The Astonishing Large Family of HSP40/DnaJ Proteins Existing in Leishmania
Abstract
:1. Introduction
2. The Stressful Life of Leishmania Parasites
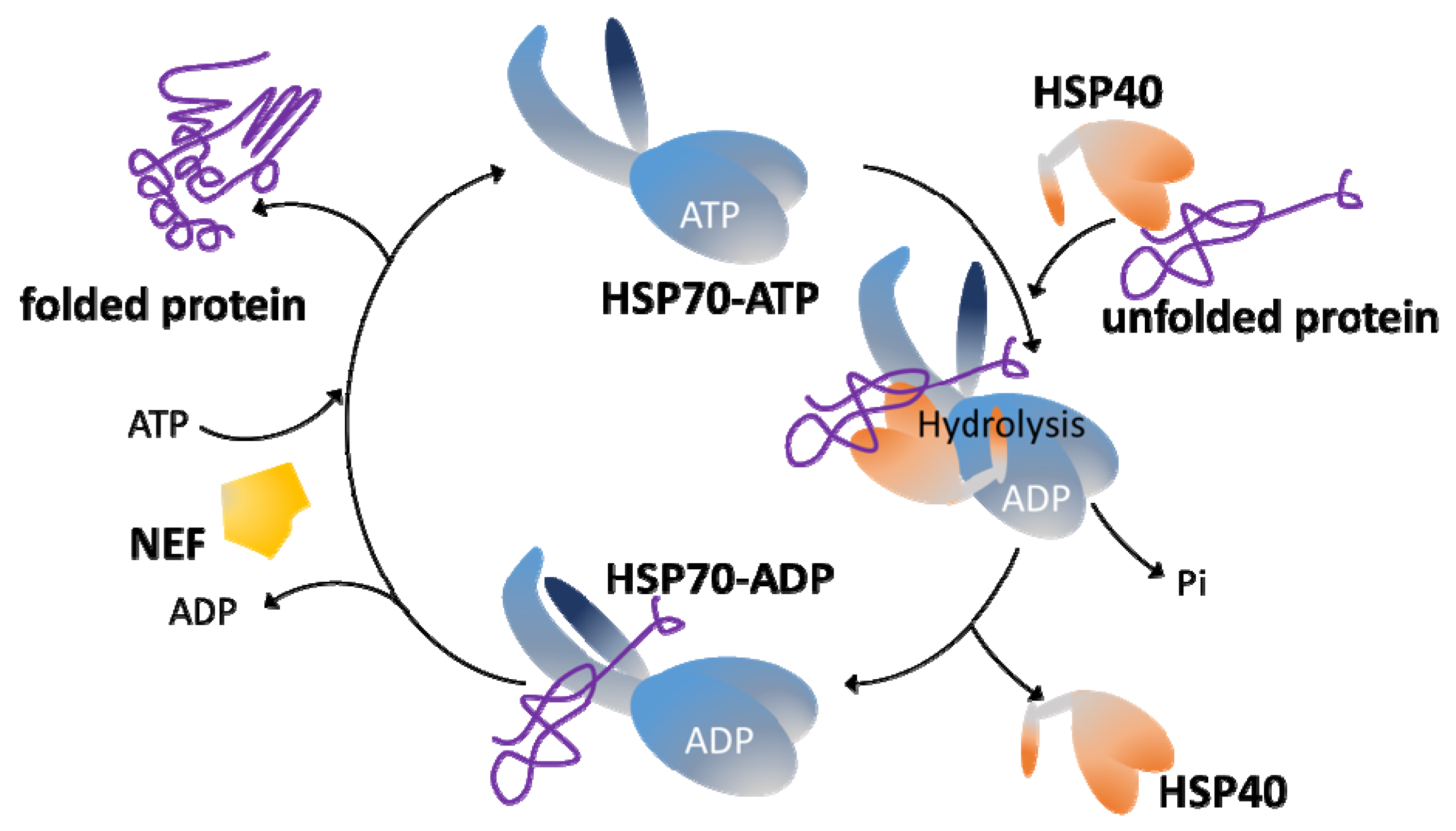
3. The HSP70/HSP40 Chaperone System
4. J-Domain Proteins
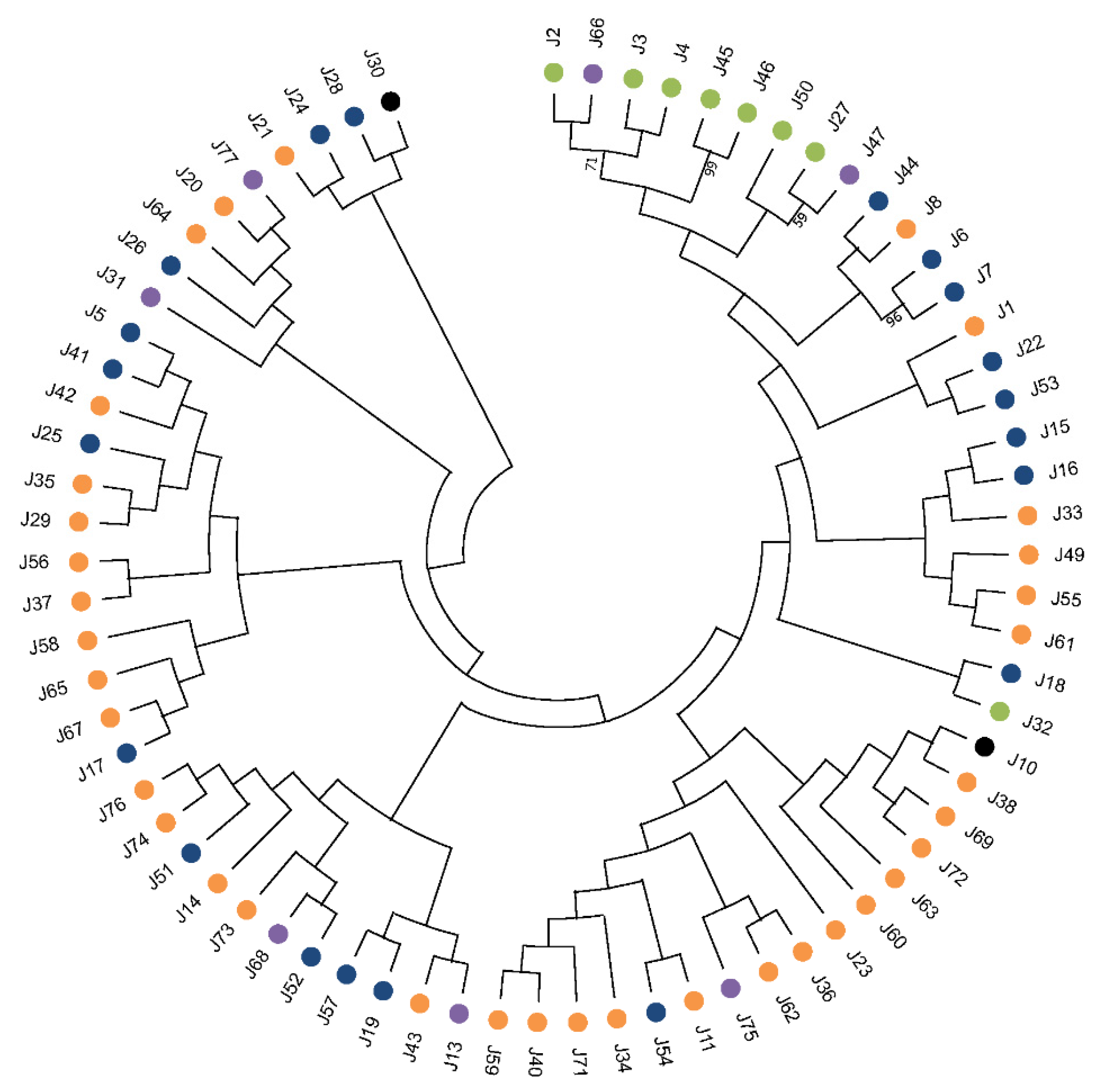
5. Appraisal and Updating of the Compendium of HSP40s in L. infantum
6. Concluding Remarks and Future Work
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Fernández-Fernández, M.R.; Valpuesta, J.M. Hsp70 chaperone: A master player in protein homeostasis. F1000Research 2018, 7, 1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, E.A.; Marszalek, J. How Do J-Proteins Get Hsp70 to Do So Many Different Things? Trends Biochem. Sci. 2017, 42, 355–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Sasidharan, S.; Saudagar, P. Leishmaniasis: Where are we and where are we heading? Parasitol. Res. 2021, 120, 1541–1554. [Google Scholar] [CrossRef]
- Requena, J.M. The Stressful Life of pathogenic Leishmania species. In Stress Response in Microbiology; Requena, J.M., Ed.; Caister Academic Press: Norfolk, UK, 2012; pp. 323–346. [Google Scholar]
- Lindquist, S.; Craig, E.A. THE HEAT-SHOCK PROTEINS. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Requena, J.M.; Montalvo, A.M.; Fraga, J. Molecular Chaperones of Leishmania: Central Players in Many Stress-Related and -Unrelated Physiological Processes. Biomed. Res. Int. 2015, 2015, 301326. [Google Scholar] [CrossRef] [Green Version]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef] [Green Version]
- Hartl, F.U.; Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009, 16, 574–581. [Google Scholar] [CrossRef]
- Mayer, M.P.; Gierasch, L.M. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 2019, 294, 2085–2097. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.S. Protein Phylogenies and Signature Sequences: A Reappraisal of Evolutionary Relationships among Archaebacteria, Eubacteria, and Eukaryotes. Microbiol. Mol. Biol. Rev. 1998, 62, 1435–1491. [Google Scholar] [CrossRef] [Green Version]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preissler, S.; Deuerling, E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 2012, 37, 274–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Rosenzweig, R. Structural and Biochemical Properties of Hsp40/Hsp70 Chaperone System. Adv. Exp. Med. Biol. 2020, 1243, 3–20. [Google Scholar] [CrossRef]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. CMLS 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Langer, T.; Lu, C.; Echols, H.; Flanagan, J.; Hayer, M.K.; Hartl, F.U. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 1992, 356, 683–689. [Google Scholar] [CrossRef]
- Hennessy, F.; Nicoll, W.S.; Zimmermann, R.; Cheetham, M.E.; Blatch, G.L. Not all J domains are created equal: Implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. A Publ. Protein Soc. 2005, 14, 1697–1709. [Google Scholar] [CrossRef] [Green Version]
- Craig, E.A.; Huang, P.; Aron, R.; Andrew, A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. In Reviews of Physiology, Biochemistry and Pharmacology; Springer: Berlin/Heidelberg, Germany, 2006; Volume 156, pp. 1–21. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [Green Version]
