High Concentration or Combined Treatment of Antisense Oligonucleotides for Spinal Muscular Atrophy Perturbed SMN2 Splicing in Patient Fibroblasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection Protocol
2.2. Treatments with ASOs
2.3. RNA Extraction and cDNA Synthesis
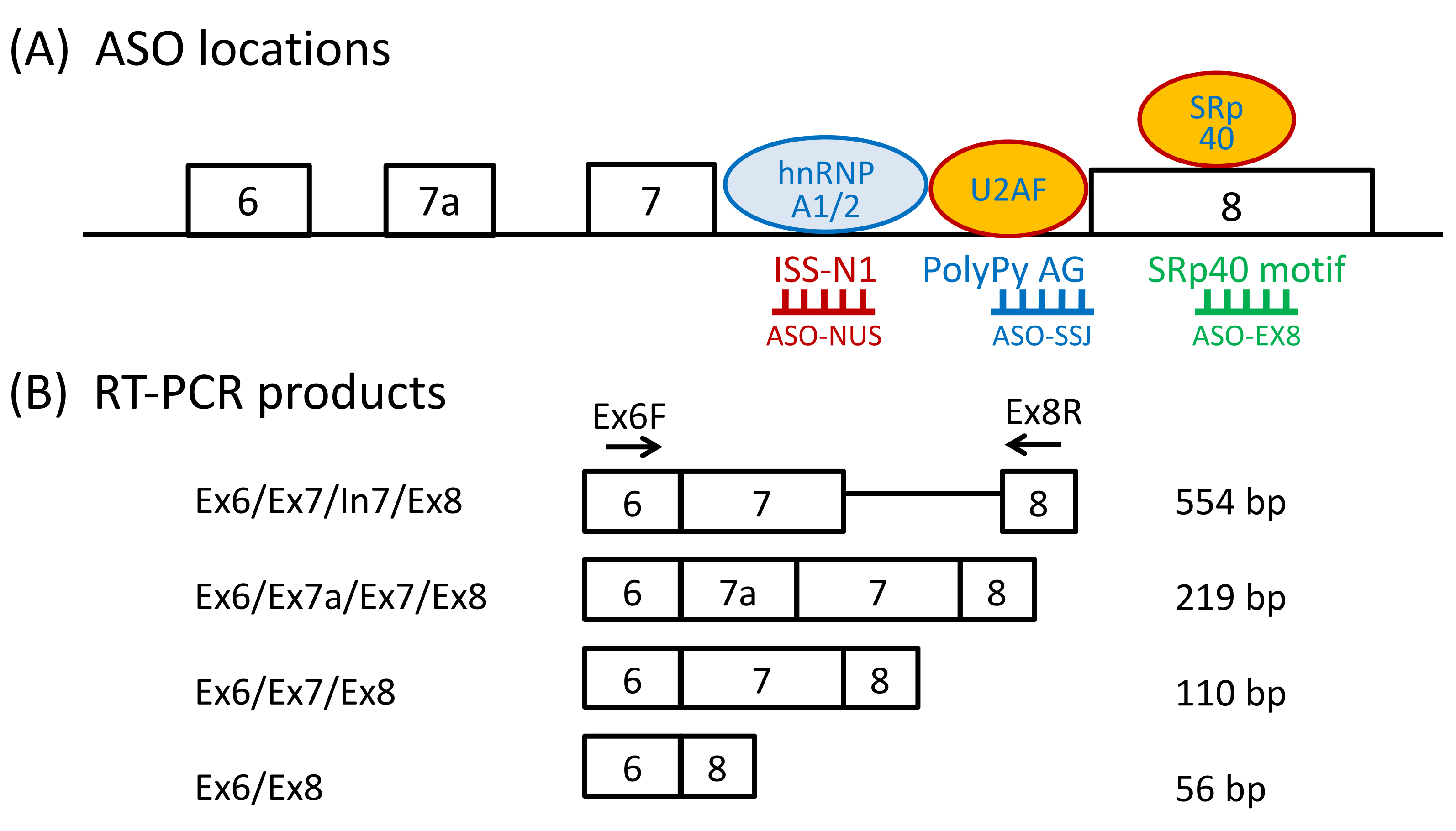
2.4. Reverse-Transcription PCR (RT-PCR) Analysis
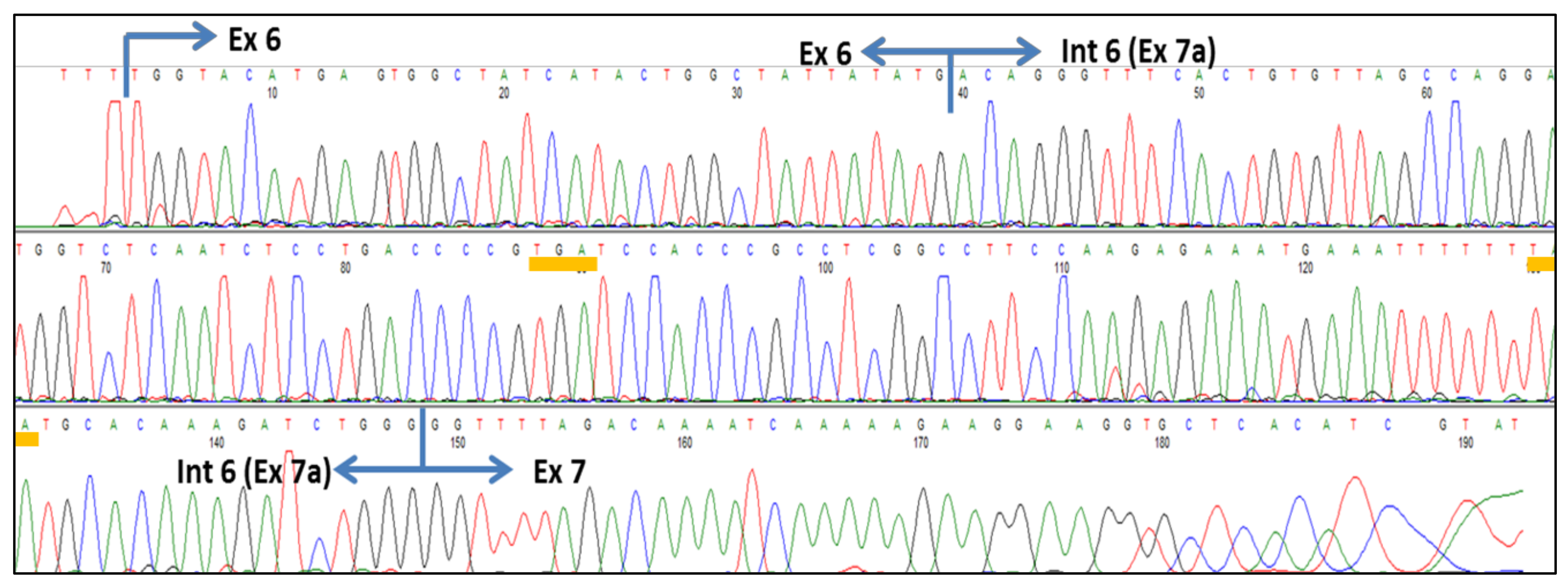
2.5. Subcloning and Sequence Analysis
2.6. Statistics
3. Results
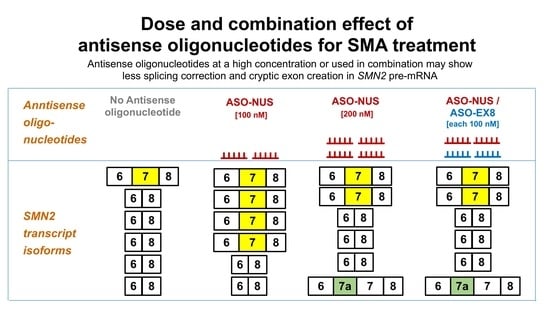
3.1. Dose Effect of ASOs
3.1.1. ASO-NUS
3.1.2. ASO-EX8
3.1.3. ASO-SSJ
3.2. Effects of ASOs in Combination
3.2.1. ASO-NUS/ASO-EX8
3.2.2. ASO-NUS/ASO-SSJ
3.2.3. ASO-EX8/ASO-SSJ
4. Discussion
4.1. ASO Targeting ISS-N1 Site
4.2. ASO Targeting SMN2 Exon 8
4.3. Activation of Cryptic Splice Sites in SMN2 Intron 6 by ASOs
4.4. Research Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnold, W.D.; Kassar, D.; Kissel, J.T. Spinal muscular atrophy: Diagnosis and management in a new therapeutic era. Muscle Nerve 2015, 51, 157–167. [Google Scholar] [CrossRef]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M.; et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef]
- Lefebvre, S.; Burlet, P.; Liu, Q.; Bertrandy, S.; Clermont, O.; Munnich, A.; Dreyfuss, G.; Melki, J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997, 16, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Schrank, B.; Götz, R.; Gunnersen, J.M.; Ure, J.M.; Toyka, K.V.; Smith, A.G.; Sendtner, M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA 1997, 94, 9920–9925. [Google Scholar] [CrossRef] [PubMed]
- Hsieh-li, H.M.; Chang, J.; Jong, Y.; Wu, M.; Wang, N.M.; Tsai, C.H.; Li, H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000, 24, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Vitte, J.; Fassier, C.; Tiziano, F.D.; Dalard, C.; Soave, S.; Roblot, N.; Brahe, C.; Saugier-Veber, P.; Bonnefont, J.P.; Melki, J. Refined characterization of the expression and stability of the SMN gene products. Am. J. Pathol. 2007, 171, 1269–1280. [Google Scholar] [CrossRef]
- D’Amico, A.; Mercuri, E.; Tiziano, F.D.; Bertini, E. Spinal muscular atrophy. Orphanet J. Rare Dis. 2011, 6, 71. [Google Scholar] [CrossRef]
