Genome-Wide Analysis of the LRR-RLP Gene Family in a Wild Banana (Musa acuminata ssp. malaccensis) Uncovers Multiple Fusarium Wilt Resistance Gene Candidates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval, Annotation of LRR-RLPs and Prediction of Subcellular Localization
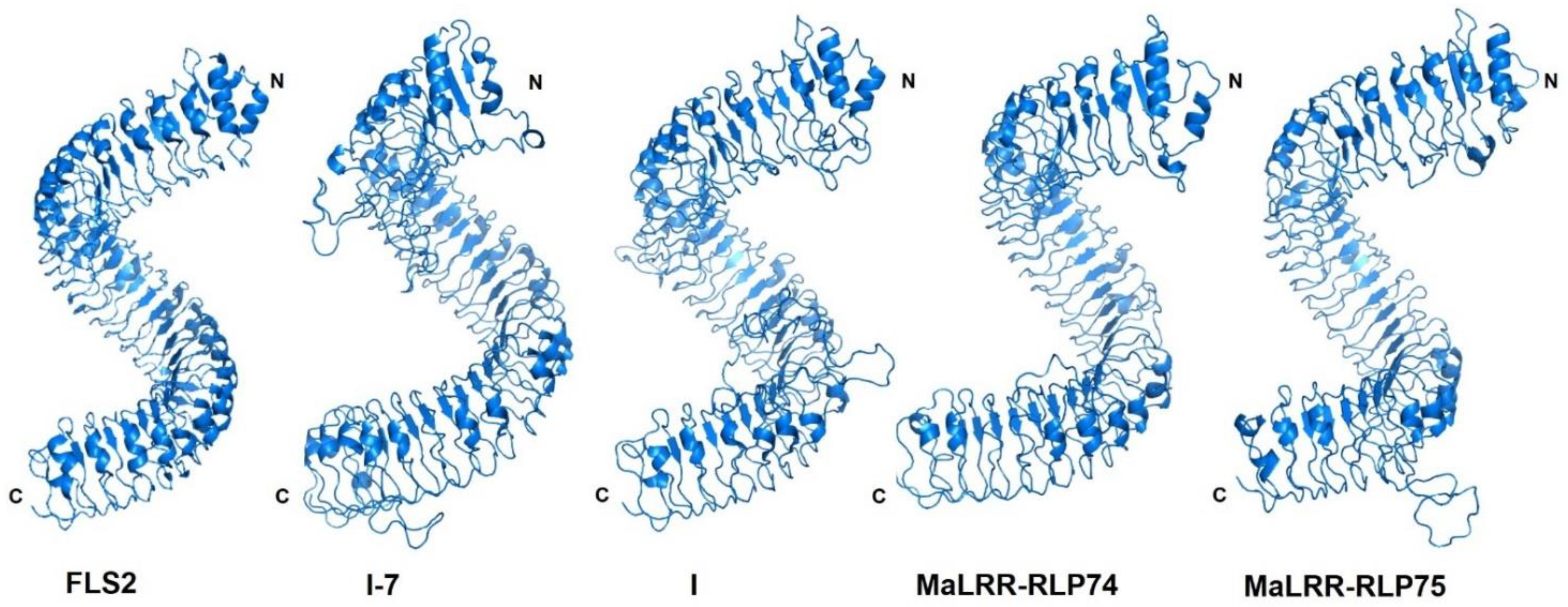
2.2. Gene Structure, Identification of Conserved Protein Motifs and 3D Protein Modeling
2.3. Chromosome Location of LRR-RLP Genes
2.4. Multiple Sequence Alignment and Phylogenetic Tree Construction