- Njunge, J.M.; Ludewig, M.H.; Boshoff, A.; Pesce, E.R.; Blatch, G.L. Hsp70s and J proteins of Plasmodium parasites infecting rodents and primates: Structure, function, clinical relevance, and drug targets. Curr. Pharm. Des. 2013, 19, 387–403. [Google Scholar] [CrossRef]
- Kityk, R.; Kopp, J.; Mayer, M.P. Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones. Mol. Cell 2018, 69, 227–237.e224. [Google Scholar] [CrossRef]
- Tsai, J.; Douglas, M.G. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 1996, 271, 9347–9354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Liang, C.; Zhou, L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. A Publ. Protein Soc. 2020, 29, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Craig, E.A. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol. Cell. Biol. 1999, 19, 7751–7758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louw, C.A.; Ludewig, M.H.; Mayer, J.; Blatch, G.L. The Hsp70 chaperones of the Tritryps are characterized by unusual features and novel members. Parasitol. Int. 2010, 59, 497–505. [Google Scholar] [CrossRef]
- Nillegoda, N.B.; Kirstein, J.; Szlachcic, A.; Berynskyy, M.; Stank, A.; Stengel, F.; Arnsburg, K.; Gao, X.; Scior, A.; Aebersold, R.; et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 2015, 524, 247–251. [Google Scholar] [CrossRef] [Green Version]
- Kampinga, H.H.; Andreasson, C.; Barducci, A.; Cheetham, M.E.; Cyr, D.; Emanuelsson, C.; Genevaux, P.; Gestwicki, J.E.; Goloubinoff, P.; Huerta-Cepas, J.; et al. Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones 2019, 24, 7–15. [Google Scholar] [CrossRef]
- Ajit Tamadaddi, C.; Sahi, C. J domain independent functions of J proteins. Cell Stress Chaperones 2016, 21, 563–570. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-de la Fuente, S.; Peiro-Pastor, R.; Rastrojo, A.; Moreno, J.; Carrasco-Ramiro, F.; Requena, J.M.; Aguado, B. Resequencing of the Leishmania infantum (strain JPCM5) genome and de novo assembly into 36 contigs. Sci. Rep. 2017, 7, 18050. [Google Scholar] [CrossRef]
- Folgueira, C.; Requena, J.M. A postgenomic view of the heat shock proteins in kinetoplastids. FEMS Microbiol. Rev. 2007, 31, 359–377. [Google Scholar] [CrossRef]
- Fan, F.; Yang, X.; Cheng, Y.; Kang, Y.; Chai, X. The DnaJ Gene Family in Pepper (Capsicum annuum L.): Comprehensive Identification, Characterization and Expression Profiles. Front. Plant Sci. 2017, 8, 689. [Google Scholar] [CrossRef] [Green Version]
- Requena, J.M.; Solana, J.C. LINF_070013700. In Mendeley Data, V2; Universidad Autonoma de Madrid: Madrid, Spain, 2022. [Google Scholar] [CrossRef]
- Requena, J.M.; Solana, J.C. LINF_170010900. In Mendeley Data, V1; Universidad Autonoma de Madrid: Madrid, Spain, 2021. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. CABIOS 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Walsh, P.; Bursać, D.; Law, Y.C.; Cyr, D.; Lithgow, T. The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 2004, 5, 567–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, P.B.; Stevens, J.R.; Gaunt, M.W.; Gidley, J.; Gibson, W.C. Trypanosomes are monophyletic: Evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int. J. Parasitol. 2004, 34, 1393–1404. [Google Scholar] [CrossRef]
- Xie, J.L.; Bohovych, I.; Wong, E.O.Y.; Lambert, J.P.; Gingras, A.C.; Khalimonchuk, O.; Cowen, L.E.; Leach, M.D. Ydj1 governs fungal morphogenesis and stress response, and facilitates mitochondrial protein import via Mas1 and Mas2. Microb. Cell 2017, 4, 342–361. [Google Scholar] [CrossRef]