- Nurputra, D.K.; Lai, P.S.; Harahap, N.I.F.; Morikawa, S.; Yamamoto, T.; Nishimura, N.; Kubo, Y.; Takeuchi, A.; Saito, T.; Takeshima, Y.; et al. Spinal muscular atrophy: From gene discovery to clinical trials. Ann. Hum. Genet. 2013, 77, 435–463. [Google Scholar] [CrossRef]
- Lim, S.R.; Hertel, K.J. Modulation of Survival Motor Neuron Pre-mRNA Splicing by Inhibition of Alternative 3′ Splice Site Pairing. J. Biol. Chem. 2001, 276, 45476–45483. [Google Scholar] [CrossRef]
- Miyajima, H.; Miyaso, H.; Okumura, M.; Kurisu, J.; Imaizumi, K. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J. Biol. Chem. 2002, 277, 23271–23277. [Google Scholar] [CrossRef]
- Cartegni, L.; Krainer, A.R. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Biol. 2003, 10, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Skordis, L.A.; Dunckley, M.G.; Yue, B.; Eperon, I.C.; Muntoni, F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl. Acad. Sci. USA 2003, 100, 4114–4119. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.D.; Dickinson, R.L.; Lucas, C.M.; Cousins, A.; Malygin, A.A.; Weldon, C.; Perrett, A.J.; Bottrill, A.R.; Searle, M.S.; Burley, G.A.; et al. A targeted oligonucleotide enhancer of SMN2 Exon 7 splicing forms competing quadruplex and protein complexes in functional conditions. Cell Rep. 2014, 9, 193–205. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coady, T.H.; Shababi, M.; Tullis, G.E.; Lorson, C.L. Restoration of SMN function: Delivery of a trans-splicing RNA Re-directs SMN2 pre-mRNA splicing. Mol. Ther. 2007, 15, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.K.; Singh, N.N.; Androphy, E.J.; Singh, R.N. Splicing of a Critical Exon of Human Survival Motor Neuron Is Regulated by a Unique Silencer Element Located in the Last Intron. Mol. Cell. Biol. 2006, 26, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Vickers, T.A.; Okunola, H.L.; Bennett, C.F.; Krainer, A.R. Antisense Masking of an hnRNP A1/A2 Intronic Splicing Silencer Corrects SMN2 Splicing in Transgenic Mice. Am. J. Hum. Genet. 2008, 82, 834–848. [Google Scholar] [CrossRef]
- Flynn, L.L.; Mitrpant, C.; Pitout, I.L.; Fletcher, S.; Wilton, S.D. Antisense Oligonucleotide-Mediated Terminal Intron Retention of the SMN2 Transcript. Mol. Ther. Nucleic Acids 2018, 11, 91–102. [Google Scholar] [CrossRef]