2.5. Transcriptomic Analysis of Banana LRR-RLP Genes and RT–qPCR
3. Results
3.1. Identification of LRR-RLP Genes in the Banana Genome
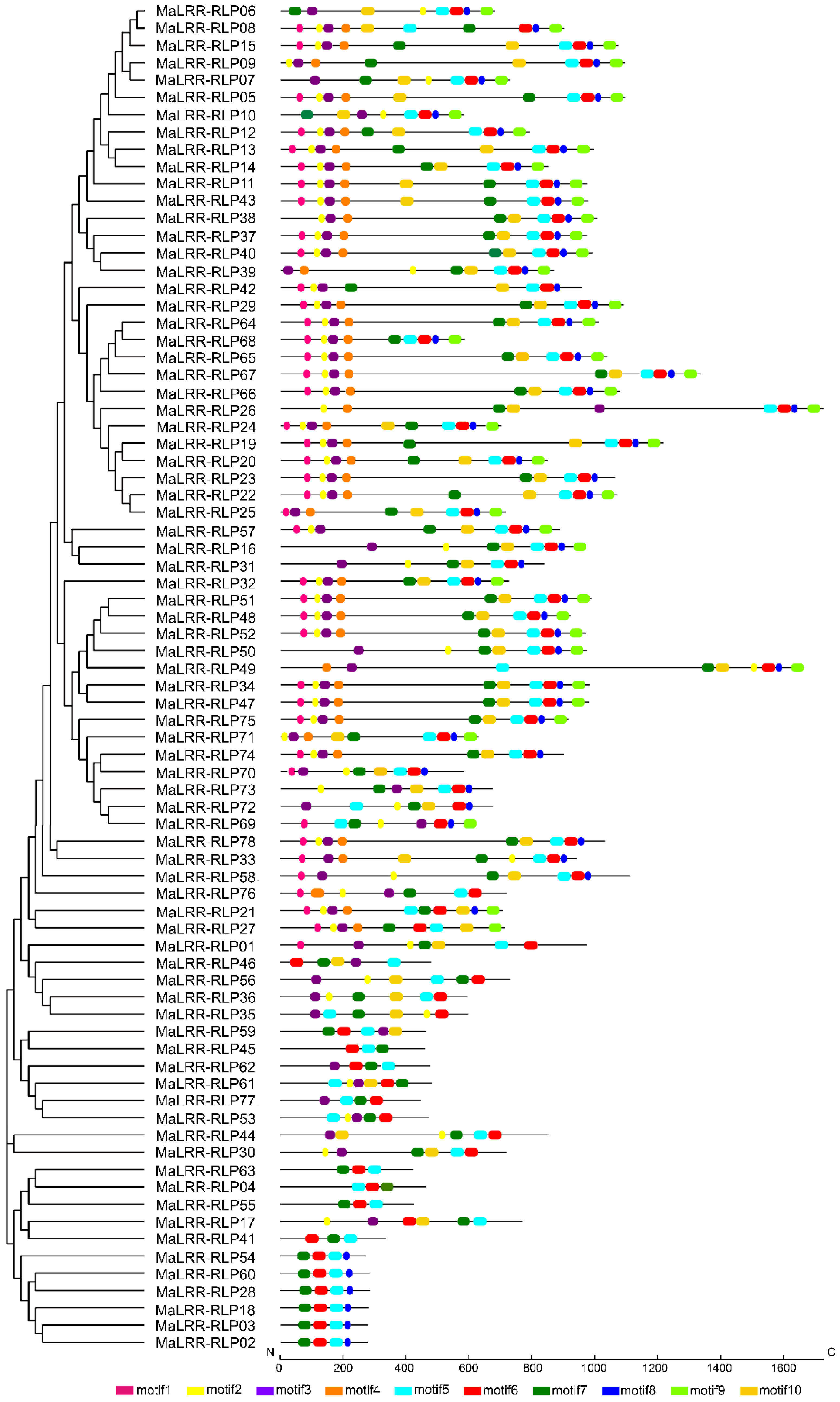
3.2. Gene Structure of MaLRR-RLPs and Identification of Conserved Motifs
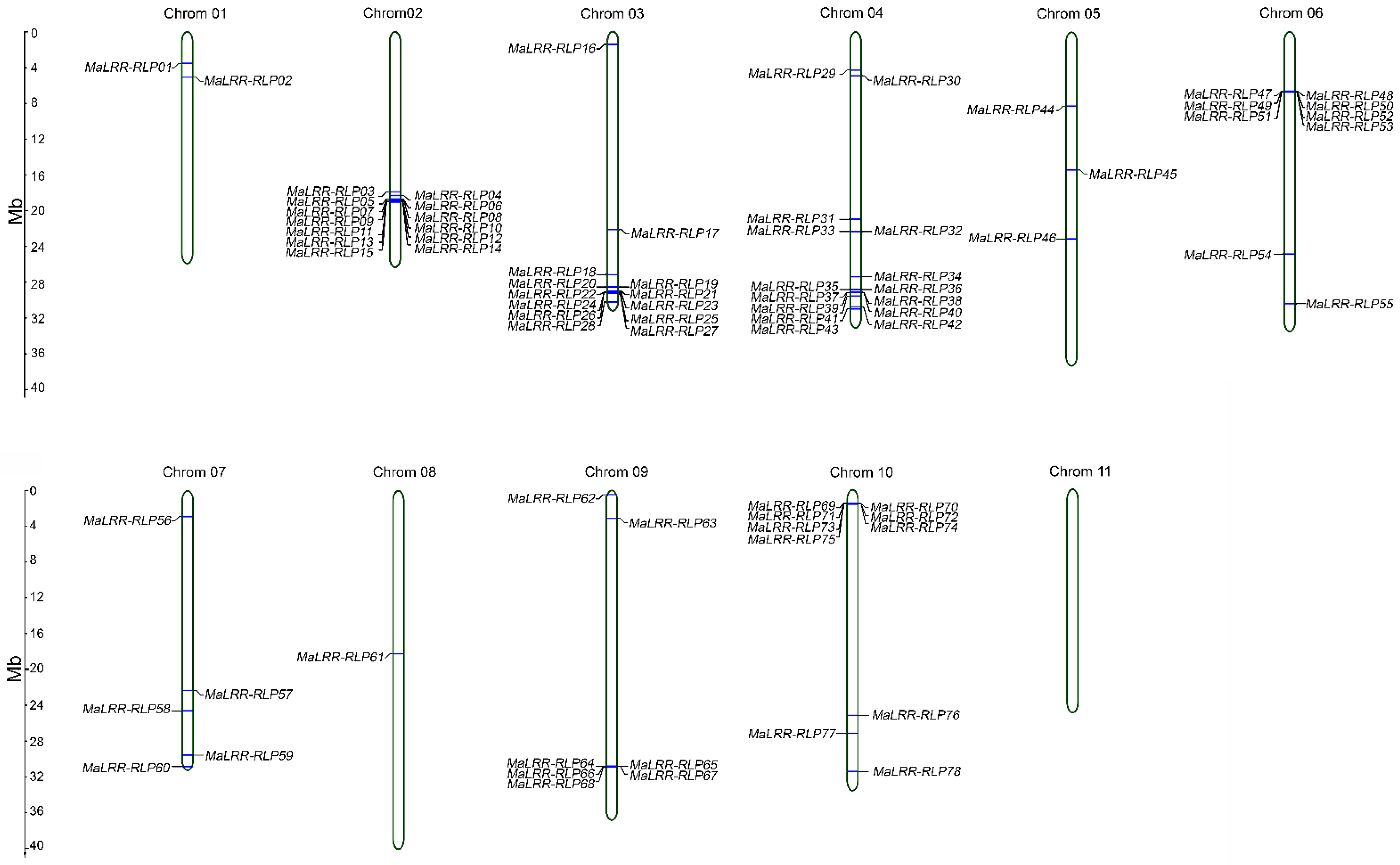
