotation of the Overlap of Differentially m6A-Modified mRNA and Differentially Expressed mRNA
3.5. The m6A Conservation and Disease Association
3.6. Gene Expression of Tight Junctions, Adherens Junctions, and Regulation of Actin Cytoskeleton, and Their Potential Regulators
3.7. Potential RNA m6A Regulators of Differentially Methylated Genes
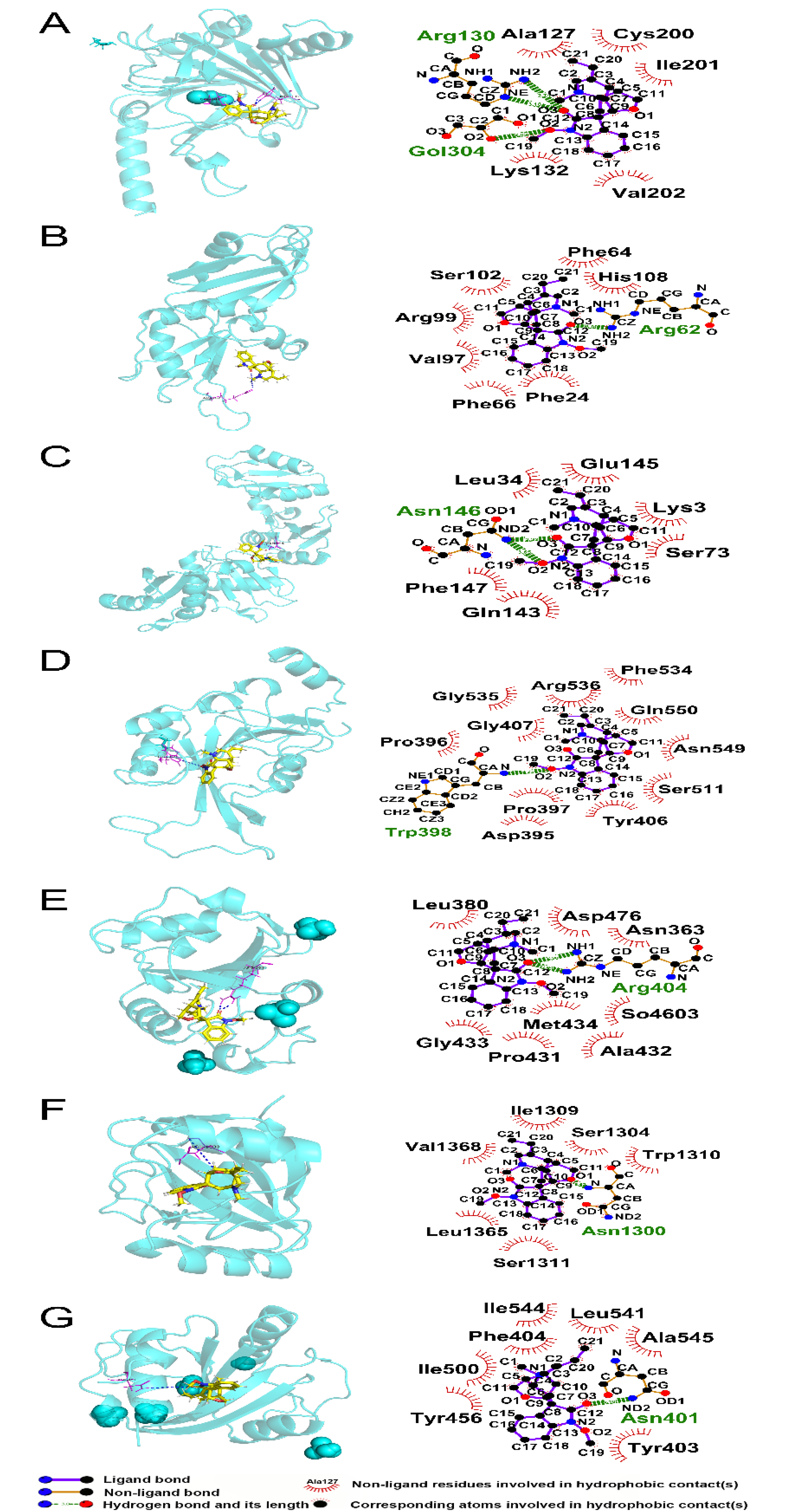
3.8. Molecular Interactions of Humantenine with Differently Expressed Regulators of RNA m6A Modification
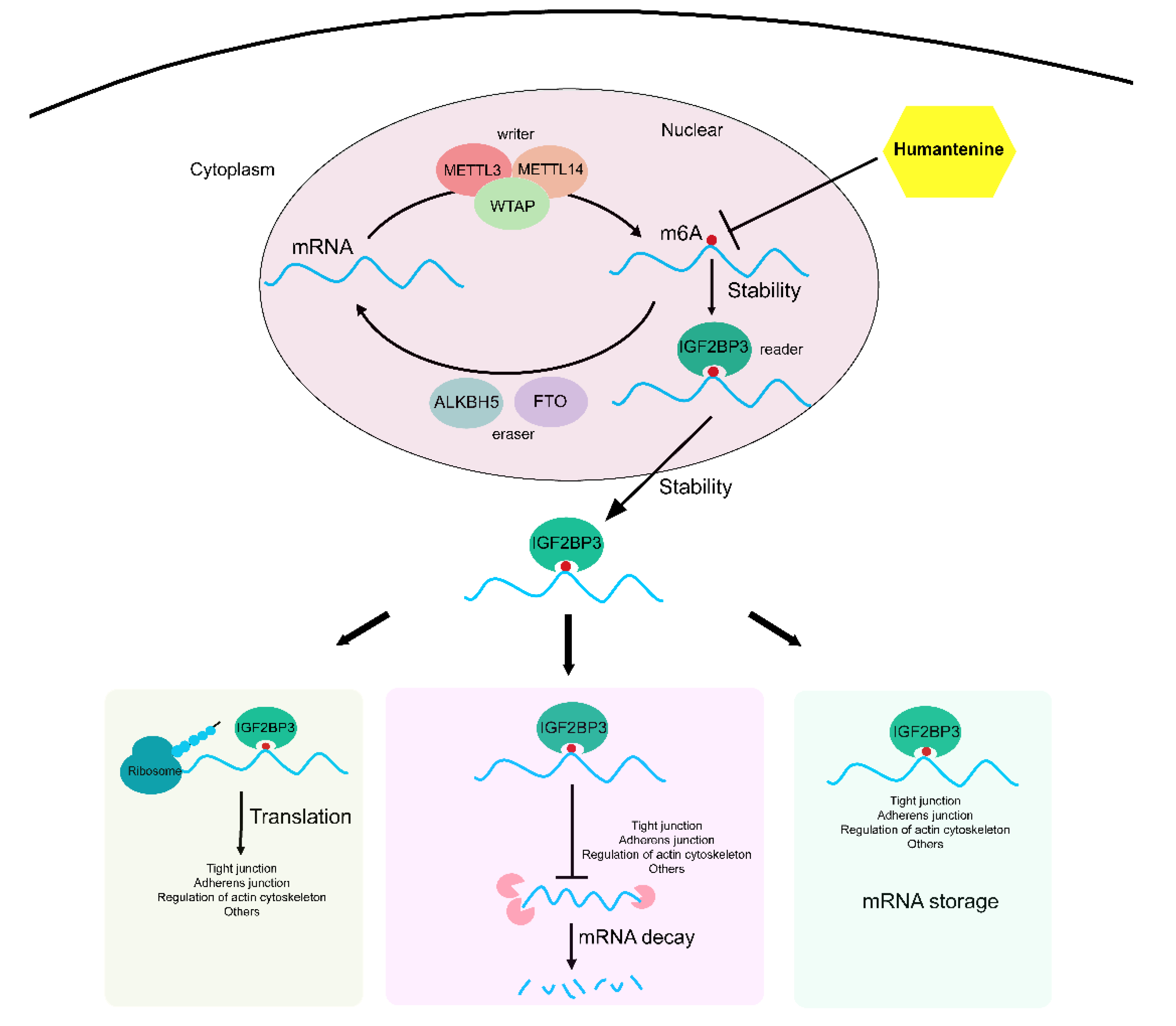
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 3′UTR | 3′ untranslated regions |
| 5′UTR | 5′ untranslated regions |
| CDS | coding sequence (CDS) |
| G. elegans | Gelsemium elegans (Gardner & Chapm.) Benth. |
| GO | Gene Ontology |
| HPLC | high-performance liquid chromatography |
| IGF2BP3 | insulin-like growth factor 2 mRNA-binding protein 3 |
| IGV | Integrative Genomics Viewer |
| IQGAP3 | IQ motif containing GTPase activating protein 3 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| m6A | N6-methyladenosine |
| MeRIP-seq | methylated RNA immunoprecipitation sequencing |
| NMR | nuclear magnetic resonance |
| PPI | protein–protein interaction |
References
- Zhao, Y.T.; Wu, S.P.; Hu, C.L.; Kang, J.; Zhao, M. Reviews on chemical compositions and pharmacological effect of Gelsemium elegans. Chin. J. Exp. Tradit. Med. Formulae 2019, 25, 200–210. [Google Scholar]
- Cao, J.J.; Yang, K.; Huang, C.Y.; Li, Y.J.; Yu, H.; Wu, Y.; Sun, Z.L.; Liu, Z.Y. Pharmacokinetic study of multiple components of Gelsemium elegans in goats by Ultra-performance liquid chromatography coupled to tandem mass spectrometry. J. Anal. Toxicol. 2020, 44, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.L.; Qiu, H.Q.; Cheng, Y.; Liu, M.B.; Chen, M.H.; Que, Y.X.; Que, W.C. Gelsemium elegans Benth: Chemical components, pharmacological effects, and toxicity mechanisms. Molecules 2021, 26, 7145. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.X.; Xun, Q.Q.; Meng, W.Q.; Cen, J.F.; Mao, G.C.; Pei, Z.P.; Xiao, K. Research progress on pharmacology and toxicological mechanism of Gelsemium elegans. J. Toxicol. 2020, 34, 336–341. [Google Scholar]