- Nicolau, S.; Waldrop, M.A.; Connolly, A.M.; Mendell, J.R. Spinal Muscular Atrophy. Semin. Pediatr. Neurol. 2021, 37, 100878. [Google Scholar] [CrossRef]
- Mercuri, E.; Darras, B.T.; Chiriboga, C.A.; Day, J.W.; Campbell, C.; Connolly, A.M.; Iannaccone, S.T.; Kirschner, J.; Kuntz, N.L.; Saito, K.; et al. Nusinersen versus sham control in later-onset spinal muscular atrophy. N. Engl. J. Med. 2018, 378, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Al-Zaidy, S.; Shell, R.; Arnold, W.D.; Rodino-Klapac, L.R.; Prior, T.W.; Lowes, L.; Alfano, L.; Berry, K.; Church, K.; et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017, 377, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Ratni, H.; Ebeling, M.; Baird, J.; Bendels, S.; Bylund, J.; Chen, K.S.; Denk, N.; Feng, Z.; Green, L.; Guerard, M.; et al. Discovery of Risdiplam, a Selective Survival of Motor Neuron-2 (SMN2) Gene Splicing Modifier for the Treatment of Spinal Muscular Atrophy (SMA). J. Med. Chem. 2018, 61, 6501–6517. [Google Scholar] [CrossRef] [PubMed]
- Darras, B.T.; Masson, R.; Mazurkiewicz-Bełdzińska, M.; Rose, K.; Xiong, H.; Zanoteli, E.; Baranello, G.; Bruno, C.; Vlodavets, D.; Wang, Y.; et al. Risdiplam-Treated Infants with Type 1 Spinal Muscular Atrophy versus Historical Controls. N. Engl. J. Med. 2021, 385, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Baranello, G.; Darras, B.T.; Day, J.W.; Deconinck, N.; Klein, A.; Masson, R.; Mercuri, E.; Rose, K.; El-Khairi, M.; Gerber, M.; et al. Risdiplam in Type 1 Spinal Muscular Atrophy. N. Engl. J. Med. 2021, 384, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Coratti, G.; Cutrona, C.; Pera, M.C.; Bovis, F.; Ponzano, M.; Chieppa, F.; Antonaci, L.; Sansone, V.; Finkel, R.; Pane, M.; et al. Motor function in type 2 and 3 SMA patients treated with Nusinersen: A critical review and meta-analysis. Orphanet J. Rare Dis. 2021, 16, 1–12. [Google Scholar] [CrossRef]
- Finkel, R.S.; Mercuri, E.; Darras, B.T.; Connolly, A.M.; Kuntz, N.L.; Kirschner, J.; Chiriboga, C.A.; Saito, K.; Servais, L.; Tizzano, E.; et al. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2017, 377, 1723–1732. [Google Scholar] [CrossRef]