3.3. Distribution of MaLRR-RLP Genes on Chromosomes
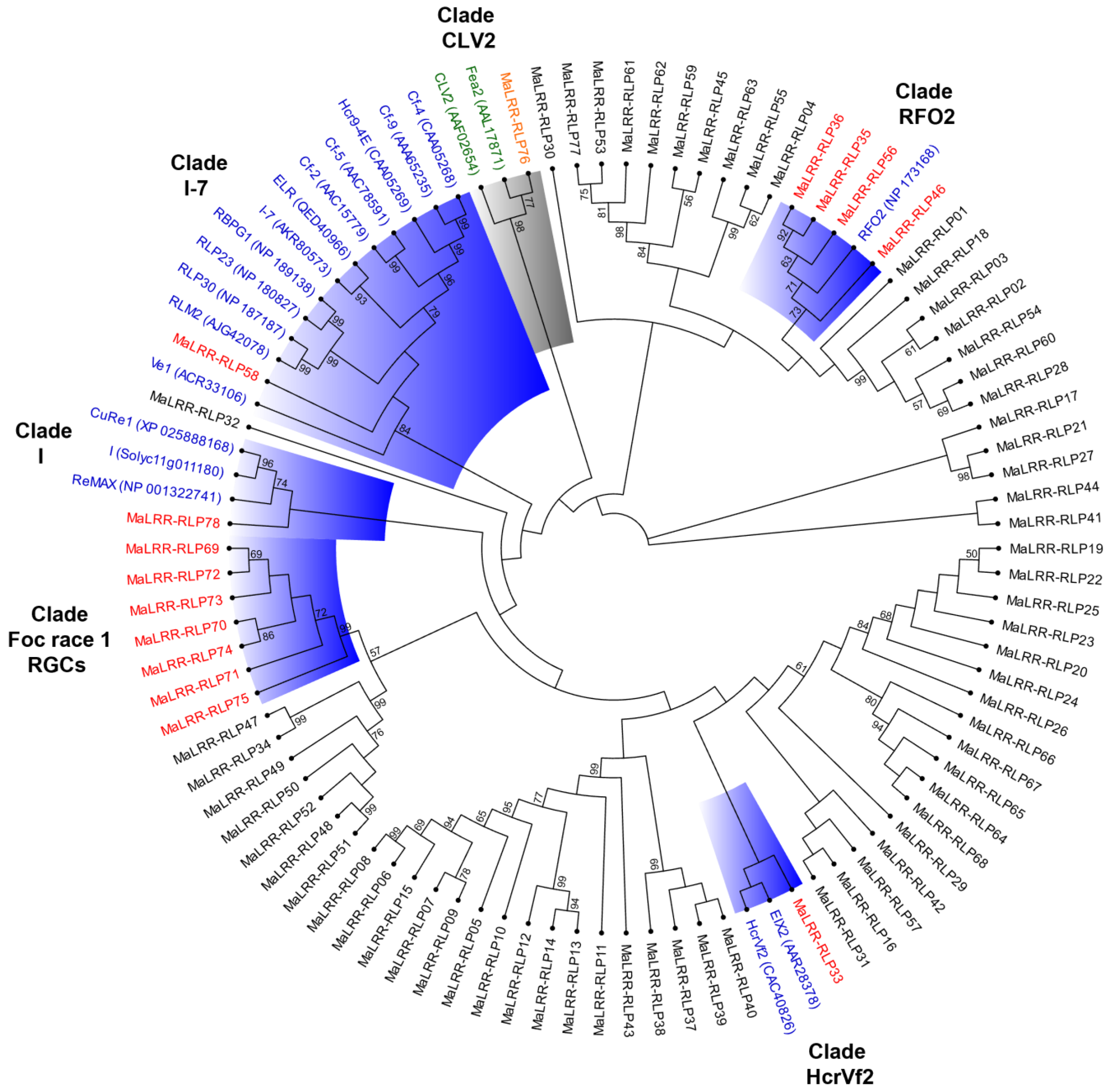
3.4. Banana LRR-RLPs with Potential for Foc Race 1 Resistance Clustered on Chromosome 10
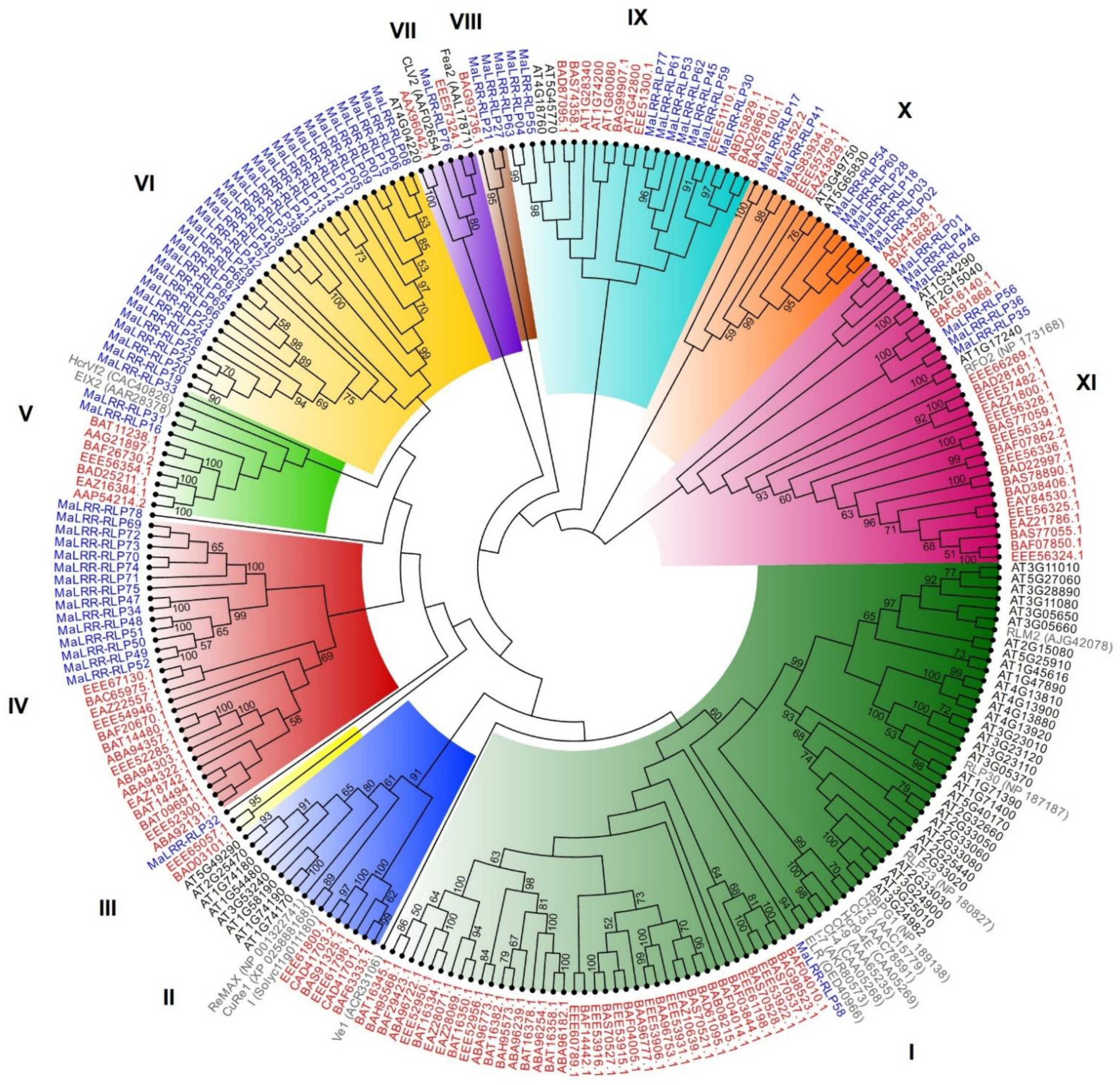
3.5. Phylogenetic Analysis of the Banana LRR-RLP Family