- Wang, R.; Ding, F.; Gao, Y.J.; Wang, X.Y.; Li, Q. Epidemiological analysis for vegetal food poisoning in China, 2004–2013. Chin. J. Food Hyg. 2016, 28, 580–584. [Google Scholar]
- Zhong, Y.X.; Xie, Y.H.; Jiang, Y.Y.; Liu, Y.P.; Shi, M.M.; Yang, W.M. Analysis of gelsemine poisoning events in the Guangxi Zhuang autonomous region during 2015–2017. Chin. J. Food Hyg. 2019, 31, 81–83. [Google Scholar]
- Liu, Y.C.; Sun, Z.L.; Liu, Z.Y. Research progress on non-alkaloids analysis for chemical constituents from Gelsemium. Chin. J. Vet. Drug 2016, 50, 55–64. [Google Scholar]
- Mu, F.Y.; Mu, F.W.; Mu, F.M.; Chen, Z.H.; Ma, X.D.; Wu, D.; Guo, X.K.; Liu, C.X. Study on chemical components and pharmacological activity of Gelsemium elegans. Sci. Technol. Innov. 2015, 3, 75–77. [Google Scholar]
- Tan, J.Q.; Qiu, C.Z.; Zheng, L.Z. Analgesic effect and no physical dependence of Gelsemium elegans Benth. Pharmacol. Clin. Chin. Mater. Med. 1988, 4, 24–28. [Google Scholar]
- Li, X.; Xie, X.L.; Gu, Y.C.; Zhang, J.M.; Song, J.; Cheng, X.F.; Gao, Y.J.; Ai, Y.L. Fat mass and obesity-associated protein regulates tumorigenesis of arecoline-promoted human oral carcinoma. Cancer Med. 2021, 10, 6402–6415. [Google Scholar] [CrossRef]
- Zhao, Z.Y.; Zeng, J.H.; Guo, Q.; Pu, K.M.; Yang, Y.; Chen, N.Z.; Zhang, G.; Zhao, M.Y.; Zheng, Q.; Tang, J.Y.; et al. Berberine suppresses stemness and tumorigenicity of colorectal cancer stem-like cells by inhibiting m6A methylation. Front. Oncol. 2021, 11, 775418. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Nachtergaele, S.; Moshitch-Moshkovitz, S.; Peer, E.; Kol, N.; Ben-Haim, M.S.; Dai, Q.; Di Segni, A.; Salmon-Divon, M.; Clark, W.C.; et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016, 530, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.M.; Song, B.W.; Wei, Z.; Huang, D.Y.; Zhang, Y.X.; Su, J.L.; de Magalhães, J.P.; Rigden, D.J.; Meng, J.; Chen, K.Q. m5C-Atlas: A comprehensive database for decoding and annotating the 5-methylcytosine (m5C) epitranscriptome. Nucleic Acids Res. 2022, 50, D196–D203. [Google Scholar] [CrossRef] [PubMed]
- Song, B.W.; Tang, Y.J.; Chen, K.Q.; Wei, Z.; Rong, R.; Lu, Z.L.; Su, J.L.; de Magalhães, J.P.; Rigden, D.J.; Meng, J. m7GHub: Deciphering the location, regulation and pathogenesis of internal mRNA N7-methylguanosine (m7G) sites in human. Bioinformatics 2020, 36, 3528–3536. [Google Scholar] [CrossRef]
- Kadumuri, R.V.; Janga, S.C. Epitranscriptomic code and its alterations in human disease. Trends Mol. Med. 2018, 24, 886–903. [Google Scholar] [CrossRef]
- Jones, J.D.; Monroe, J.; Koutmou, K.S. A molecular-level perspective on the frequency, distribution, and consequences of messenger RNA modifications. Wiley Interdiscip. Rev. RNA 2020, 11, e1586. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.J.; Luo, K.X.; Zou, Z.Y.; Qiu, M.G.Y.; Tian, J.K.; Sieh, L.; Shi, H.L.; Zou, Y.X.; Wang, G.; Morrison, J.; et al. Genetic analyses support the contribution of mRNA N6-methyladenosine (m6A) modification to human disease heritability. Nat. Genet. 2020, 52, 939–949. [Google Scholar] [CrossRef]
- Cao, G.; Li, H.B.; Yin, Z.; Flavell, R.A. Recent advances in dynamic m6A RNA modification. Open Biol. 2016, 6, 160003. [Google Scholar] [CrossRef] [Green Version]
- Erson-Bensan, A.E.; Begik, O. m6A modification and implications for microRNAs. MicroRNA 2017, 6, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, A.; Priyadharsini, J.V. Epigenetic modifications of RNA and their implications in antiviral immunity. Epigenomics 2020, 12, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Qi, X.Q.; Liu, L.N.; Ma, S.Q.; Liu, J.W.; Wu, J. Epigenetic regulation of m6a modifications in human cancer. Mol. Ther-Nucl. Acids 2020, 19, 405–412. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Cui, X.D.; Rao, M.K.; Chen, Y.D.; Huang, Y.F. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics 2013, 29, 1565–1567. [Google Scholar] [CrossRef]