- De Vivo, D.C.; Bertini, E.; Swoboda, K.J.; Hwu, W.L.; Crawford, T.O.; Finkel, R.S.; Kirschner, J.; Kuntz, N.L.; Parsons, J.A.; Ryan, M.M.; et al. Nusinersen initiated in infants during the presymptomatic stage of spinal muscular atrophy: Interim efficacy and safety results from the Phase 2 NURTURE study. Neuromuscul. Disord. 2019, 29, 842–856. [Google Scholar] [CrossRef]
- Maggi, L.; Bello, L.; Bonanno, S.; Govoni, A.; Caponnetto, C.; Passamano, L.; Grandis, M.; Trojsi, F.; Cerri, F.; Ferraro, M.; et al. Nusinersen safety and effects on motor function in adult spinal muscular atrophy type 2 and 3. J. Neurol. Neurosurg. Psychiatry 2020, 91, 1166–1174. [Google Scholar] [CrossRef]
- Schorling, D.C.; Pechmann, A.; Kirschner, J. Advances in Treatment of Spinal Muscular Atrophy - New Phenotypes, New Challenges, New Implications for Care. J. Neuromuscul. Dis. 2020, 7, 1–13. [Google Scholar] [CrossRef]
- Ottesen, E.W.; Luo, D.; Singh, N.N.; Singh, R.N. High concentration of an iss-n1-targeting antisense oligonucleotide causes massive perturbation of the transcriptome. Int. J. Mol. Sci. 2021, 22, 8378. [Google Scholar] [CrossRef]
- Pao, P.W.; Wee, K.B.; Yee, W.C.; Dwipramono, Z.A. Dual masking of specific negative splicing regulatory elements resulted in maximal exon 7 inclusion of SMN2 gene. Mol. Ther. 2014, 22, 854–861. [Google Scholar] [CrossRef]
- Harada, Y.; Rao, V.K.; Arya, K.; Kuntz, N.L.; DiDonato, C.J.; Napchan-Pomerantz, G.; Agarwal, A.; Stefans, V.; Katsuno, M.; Veerapandiyan, A. Combination molecular therapies for type 1 spinal muscular atrophy. Muscle Nerve 2020, 62, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, S.; Harahap, N.I.F.; Hamamura, Y.; Ar Rochmah, M.; Shima, A.; Morisada, N.; Shinohara, M.; Saito, T.; Saito, K.; Lai, P.S.; et al. Alternative splicing of a cryptic exon embedded in intron 6 of SMN1 and SMN2. Hum. Genome Var. 2016, 3, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.X.; Zhang, M.; Krainer, A.R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998, 12, 1998–2012. [Google Scholar] [CrossRef] [PubMed]
- Cartegni, L.; Wang, J.; Zhu, Z.; Zhang, M.Q.; Krainer, A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003, 31, 3568–3571. [Google Scholar] [CrossRef]
- Graveley, B.R.; Hertel, K.J.; Maniatis, T. The role of U2AF 35 and U2AF 65 in enhancer-dependent splicing. RNA 2001, 7, 806–818. [Google Scholar] [CrossRef]
