Euarchontoglires Challenged by Incomplete Lineage Sorting
Abstract
1. Introduction
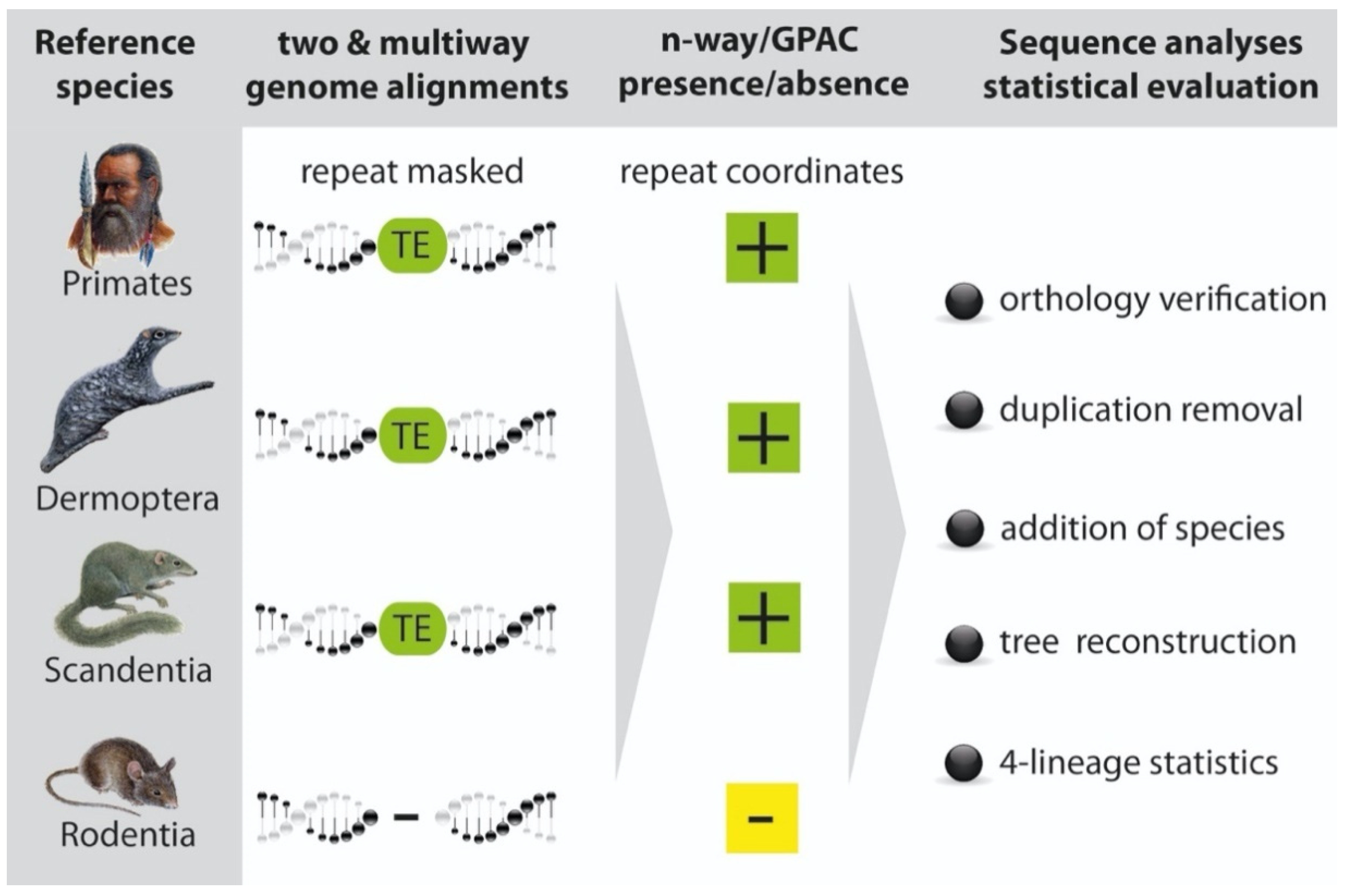
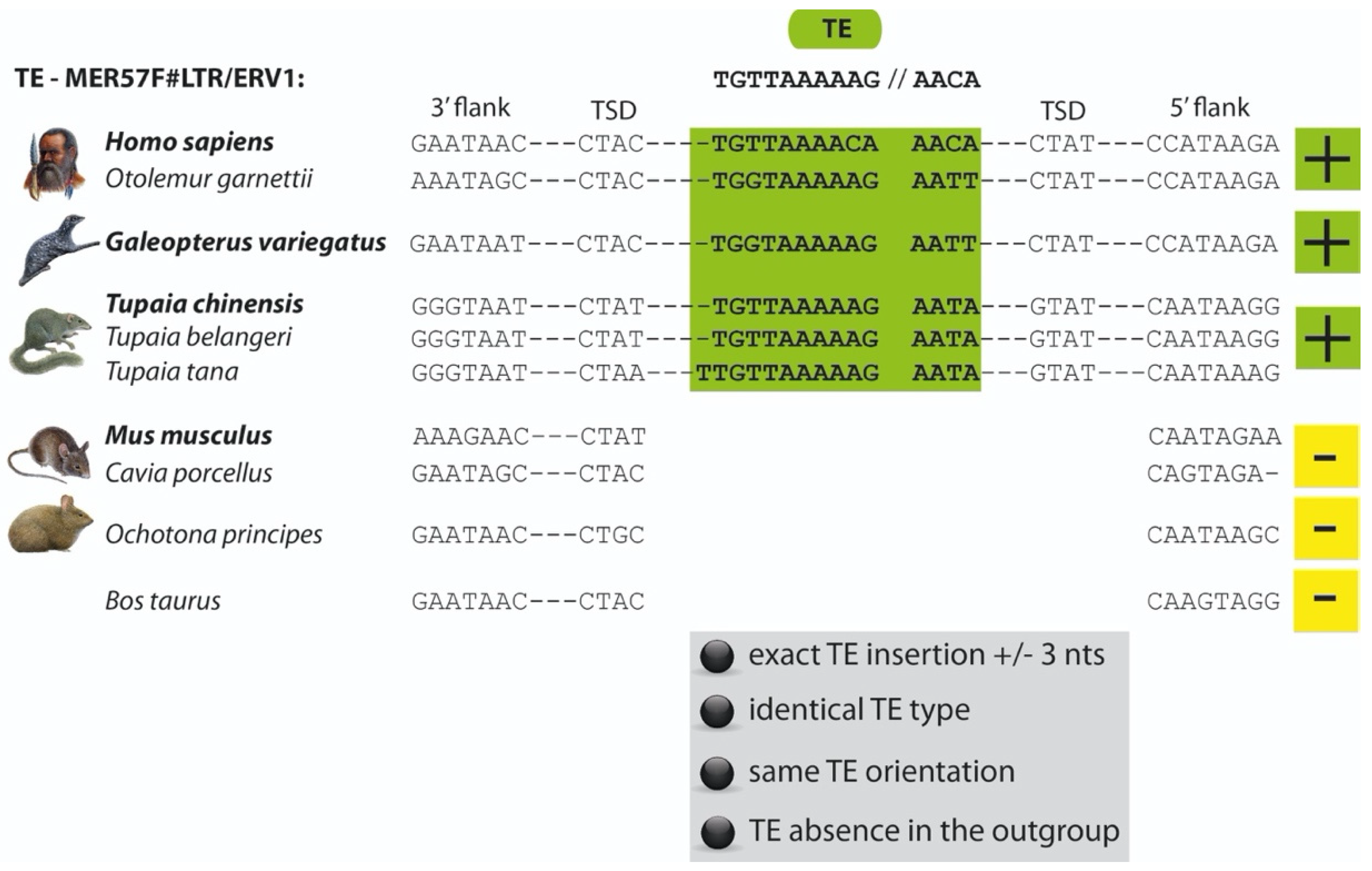
2. Materials and Methods
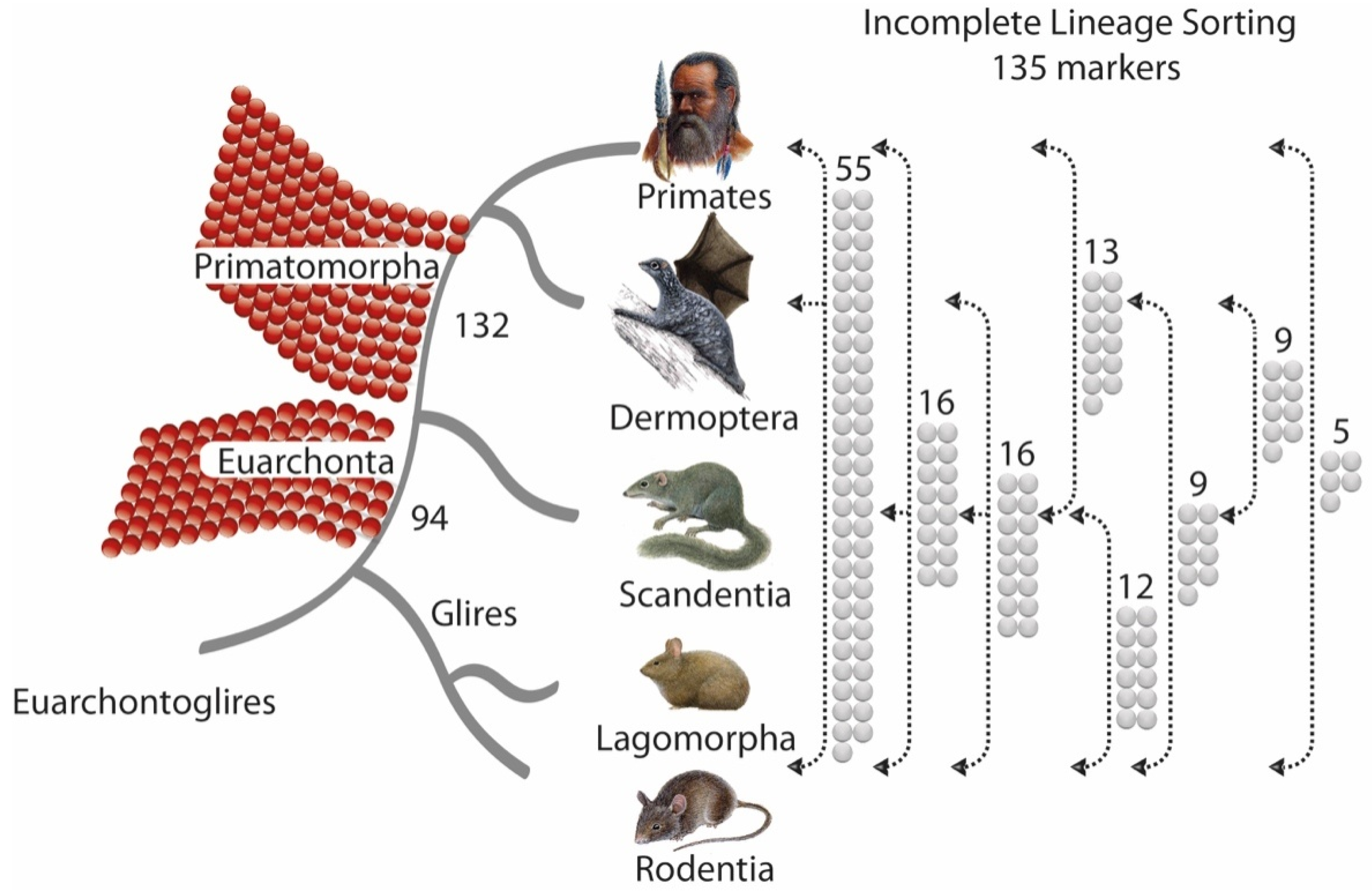
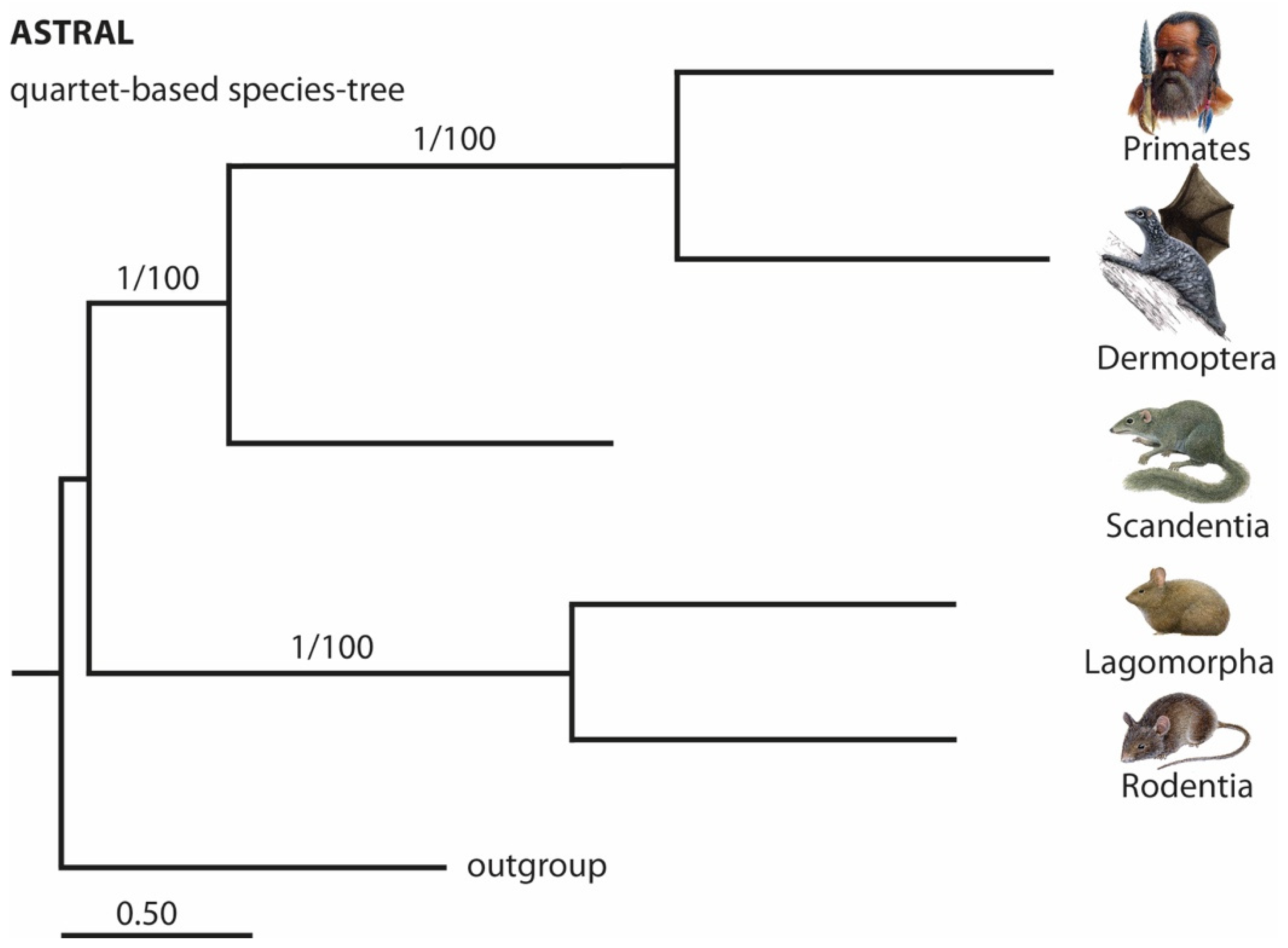
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murphy, W.J.; Eizirik, E.; Johnson, W.E.; Zhang, Y.P.; Ryder, O.A.; O’Brien, S.J. Molecular phylogenetics and the origins of placental mammals. Nature 2001, 409, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Waddell, P.J.; Kishino, H.; Ota, R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 2001, 12, 141–154. [Google Scholar] [PubMed]
- Kriegs, J.O.; Churakov, G.; Kiefmann, M.; Jordan, U.; Brosius, J.; Schmitz, J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006, 4, e91. [Google Scholar] [CrossRef]
- Mason, V.C.; Li, G.; Minx, P.; Schmitz, J.; Churakov, G.; Doronina, L.; Melin, A.D.; Dominy, N.J.; Lim, N.T.L.; Springer, M.S.; et al. Genomic analysis reveals hidden biodiversity within colugos, the sister group to primates. Sci. Adv. 2016, 2, e1600633. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Liu, L.; Edwards, S.V.; Wu, S. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc. Natl. Acad. Sci. USA 2012, 109, 14942–14947. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Huang, Z.Y.; Cao, C.C.; Chen, C.S.; Chen, Y.X.; Fan, D.D.; He, J.; Hou, H.L.; Hu, L.; Hu, X.T.; et al. Genome of the Chinese tree shrew. Nat. Commun. 2013, 4, 1426. [Google Scholar] [CrossRef]
- Kumar, V.; Hallström, B.M.; Janke, A. Coalescent-based genome analyses resolve the early branches of the euarchontoglires. PLoS ONE 2013, 8, e60019. [Google Scholar] [CrossRef]
- Nishihara, H.; Terai, Y.; Okada, N. Characterization of novel Alu- and tRNA-related SINEs from the tree shrew and evolutionary implications of their origins. Mol. Biol. Evol. 2002, 19, 1964–1972. [Google Scholar] [CrossRef][Green Version]
- Schmitz, J.; Ohme, M.; Zischler, H. The complete mitochondrial genome of Tupaia belangeri and the phylogenetic affiliation of Scandentia to other eutherian orders. Mol. Biol. Evol. 2000, 17, 1334–1343. [Google Scholar] [CrossRef]
- Xu, L.; Chen, S.Y.; Nie, W.H.; Jiang, X.L.; Yao, Y.G. Evaluating the phylogenetic position of Chinese tree shrew (Tupaia belangeri chinensis) based on complete mitochondrial genome: Implication for using tree shrew as an alternative experimental animal to primates in biomedical research. JGG 2012, 39, 131–137. [Google Scholar] [CrossRef]