- Tang, Y.J.; Chen, K.Q.; Song, B.W.; Ma, J.M.; Wu, X.Y.; Xu, Q.R.; Wei, Z.; Su, J.L.; Liu, G.; Rong, R.; et al. m6A-Atlas: A comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res. 2021, 49, D134–D143. [Google Scholar] [CrossRef]
- Bailey, T.L. STREME: Accurate and versatile sequence motif discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef]
- Bu, D.C.; Luo, H.T.; Huo, P.P.; Wang, Z.H.; Zhang, S.; He, Z.H.; Wu, Y.; Zhao, L.H.; Liu, J.J.; Guo, J.C.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.L.; Sherman, B.T.; da Huang, W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chen, K.Q.; Wei, Z.; Coenen, F.; Su, J.L.; Meng, J. MetaTX: Deciphering the distribution of mRNA-related features in the presence of isoform ambiguity, with applications in epitranscriptome analysis. Bioinformatics 2021, 37, 1285–1291. [Google Scholar] [CrossRef] [PubMed]
- Song, B.W.; Chen, K.Q.; Tang, Y.J.; Wei, Z.; Su, J.L.; de Magalhães, J.P.; Rigden, D.J.; Meng, J. ConsRM: Collection and large-scale prediction of the evolutionarily conserved RNA methylation sites, with implications for the functional epitranscriptome. Brief. Bioinform. 2021, 22, bbab088. [Google Scholar] [CrossRef]
- Chen, K.Q.; Song, B.W.; Tang, Y.J.; Wei, Z.; Xu, Q.R.; Su, J.L.; de Magalhães, J.P.; Rigden, D.J.; Meng, J. RMDisease: A database of genetic variants that affect RNA modifications, with implications for epitranscriptome pathogenesis. Nucleic Acids Res. 2021, 49, D1396–D1404. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Zhang, L.; Zhu, A.; Liu, X.Y.; Wang, T.X.; Wan, M.Q.; Yang, X.W.; Zhang, Y.T.; Zhang, Y.B. Metabolic profiling of nuciferine in vivo and in vitro. J. Agric. Food Chem. 2020, 68, 14135–14147. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.Q.; Zhang, X.; Shen, X.F.; Wang, S.; Wang, Q.; Yang, X.W. Computational and experimental characterization of isomers of escin-induced renal cytotoxicity by inhibiting heat shock proteins. Eur. J. Pharmacol. 2021, 908, 174372. [Google Scholar] [CrossRef]
- Zhu, A.; Sun, Y.Q.; Zhong, Q.W.; Yang, J.L.; Zhang, T.; Zhao, J.W.; Wang, Q. Effect of euphorbia factor L1 on oxidative stress, apoptosis, and autophagy in human gastric epithelial cells. Phytomedicine 2019, 64, 152929. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Harper, J.E.; Miceli, S.M.; Roberts, R.J.; Manley, J.L. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 1990, 18, 5735–5741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.M.; Zhou, C.X.; Sun, Y.Y.; He, X.Z.; Xue, D. m6A RNA modification modulates gene expression and cancer-related pathways in clear cell renal cell carcinoma. Epigenomics 2020, 12, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.Z.; MacQueen, A.; Zheng, G.Q.; Duan, H.C.; Dore, L.C.; Lu, Z.K.; Liu, J.; Chen, K.; Jia, G.F.; Bergelson, J.; et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014, 5, 5630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Lu, Z.K.; Gomez, A.; Hon, G.C.; Yue, Y.N.; Han, D.L.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.F.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Lee, A.; White, N.; van der Walle, C.F. The intestinal zonula occludens toxin (ZOT) receptor recognises non-native ZOT conformers and localises to the intercellular contacts. FEBS Lett. 2003, 555, 638–642. [Google Scholar] [CrossRef] [Green Version]
- Farhadi, A.; Banan, A.; Fields, J.; Keshavarzian, A. Intestinal barrier: An interface between health and disease. J. Gastroenterol. Hepatol. 2003, 18, 479–497. [Google Scholar] [CrossRef]