- Kim Chiaw, P.; Hantouche, C.; Wong, M.J.H.; Matthes, E.; Robert, R.; Hanrahan, J.W.; Shrier, A.; Young, J.C. Hsp70 and DNAJA2 limit CFTR levels through degradation. PLoS ONE 2019, 14, e0220984. [Google Scholar] [CrossRef] [Green Version]
- Schlenstedt, G.; Harris, S.; Risse, B.; Lill, R.; Silver, P.A. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s. J. Cell Biol. 1995, 129, 979–988. [Google Scholar] [CrossRef]
- Elwi, A.N.; Lee, B.; Meijndert, H.C.; Braun, J.E.; Kim, S.W. Mitochondrial chaperone DnaJA3 induces Drp1-dependent mitochondrial fragmentation. Int. J. Biochem. Cell Biol. 2012, 44, 1366–1376. [Google Scholar] [CrossRef]
- Deloche, O.; Liberek, K.; Zylicz, M.; Georgopoulos, C. Purification and biochemical properties of Saccharomyces cerevisiae Mdj1p, the mitochondrial DnaJ homologue. J. Biol. Chem. 1997, 272, 28539–28544. [Google Scholar] [CrossRef] [Green Version]
- Zhong, T.; Arndt, K.T. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell 1993, 73, 1175–1186. [Google Scholar] [CrossRef]
- Klaips, C.L.; Gropp, M.H.M.; Hipp, M.S.; Hartl, F.U. Sis1 potentiates the stress response to protein aggregation and elevated temperature. Nat. Commun. 2020, 11, 6271. [Google Scholar] [CrossRef] [PubMed]
- Young, B.P.; Craven, R.A.; Reid, P.J.; Willer, M.; Stirling, C.J. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001, 20, 262–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldshmidt, H.; Sheiner, L.; Butikofer, P.; Roditi, I.; Uliel, S.; Gunzel, M.; Engstler, M.; Michaeli, S. Role of protein translocation pathways across the endoplasmic reticulum in Trypanosoma brucei. J. Biol. Chem. 2008, 283, 32085–32098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daverkausen-Fischer, L.; Pröls, F. Dual topology of co-chaperones at the membrane of the endoplasmic reticulum. Cell Death Discov. 2021, 7, 203. [Google Scholar] [CrossRef]
- Nishikawa, S.; Endo, T. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J. Biol. Chem. 1997, 272, 12889–12892. [Google Scholar] [CrossRef] [Green Version]
- Webb, T.R.; Cross, S.H.; McKie, L.; Edgar, R.; Vizor, L.; Harrison, J.; Peters, J.; Jackson, I.J. Diphthamide modification of eEF2 requires a J-domain protein and is essential for normal development. J. Cell Sci. 2008, 121, 3140–3145. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Ziegelhoffer, T.; Delewski, W.; Berger, S.E.; Sabat, G.; Craig, E.A. Pathway of Hsp70 interactions at the ribosome. Nat. Commun. 2021, 12, 5666. [Google Scholar] [CrossRef]
- Ng, A.C.; Baird, S.D.; Screaton, R.A. Essential role of TID1 in maintaining mitochondrial membrane potential homogeneity and mitochondrial DNA integrity. Mol. Cell. Biol. 2014, 34, 1427–1437. [Google Scholar] [CrossRef] [Green Version]
- Ioakeimidis, F.; Ott, C.; Kozjak-Pavlovic, V.; Violitzi, F.; Rinotas, V.; Makrinou, E.; Eliopoulos, E.; Fasseas, C.; Kollias, G.; Douni, E. A splicing mutation in the novel mitochondrial protein DNAJC11 causes motor neuron pathology associated with cristae disorganization, and lymphoid abnormalities in mice. PLoS ONE 2014, 9, e104237. [Google Scholar] [CrossRef] [Green Version]
- Voisine, C.; Cheng, Y.C.; Ohlson, M.; Schilke, B.; Hoff, K.; Beinert, H.; Marszalek, J.; Craig, E.A. Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2001, 98, 1483–1488. [Google Scholar] [CrossRef] [Green Version]