- Murphy, W.J.; Eizirik, E.; O’Brien, S.J.; Madsen, O.; Scally, M.; Douady, C.J.; Teeling, E.; Ryder, O.A.; Stanhope, M.J.; de Jong, W.W.; et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 2001, 294, 2348–2351. [Google Scholar] [CrossRef] [PubMed]
- Adkins, R.M.; Honeycutt, R.L. Molecular phylogeny of the superorder Archonta. Proc. Natl. Acad. Sci. USA 1991, 88, 10317–10321. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sun, F.; Xu, S.; Yang, G.; Li, M. The position of tree shrews in the mammalian tree: Comparing multi-gene analyses with phylogenomic results leaves monophyly of Euarchonta doubtful. Integr. Zool. 2015, 10, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, H.; Satta, Y.; Nikaido, M.; Thewissen, J.G.M.; Stanhope, M.J.; Okada, N. A retroposon analysis of Afrotherian phylogeny. Mol. Biol. Evol. 2005, 22, 1823–1833. [Google Scholar] [CrossRef]
- Doronina, L.; Churakov, G.; Kuritzin, A.; Shi, J.; Baertsch, R.; Clawson, H.; Schmitz, J. Speciation network in Laurasiatheria: Retrophylogenomic signals. Genome Res. 2017, 27, 997–1003. [Google Scholar] [CrossRef]
- Suh, A.; Paus, M.; Kiefmann, M.; Churakov, G.; Franke, F.A.; Brosius, J.; Kriegs, J.O.; Schmitz, J. Mesozoic retroposons reveal parrots as the closest living relatives of passerine birds. Nat. Commun. 2011, 2, 443. [Google Scholar] [CrossRef]