- Tervonen, A.; Ihalainen, T.O.; Nymark, S.; Hyttinen, J. Structural dynamics of tight junctions modulate the properties of the epithelial barrier. PLoS ONE 2019, 14, e0214876. [Google Scholar] [CrossRef]
- Kumar, A.; Chatterjee, I.; Anbazhagan, A.N.; Jayawardena, D.; Priyamvada, S.; Alrefai, W.A.; Sun, J.; Borthakur, A.; Dudeja, P.K. Cryptosporidium parvum disrupts intestinal epithelial barrier function via altering expression of key tight junction and adherens junction proteins. Cell. Microbiol. 2018, 20, e12830. [Google Scholar] [CrossRef]
- Guttman, J.A.; Finlay, B.B. Tight junctions as targets of infectious agents. Biochim. Biophys. Acta 2009, 1788, 832–841. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Reytor, D.; Jaña, V.; Pavez, L.; Navarrete, P.; García, K. Accessory toxins of Vibrio pathogens and their role in epithelial disruption during infection. Front Microbiol. 2018, 9, 2248. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.C.; Shi, Y.F.; Shen, H.F.; Xie, W.Z. m6A-binding proteins: The emerging crucial performers in epigenetics. J. Hematol. Oncol. 2020, 13, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell. Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Wang, S.; Kaibuchi, K. IQGAPs as key regulators of actin-cytoskeleton dynamics. Cell Struct. Funct. 2015, 40, 69–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]



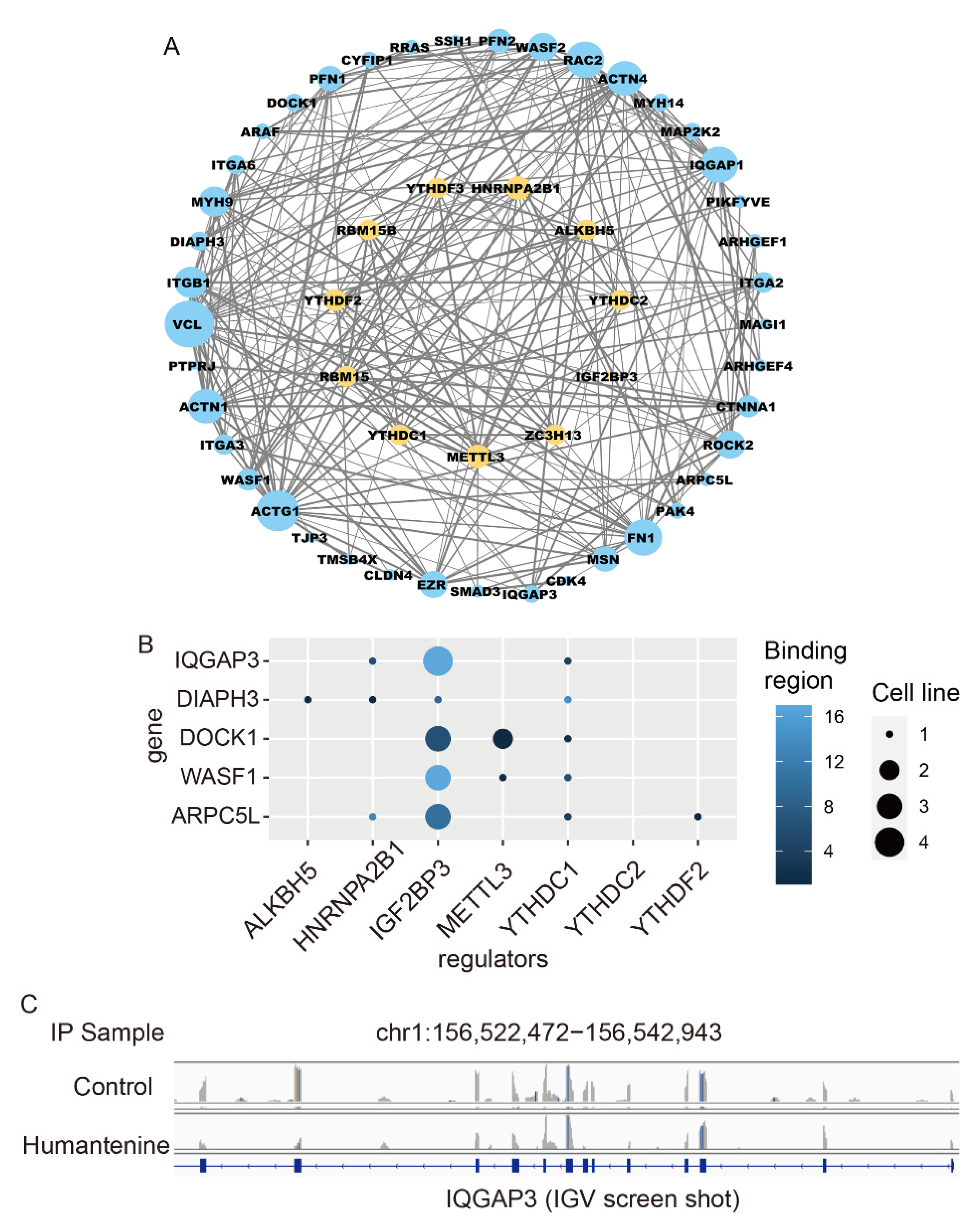
| Gene | log2FoldChange | padj | Writer | Reader | Eraser | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| METTL3 | HNRNPA2B1 | IGF2BP3 | YTHDC1 | YTHDC2 | YTHDF2 | ALKBH5 | ||||
| Tight junction | CLDN4 | 0.5708 | 8.04 × 10−32 | N | N | Y | N | N | N | N |
| CDK4 | 0.4725 | 4.43 × 10−26 | Y | Y | Y | Y | N | N | N | |
| TJP3 | −0.9036 | 1.84 × 10−67 | N | N | Y | N | N | N | N | |
| MAGI1 | −0.4761 | 6.83 × 10−16 | Y | Y | Y | Y | N | N | N | |
| Adherens junction | ACTN1 | 0.2529 | 3.31 × 10−11 | Y | Y | Y | Y | N | N | N |
| NECTIN2 | 0.2513 | 1.58 × 10−5 | N | Y | Y | Y | N | N | N | |