- Maio, N.; Ghezzi, D.; Verrigni, D.; Rizza, T.; Bertini, E.; Martinelli, D.; Zeviani, M.; Singh, A.; Carrozzo, R.; Rouault, T.A. Disease-Causing SDHAF1 Mutations Impair Transfer of Fe-S Clusters to SDHB. Cell Metab. 2016, 23, 292–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Silva, P.D.; Schilke, B.; Walter, W.; Andrew, A.; Craig, E.A. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. USA 2003, 100, 13839–13844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaschner, L.A.; Sharma, R.; Shrestha, O.K.; Meyer, A.E.; Craig, E.A. A conserved domain important for association of eukaryotic J-protein co-chaperones Jjj1 and Zuo1 with the ribosome. Biochim. Biophys. Acta 2015, 1853, 1035–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammond, C.M.; Bao, H.; Hendriks, I.A.; Carraro, M.; García-Nieto, A.; Liu, Y.; Reverón-Gómez, N.; Spanos, C.; Chen, L.; Rappsilber, J.; et al. DNAJC9 integrates heat shock molecular chaperones into the histone chaperone network. Mol. Cell 2021, 81, 2533–2548.e2539. [Google Scholar] [CrossRef] [PubMed]
- Fujibayashi, A.; Taguchi, T.; Misaki, R.; Ohtani, M.; Dohmae, N.; Takio, K.; Yamada, M.; Gu, J.; Yamakami, M.; Fukuda, M.; et al. Human RME-8 is involved in membrane trafficking through early endosomes. Cell Struct. Funct. 2008, 33, 35–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamdhade, M.D.; Pawar, H.; Chavan, S.; Sathe, G.; Umasankar, P.K.; Mahale, K.N.; Dixit, T.; Madugundu, A.K.; Prasad, T.S.; Gowda, H.; et al. Comprehensive proteomics analysis of glycosomes from Leishmania donovani. Omics 2015, 19, 157–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, S.; Morris, M.T. Glycosome biogenesis in trypanosomes and the de novo dilemma. PLoS Negl. Trop. Dis. 2017, 11, e0005333. [Google Scholar] [CrossRef]
- Yamamoto, Y.H.; Kimura, T.; Momohara, S.; Takeuchi, M.; Tani, T.; Kimata, Y.; Kadokura, H.; Kohno, K. A novel ER J-protein DNAJB12 accelerates ER-associated degradation of membrane proteins including CFTR. Cell Struct. Funct. 2010, 35, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Liu, Z.; Zhang, S.; Li, Y.; Xia, W.; Wang, C.; Xiang, H.; Liu, Z.; Tan, L.; Fang, Y.; et al. Hsp40 proteins phase separate to chaperone the assembly and maintenance of membraneless organelles. Proc. Natl. Acad. Sci. USA 2020, 117, 31123–31133. [Google Scholar] [CrossRef]
- Deng, S.; Marmorstein, R. Protein N-terminal Acetylation: Structural Basis, Mechanism, Versatility, and Regulation. Trends Biochem. Sci. 2021, 46, 15–27. [Google Scholar] [CrossRef]
- Ito, N.; Kamiguchi, K.; Nakanishi, K.; Sokolovskya, A.; Hirohashi, Y.; Tamura, Y.; Murai, A.; Yamamoto, E.; Kanaseki, T.; Tsukahara, T.; et al. A novel nuclear DnaJ protein, DNAJC8, can suppress the formation of spinocerebellar ataxia 3 polyglutamine aggregation in a J-domain independent manner. Biochem. Biophys. Res. Commun. 2016, 474, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, N.S.; Bruinsma, E.; Uhl, C.; Obermann, W.M.; Toft, D. Role of the cochaperone Tpr2 in Hsp90 chaperoning. Biochemistry 2008, 47, 8203–8213. [Google Scholar] [CrossRef] [PubMed]
- Tsigankov, P.; Gherardini, P.F.; Helmer-Citterich, M.; Späth, G.F.; Zilberstein, D. Phosphoproteomic analysis of differentiating Leishmania parasites reveals a unique stage-specific phosphorylation motif. J. Proteome Res. 2013, 12, 3405–3412. [Google Scholar] [CrossRef] [PubMed]