- Kuritzin, A.; Kischka, T.; Schmitz, J.; Churakov, G. Incomplete lineage sorting and hybridization statistics for large-scale retroposon insertion data. PLoS Comput. Biol. 2016, 12, e1004812. [Google Scholar] [CrossRef]
- Doronina, L.; Feigin, C.Y.; Schmitz, J. Reunion of Australasian possums by shared SINE insertions. Syst. Biol. 2022, syac025. [Google Scholar] [CrossRef]
- Degnan, J.H.; Rosenberg, N.A. Gene tree discordance, phylogenetic inference and the multispecies coalescent. TREE 2009, 34, 332–340. [Google Scholar] [CrossRef]
- Doronina, L.; Reising, O.; Clawson, H.; Ray, D.A.; Schmitz, J. True homoplasy of retrotransposon insertions in primates. Syst. Biol. 2019, 68, 482–493. [Google Scholar] [CrossRef]
- Doronina, L.; Reising, O.; Schmitz, J. Gene conversion amongst Alu SINE elements. Genes 2021, 12, 905. [Google Scholar] [CrossRef] [PubMed]
- Churakov, G.; Grundmann, N.; Kuritzin, A.; Brosius, J.; Makalowski, W.; Schmitz, J. A novel web-based TinT application and the chronology of the Primate Alu retroposon activity. BMC Evol. Biol. 2010, 10, 376. [Google Scholar] [CrossRef] [PubMed]
- Noll, A.; Grundmann, N.; Churakov, G.; Brosius, J.; Makalowski, W.; Schmitz, J. GPAC—Genome presence/absence compiler: A web application to comparatively visualize multiple genome-level changes. Mol. Biol. Evol. 2015, 32, 275–286. [Google Scholar] [CrossRef]