- Singh, N.N.; Howell, M.D.; Androphy, E.J.; Singh, R.N. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017, 24, 520–526. [Google Scholar] [CrossRef]
- Ottesen, E.W. ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl. Neurosci. 2017, 8, 1–6. [Google Scholar] [CrossRef]
- Seo, J.; Singh, N.N.; Ottesen, E.W.; Lee, B.M.; Singh, R.N. A novel human-specific splice isoform alters the critical C-terminus of Survival Motor Neuron protein. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Buratti, E.; Baralle, F.E. Influence of RNA Secondary Structure on the Pre-mRNA Splicing Process. Mol. Cell. Biol. 2004, 24, 10505–10514. [Google Scholar] [CrossRef]
- Hua, Y.; Sahashi, K.; Hung, G.; Rigo, F.; Passini, M.A.; Bennett, C.F.; Krainer, A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010, 24, 1634–1644. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wijaya, Y.O.S.; Niba, E.T.E.; Nishio, H.; Okamoto, K.; Awano, H.; Saito, T.; Takeshima, Y.; Shinohara, M. High Concentration or Combined Treatment of Antisense Oligonucleotides for Spinal Muscular Atrophy Perturbed SMN2 Splicing in Patient Fibroblasts. Genes 2022, 13, 685. https://doi.org/10.3390/genes13040685
Wijaya YOS, Niba ETE, Nishio H, Okamoto K, Awano H, Saito T, Takeshima Y, Shinohara M. High Concentration or Combined Treatment of Antisense Oligonucleotides for Spinal Muscular Atrophy Perturbed SMN2 Splicing in Patient Fibroblasts. Genes. 2022; 13(4):685. https://doi.org/10.3390/genes13040685
Chicago/Turabian StyleWijaya, Yogik Onky Silvana, Emma Tabe Eko Niba, Hisahide Nishio, Kentaro Okamoto, Hiroyuki Awano, Toshio Saito, Yasuhiro Takeshima, and Masakazu Shinohara. 2022. "High Concentration or Combined Treatment of Antisense Oligonucleotides for Spinal Muscular Atrophy Perturbed SMN2 Splicing in Patient Fibroblasts" Genes 13, no. 4: 685. https://doi.org/10.3390/genes13040685
APA StyleWijaya, Y. O. S., Niba, E. T. E., Nishio, H., Okamoto, K., Awano, H., Saito, T., Takeshima, Y., & Shinohara, M. (2022). High Concentration or Combined Treatment of Antisense Oligonucleotides for Spinal Muscular Atrophy Perturbed SMN2 Splicing in Patient Fibroblasts. Genes, 13(4), 685. https://doi.org/10.3390/genes13040685