- Douanne, N.; Dong, G.; Douanne, M.; Olivier, M.; Fernandez-Prada, C. Unravelling the proteomic signature of extracellular vesicles released by drug-resistant Leishmania infantum parasites. PLoS Negl. Trop. Dis. 2020, 14, e0008439. [Google Scholar] [CrossRef]
- Martin, J.L.; Yates, P.A.; Soysa, R.; Alfaro, J.F.; Yang, F.; Burnum-Johnson, K.E.; Petyuk, V.A.; Weitz, K.K.; Camp, D.G., 2nd; Smith, R.D.; et al. Metabolic reprogramming during purine stress in the protozoan pathogen Leishmania donovani. PLoS Pathog. 2014, 10, e1003938. [Google Scholar] [CrossRef] [Green Version]
- Brotherton, M.C.; Racine, G.; Foucher, A.L.; Drummelsmith, J.; Papadopoulou, B.; Ouellette, M. Analysis of stage-specific expression of basic proteins in Leishmania infantum. J. Proteome Res. 2010, 9, 3842–3853. [Google Scholar] [CrossRef]
- Sanchiz, A.; Morato, E.; Rastrojo, A.; Camacho, E.; Gonzalez-de la Fuente, S.G.; Marina, A.; Aguado, B.; Requena, J.M. The Experimental Proteome of Leishmania infantum Promastigote and Its Usefulness for Improving Gene Annotations. Genes 2020, 11, 1036. [Google Scholar] [CrossRef]
- Beneke, T.; Demay, F.; Hookway, E.; Ashman, N.; Jeffery, H.; Smith, J.; Valli, J.; Becvar, T.; Myskova, J.; Lestinova, T.; et al. Genetic dissection of a Leishmania flagellar proteome demonstrates requirement for directional motility in sand fly infections. PLoS Pathog. 2019, 15, e1007828. [Google Scholar] [CrossRef] [Green Version]
- Blatch, G.L.; Lässle, M. The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. BioEssays News Rev. Mol. Cell. Dev. Biol. 1999, 21, 932–939. [Google Scholar] [CrossRef]
- Scheufler, C.; Brinker, A.; Bourenkov, G.; Pegoraro, S.; Moroder, L.; Bartunik, H.; Hartl, F.U.; Moarefi, I. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 2000, 101, 199–210. [Google Scholar] [CrossRef]
- Rodríguez-Vega, A.; Losada-Barragán, M.; Berbert, L.R.; Mesquita-Rodrigues, C.; Bombaça, A.C.S.; Menna-Barreto, R.; Aquino, P.; Carvalho, P.C.; Padrón, G.; de Jesus, J.B.; et al. Quantitative analysis of proteins secreted by Leishmania (Viannia) braziliensis strains associated to distinct clinical manifestations of American Tegumentary Leishmaniasis. J. Proteom. 2021, 232, 104077. [Google Scholar] [CrossRef] [PubMed]
- Tsigankov, P.; Gherardini, P.F.; Helmer-Citterich, M.; Späth, G.F.; Myler, P.J.; Zilberstein, D. Regulation dynamics of Leishmania differentiation: Deconvoluting signals and identifying phosphorylation trends. Mol. Cell. Proteom. 2014, 13, 1787–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales, M.A.; Watanabe, R.; Dacher, M.; Chafey, P.; Osorio y Fortéa, J.; Scott, D.A.; Beverley, S.M.; Ommen, G.; Clos, J.; Hem, S.; et al. Phosphoproteome dynamics reveal heat-shock protein complexes specific to the Leishmania donovani infectious stage. Proc. Natl. Acad. Sci. USA 2010, 107, 8381–8386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenzweig, D.; Smith, D.; Myler, P.J.; Olafson, R.W.; Zilberstein, D. Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics 2008, 8, 1843–1850. [Google Scholar] [CrossRef]
- Sprung, R.; Chen, Y.; Zhang, K.; Cheng, D.; Zhang, T.; Peng, J.; Zhao, Y. Identification and validation of eukaryotic aspartate and glutamate methylation in proteins. J. Proteome Res. 2008, 7, 1001–1006. [Google Scholar] [CrossRef] [Green Version]