- Churakov, G.; Zhang, F.; Grundmann, N.; Makalowski, W.; Noll, A.; Doronina, L.; Schmitz, J. The multicomparative 2-n-way genome suite. Genome Res. 2020, 30, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Churakov, G.; Kuritzin, A.; Chukharev, K.; Zhang, F.; Wünnemann, F.; Ulyantsev, V.; Schmitz, J. A 4-lineage statistical suite to evaluate the support of large-scale retrotransposon insertion data to reconstruct evolutionary trees. BioRxiv 2020. [Google Scholar] [CrossRef]
- Springer, M.S.; Molloy, E.K.; Sloan, D.B.; Simmons, M.P.; Gatesy, J. ILS-aware analysis of low-homoplasy retroelement insertions: Inference of species trees and introgression using quartets. J. Hered. 2020, 111, 147–168. [Google Scholar] [CrossRef]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018, 19, 153. [Google Scholar] [CrossRef]
- Mirarab, S.; Reaz, R.; Bayzid, M.S.; Zimmermann, T.; Swenson, M.S.; Warnow, T. ASTRAL: Genome-scale coalescent-based species tree estimation. Bioinformatics 2014, 30, i541–i548. [Google Scholar] [CrossRef]
- Kriegs, J.O.; Churakov, G.; Jurka, J.; Brosius, J.; Schmitz, J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007, 23, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.L.; Li, M.L.; Ayoola, A.O.; Murphy, R.W.; Wu, D.D.; Shao, Y. Conserved