| WASF1 | −0.4224 | 9.38 × 10−5 | Y | N | Y | Y | N | N | N | |
| Regulation of actin cytoskeleton | MYH9 | 0.3207 | 2.97 × 10−22 | Y | Y | Y | Y | N | N | N |
| IQGAP3 | −0.8892 | 3.82 × 10−16 | N | Y | Y | Y | N | N | N | |
| DIAPH3 | −0.6702 | 2.45 × 10−9 | N | Y | Y | Y | N | N | Y | |
| WASF1 | −0.4224 | 9.38 × 10−5 | Y | N | Y | Y | N | N | N | |
| ARPC5L | −0.3197 | 1.42 × 10−4 | N | Y | Y | Y | N | Y | N | |
| DOCK1 | −0.3172 | 8.64 × 10−6 | Y | N | Y | Y | N | N | N | |
| Gene | Regulation | Base Mean | log2FoldChange | padj |
|---|---|---|---|---|
| ALKBH5 | eraser | 5355.58 | 0.5337 | 2.43 × 10−18 |
| HNRNPA2B1 | reader | 31,317.46 | −0.2827 | 3.61 × 10−13 |
| YTHDF2 | reader | 5827.43 | −0.3176 | 1.44 × 10−7 |
| YTHDC2 | reader | 1996.91 | −0.3977 | 9.40 × 10−5 |
| IGF2BP3 | reader | 7360.70 | −0.1831 | 5.89 × 10−4 |
| RBM15B | writer | 4931.66 | −0.2692 | 5.93 × 10−4 |
| YTHDF3 | reader | 4575.69 | 0.2471 | 2.44 × 10−3 |
| YTHDC1 | reader | 4745.43 | −0.1911 | 3.73 × 10−3 |
| RBM15 | writer | 1909.31 | 0.3036 | 6.91 × 10−3 |
| METTL3 | writer | 1801.37 | 0.2508 | 2.14 × 10−2 |
| ZC3H13 | writer | 8932.93 | −0.1169 | 2.65 × 10−2 |
| VIRMA | writer | 6469.09 | 0.1091 | 1.41 × 10−1 |
| FTO | eraser | 2646.90 | 0.1141 | 2.69 × 10−1 |
| IGF2BP1 | reader | 65.30 | 0.5237 | 3.67 × 10−1 |
| YTHDF1 | reader | 5181.16 | 0.0458 | 5.94 × 10−1 |
| HNRNPC | reader | 24,691.72 | −0.0436 | 6.68 × 10−1 |
| METTL14 | writer | 2711.17 | 0.0432 | 7.66 × 10−1 |
| METTL5 | writer | 1640.50 | 0.0410 | 7.80 × 10−1 |
| CBLL1 | writer | 2958.85 | −0.0289 | 8.24 × 10−1 |
| IGF2BP2 | reader | 8415.70 | −0.0156 | 8.55 × 10−1 |
| FMR1 | reader | 2456.26 | −0.0059 | 9.70 × 10−1 |
| WTAP | writer | 2760.03 | 0.0004 | 9.98 × 10−1 |
| Protein | PDB ID | Total Score | Crach | Polar | H-Bond Number | Residues Involved in H-Bond Formation | Hydrophobic Contacts Number | Residues Involved in Hydrophobic Contacts |
|---|---|---|---|---|---|---|---|---|