- Dutta, T.; Pesce, E.R.; Maier, A.G.; Blatch, G.L. Role of the J Domain Protein Family in the Survival and Pathogenesis of Plasmodium falciparum. Adv. Exp. Med. Biol. 2021, 1340, 97–123. [Google Scholar] [CrossRef]
- Yagoubat, A.; Corrales, R.M.; Bastien, P.; Lévêque, M.F.; Sterkers, Y. Gene Editing in Trypanosomatids: Tips and Tricks in the CRISPR-Cas9 Era. Trends Parasitol. 2020, 36, 745–760. [Google Scholar] [CrossRef]
- Baker, N.; Catta-Preta, C.M.C.; Neish, R.; Sadlova, J.; Powell, B.; Alves-Ferreira, E.V.C.; Geoghegan, V.; Carnielli, J.B.T.; Newling, K.; Hughes, C.; et al. Systematic functional analysis of Leishmania protein kinases identifies regulators of differentiation or survival. Nat. Commun. 2021, 12, 1244. [Google Scholar] [CrossRef]
- Rastrojo, A.; Garcia-Hernandez, R.; Vargas, P.; Camacho, E.; Corvo, L.; Imamura, H.; Dujardin, J.C.; Castanys, S.; Aguado, B.; Gamarro, F.; et al. Genomic and transcriptomic alterations in Leishmania donovani lines experimentally resistant to antileishmanial drugs. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 246–264. [Google Scholar] [CrossRef]
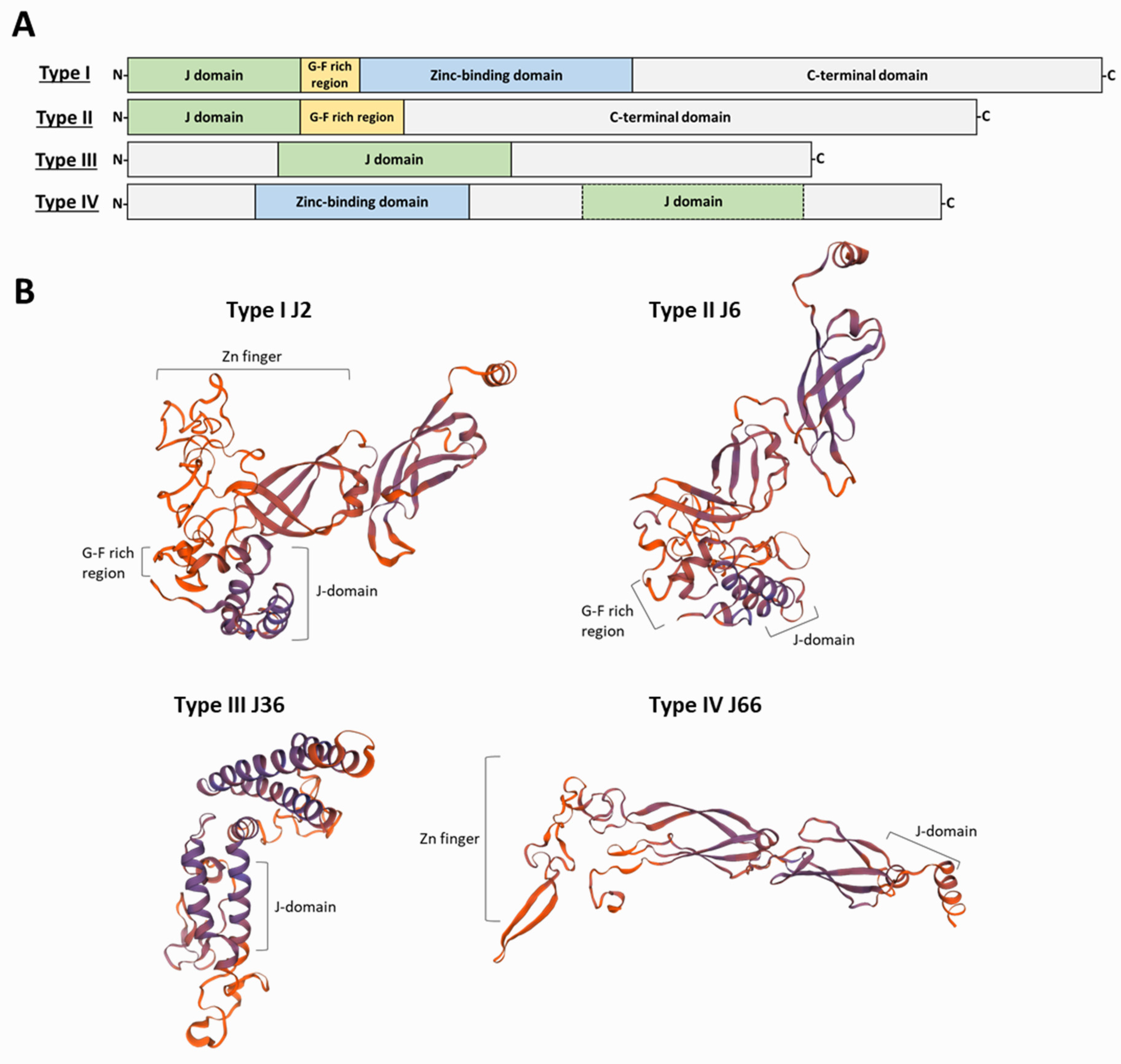
| Name | Gene ID | Size | J-Domain | G/F Rich | Zn-Finger | Type | Remarks a |
|---|---|---|---|---|---|---|---|
| J1 | LINF_320037800 | 329 | 14–80 | - | - | III | - |
| J2 | LINF_270032200 | 396 | 6–72 | 78–94 | 120–205 | I | - |
| J3 | LINF_210010300 | 453 | 6–72 | 83–99 | 125–210 | I | - |
| J4 | LINF_150019800 | 478 | 6–72 | 81–97 | 144–227 | I | - |
| J5 | LINF_360019000 | 364 | 86–180 | 161–183 | - | II | Transmembrane domains (192–215, 229–252). |
| J6 | LINF_360072700 | 345 | 4–70 | 124–140 | - | II | - |
| J7 | LINF_320025300 | 323 | 9–75 | 91–107 | - | II | - |
| J8 | LINF_240010000 | 794 | 42–108 | - | - | III | Signal peptide (1–24). |
| J10 | LINF_170010900 | 157 | - | - | - | - | Possibly mis-annotated (see text). HPD motif (5–7). Transmembrane domains (76–99, 113–136). |
| J11 | LINF_040012900 | 576 | 9–75 | - | - | III | Transmembrane domains (90–113, 127–150, 162–185, 194–217, 294–317, 346–369, 422–442, 461–484). |
| J13 | LINF_180020400 | 184 | 14–80 | - | III | - | |
| J14 | LINF_080014500 | 326 | 21–79/157–223 | - | - | III | Two J-domains. |
| J15 | LINF_190005600 | 432 | 5–71 | 128–149 | - | II | - |
| J16 | LINF_200016400 | 653 | 140–206 | 228–240 | - | II | SANT/Myb SANT/Myb domain (513–567). |
| J17 | LINF_120015200 | 608 | 4–70 | 64–126 | - | II | - |
| J18 | LINF_270009200 | 378 | 73–139 | - | - | III | Transmembrane domains (330–353, 359–377). |
| J19 | LINF_340048600 | 266 | 24–85 | 94–142 | - | II | Transmembrane domain (129–152). |
| J20 | LINF_360011700 | 260 | 171–237 | - | - | III | J-domain at C-terminus. |
| J21 | LINF_260019100 | 536 | 405–471 | - | - | III | J-domain at C-terminus. Transmembrane domain (491–514). |
| J22 | LINF_360028100 | 286 | 21–87 | 84–139 | - | II | Transmembrane domain (149–172). |
| J23 | LINF_180008300 | 244 | 49–115 | - | - | III | Transmembrane domain (185–208). |
| J24 | LINF_300022800 | 740 | 27–93 | 135–179 | - | II | - |
| J25 | LINF_260017700 | 898 | 459–525 | 535–551 | - | II | J-domain in the middle. |
| J26 | LINF_170005500 | 262 | 69–134 | - | - | III | Transmembrane domain (170–193). |
| J27 | LINF_040014400 | 493 | 92–158 | 164–194 | 253–331 | I | - |
| J28 | LINF_260017000 | 652 | 282–348 | 342–396 | - | II | J-domain in the middle. Transmembrane domains (12–35, 110–133, 139–162). |