sequences identify the closest living relatives of primates. Zool. Res. 2019, 40, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Douzery, E.J.P.; Huchon, D. Rabbits, if anything, are likely Glires. Mol. Phylogenet. Evol. 2004, 33, 922–935. [Google Scholar] [CrossRef]
- McCormack, J.E.; Faircloth, B.C.; Crawford, N.G.; Gowaty, P.A.; Brumfield, R.T.; Glenn, T.C. Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis. Genome Res. 2012, 22, 746–754. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doronina, L.; Reising, O.; Clawson, H.; Churakov, G.; Schmitz, J. Euarchontoglires Challenged by Incomplete Lineage Sorting. Genes 2022, 13, 774. https://doi.org/10.3390/genes13050774
Doronina L, Reising O, Clawson H, Churakov G, Schmitz J. Euarchontoglires Challenged by Incomplete Lineage Sorting. Genes. 2022; 13(5):774. https://doi.org/10.3390/genes13050774
Chicago/Turabian StyleDoronina, Liliya, Olga Reising, Hiram Clawson, Gennady Churakov, and Jürgen Schmitz. 2022. "Euarchontoglires Challenged by Incomplete Lineage Sorting" Genes 13, no. 5: 774. https://doi.org/10.3390/genes13050774
APA StyleDoronina, L., Reising, O., Clawson, H., Churakov, G., & Schmitz, J. (2022). Euarchontoglires Challenged by Incomplete Lineage Sorting. Genes, 13(5), 774. https://doi.org/10.3390/genes13050774