| ALKBH5 | 4NJ4 | 6.7567 | −0.8591 | 1.9259 | 3 | Arg130 (2 hydrogen bonds), Gol304 | 5 | Ala127, Lys132, Cys200, Ile201, Val202 |
| HNRNPA2B1 | 5HO4 | 5.1469 | −0.5213 | 0.9835 | 1 | Arg62 | 7 | Phe24, Phe64, Phe66, Val97, Arg99, Ser102, His108 |
| IGF2BP3 | 6FQ1 | 6.5409 | −0.9417 | 1.2075 | 2 | Asn146 (2 hydrogen bonds) | 6 | Lys3, Leu34, Ser73, Gln143, Glu145, Phe147 |
| METTL3 | 5IL2 | 6.09 | −1.0199 | 0.001 | 1 | Trp398 | 11 | Asp395, Pro396, Pro397, Tyr406, Gly407, Ser511, Phe534, Gly535, Arg536, Asn549, Gln550 |
| YTHDC1 | 6ZCN | 5.3546 | −0.3608 | 2.0979 | 2 | Arg404 (2 hydrogen bonds) | 8 | Asn363, Leu380, Pro431, Ala432, Gly433, Met434, Asp476, So4603 |
| YTHDC2 | 6K6U | 6.2361 | −0.9342 | 0 | 1 | Asn1300 | 6 | Ser1304, Ile1309, Trp1310, Ser1311, Leu1365, Val1368 |
| YTHDF2 | 7R5F | 6.2342 | −0.7086 | 0 | 1 | Asn401 | 7 | Tyr403, Phe404, Tyr456, Ile500, Leu541, Ile544, Ala545 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Chen, X.; Bao, W.; Hong, X.; Li, C.; Lu, J.; Zhang, D.; Zhu, A. Effect of Humantenine on mRNA m6A Modification and Expression in Human Colon Cancer Cell Line HCT116. Genes 2022, 13, 781. https://doi.org/10.3390/genes13050781
Wu Y, Chen X, Bao W, Hong X, Li C, Lu J, Zhang D, Zhu A. Effect of Humantenine on mRNA m6A Modification and Expression in Human Colon Cancer Cell Line HCT116. Genes. 2022; 13(5):781. https://doi.org/10.3390/genes13050781
Chicago/Turabian StyleWu, Yajiao, Xiaoying Chen, Wenqiang Bao, Xinyu Hong, Chutao Li, Jiatong Lu, Dongcheng Zhang, and An Zhu. 2022. "Effect of Humantenine on mRNA m6A Modification and Expression in Human Colon Cancer Cell Line HCT116" Genes 13, no. 5: 781. https://doi.org/10.3390/genes13050781
APA StyleWu, Y., Chen, X., Bao, W., Hong, X., Li, C., Lu, J., Zhang, D., & Zhu, A. (2022). Effect of Humantenine on mRNA m6A Modification and Expression in Human Colon Cancer Cell Line HCT116. Genes, 13(5), 781. https://doi.org/10.3390/genes13050781