| J29 | LINF_240016000 | 435 | 371–434 | - | - | III | J-domain at C-terminus. Transmembrane domain (268–291). |
| J30 | LINF_070013700 | 304 | - | - | - | - | Truncated. Lacking first 242 aa. |
| J31 | LINF_260014300 | 843 | 36–102 | - | - | IV | - |
| J32 | LINF_250029000 | 377 | 8–74 | 81–102 | 322–346 | I | - |
| J33 | LINF_360054000 | 275 | 7–73 | - | - | III | - |
| J34 | LINF_350052100 | 491 | 134–200 | - | - | III | Transmembrane domains (12–35, 112–132, 218–241). |
| J35 | LINF_140006000 | 523 | 377–443 | - | - | III | J-domain at C-terminus. |
| J36 | LINF_250023500 | 278 | 61–156 | - | - | III | - |
| J37 | LINF_180019800 | 1121 | 5–71 | - | - | III | - |
| J38 | LINF_300029900 | 336 | 15–81 | - | - | III | - |
| J40 | LINF_100017600 | 275 | 61–156 | - | - | III | - |
| J41 | LINF_310010500 | 603 | 288–354 | 373–396 | - | II | J-domain at the middle. |
| J42 | LINF_180022200 | 580 | 518–577 | - | - | III | J-domain at C-terminus. Tetratricopeptide (TPR)-like helical domain (135–272). |
| J43 | LINF_350045900 | 386 | 4–70 | - | - | III | - |
| J44 | LINF_310039900 | 217 | 18–84 | 105–148 | - | II | Transmembrane domains (121–144). |
| J45 | LINF_320040500 | 400 | 57–123 | 121–150 | 176–259 | I | Transmembrane domain (12–32). |
| J46 | LINF_250017100 | 396 | 57–123 | 123–150 | 175–258 | I | Transmembrane domains (8–31). |
| J47 | LINF_200010900 | 545 | 68–134 | - | 257–335 | IV | - |
| J49 | LINF_300015900 | 423 | 70–136 | - | - | III | Prokaryotic lipoprotein domain (1–33). |
| J50 | LINF_350035100 | 478 | 47–113 | 119–159 | 185–270 | I | - |
| J51 | LINF_340029700 | 808 | 700–766 | 779–807 | - | II | J-domain at C-terminus. TPR region (345–471, 572–677). TPR domain (345–378, 384–417, 610–643, 644–677). |
| J52 | LINF_360010300 | 510 | 386–452 | 461–509 | - | II | J-domain at C-terminus. TPR region (17–118, 254–359). TPR domain (17–50, 51–84, 208–241, 254–287, 326–359). |
| J53 | LINF_140019700 | 574 | 433–499 | 525–555 | - | II | J-domain at C-terminus. Transmembrane domain (20–43). TPR region (51–120, 223–290). |
| J54 | LINF_330016300 | 581 | 3–69 | - | - | II | Transmembrane domains (90–113, 129–152, 156–179, 198–221, 240–263, 269–292, 416–439, 459–482). |
| J55 | LINF_280018500 | 470 | 9–75 | - | - | III | - |
| J56 | LINF_330036200 | 266 | 77–133 | - | - | III | - |
| J57 | LINF_290026500 | 396 | 9–67 | 144–193 | - | II | - |
| J58 | LINF_240018200 | 808 | 5–68 | - | - | III | - |
| J59 | LINF_300027500 | 2451 | 1384–1450 | - | - | III | 2 GYF domain 2 (1059–1109). |
| J60 | LINF_090022000 | 413 | 42–108 | - | - | III | Prokaryotic lipoprotein domain (1–28). |
| J61 | LINF_080011700 | 296 | 3–69 | - | - | III | - |
| J62 | LINF_340005300 | 679 | 95–161 | - | - | III | Transmembrane domains (66–89, 174–197, 209–232). |
| J63 | LINF_320011200 | 316 | 42–108 | - | - | III | Transmembrane domain (278–301). |
| J64 | LINF_070013600 | 417 | 170–252 | - | - | III | J-domain in the middle. Transmembrane domain (362–385). |
| J65 | LINF_340046400 | 690 | 616–682 | - | - | III | J-domain at C-terminus. TPR-like helical domain (449–560). |
| J66 | LINF_220005800 | 331 | 185–275 | - | 52–139 | IV | - |
| J67 | LINF_360013200 | 850 | 733–843 | - | - | III | J-domain at C-terminus. TPR region (232–333, 569–640). TPR domain (232–265, 569–602, 607–640). |
| J68 | LINF_240025300 | 121 | 54–120 | - | - | IV | - |
| J69 | LINF_360059200 | 346 | 19–77 | - | - | III | - |
| J71 | LINF_240005500 | 439 | 50–105 | - | - | III | Transmembrane domain (318–337). |
| J72 | LINF_350036200 | 428 | 20–86 | - | - | III | Transmembrane domain (147–170). |
| J73 | LINF_260031000 | 488 | 31–84 | - | - | III | - |
| J74 | LINF_280025400 | 578 | 66–119 | - | - | III | - |
| J75 | LINF_350007400 | 368 | 216–282 | - | 37–60 | IV | J-domain at C-terminus. C3H1-type zinc finger (37–60). |
| J76 | LINF_140005800 | 384 | 60–123 | - | - | III | TPR region (1–31). |
| J77 | LINF_250010800 | 197 | 79–148 | - | - | IV | Transmembrane domain (160–183). |
| Name | Ortholog [References] | Suggested Cellular Location a | Identity (%) |
|---|---|---|---|
| J2 | DNAJA1, DNAJA4, mas5 [39,63] | Glycosome, nucleolus | 44.70-44.02 |
| J3 | DNAJA2 [40] | Glycosome | 42.35 |
| J4 | DNAJA1, DNAJA4 [39,63] | Nucleolus | 29.76, 31.97 |
| J6 | SIS1, DNAJB1, DNAJB4, DNAJB5 [44,45,64] | Glycosome, nucleus | 35.03-38.83 |
| J7 | DNAJB4, DNAJB5, DNAJB1 [44,45] | - | 33.24-32.16 |
| J8 | - | Glycosome | - |
| J10 | Sec63 [46,47] | Endoplasmic reticulum (ER), nucleus | 3.69 |
| J11 | - | Ciliary pocket | - |
| J13 | DNAJC24 [50] | Cytoskeleton | 27.38 |
| J14 | DNAJC8, SPF31 [65] | Nucleus | 40.35, 29.41 |
| J16 | DNAJC2, zuotin [51] | Ribosome-associated complex | 27.22, 35.89 |
| J22 | pi041, C17A3.05c, DNAJB12 [62] | ER membrane | 27.51-28.57 |
| J27 | DNAJA3 [42,43,52] | Mitochondrion, glycosome | 33.77 |
| J31 | DNAJC11, SPCC63.03 [53] | Mitochondrion | 32.20, 27.27 |
| J32 | DNAJC21, JJJ1 [57] | - | 40.34, 31.46 |
| J33 | DNAJC9, C1071.09c [58] | Nucleus, histone-related function | 29.92, 31.25 |
| J34 | Sec63 [46,47] | ER membrane | 31.10 |
| J36 | DNAJC20/HscB, JAC1 [54,55] | Mitochondrion | 26.56, 23.61 |
| J45 | DNAJA2, SCJ1 [40,41] | ER | 31.69, 31.40 |
| J46 | DNAJA2, SCJ1 [40,41] | ER | 36.36, 30.66 |
| J47 | DNAJA3, MDJ1 [42,43] | Mitochondrion | 24.79, 27.42 |
| J50 | DNAJA2, SPJ1, SCJ1 [40,41] | Glycosome | 37.57-30.65 |
| J51 | DNAJC7 [66] | Ciliary basal body | 28.07 |
| J52 | DNAJC7 [66] | - | 32.77 |
| J53 | DNAJC3 (ERdj6), JEM1 [48,49] | ER | 28.71, 38.94 |
| J54 | - | Flagellum, Glycosome | - |
| J56 | DNAJC8 [65] | Nucleus | 36.17 |
| J59 | DNAJC13, RME-8 [59] | Endosome | 34.63, 34.63 |
| J60 | JJJ2v | - | 38.89 |
| J66 | DNAJA2 [40] | ER | 43.46 |
| J68 | DNAJC15 (tim complex) [56] | Mitochondrion | 29.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solana, J.C.; Bernardo, L.; Moreno, J.; Aguado, B.; Requena, J.M. The Astonishing Large Family of HSP40/DnaJ Proteins Existing in Leishmania. Genes 2022, 13, 742. https://doi.org/10.3390/genes13050742
Solana JC, Bernardo L, Moreno J, Aguado B, Requena JM. The Astonishing Large Family of HSP40/DnaJ Proteins Existing in Leishmania. Genes. 2022; 13(5):742. https://doi.org/10.3390/genes13050742
Chicago/Turabian StyleSolana, Jose Carlos, Lorena Bernardo, Javier Moreno, Begoña Aguado, and Jose M. Requena. 2022. "The Astonishing Large Family of HSP40/DnaJ Proteins Existing in Leishmania" Genes 13, no. 5: 742. https://doi.org/10.3390/genes13050742
APA StyleSolana, J. C., Bernardo, L., Moreno, J., Aguado, B., & Requena, J. M. (2022). The Astonishing Large Family of HSP40/DnaJ Proteins Existing in Leishmania. Genes, 13(5), 742. https://doi.org/10.3390/genes13050742