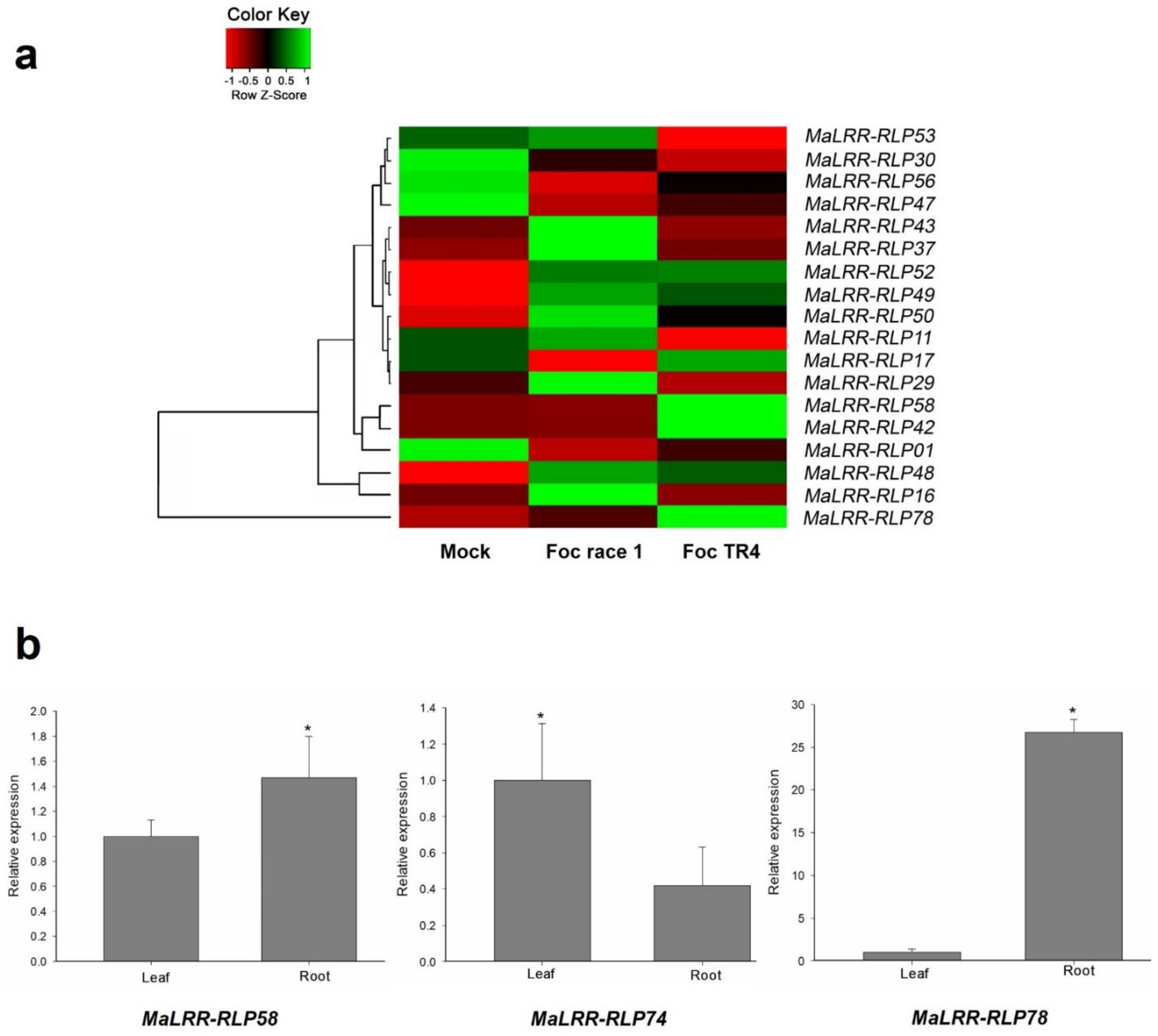
3.6. Expression Profiles of Banana LRR-RLP Genes in Plants Infected with Foc Race 1 and Foc TR4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Global Programme on Banana Fusarium Wilt Disease. 2017. Available online: https://www.fao.org/publications/card/en/c/29d4ef48-7454-4f98-8260-e5f19249d246 (accessed on 21 February 2022).
- Ploetz, R.C. Fusarium wilt of banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arango Isaza, R.E.; Diaz-Trujillo, C.; Dhillon, B.; Aerts, A.; Carlier, J.; Crane, C.F.; de Jong, T.V.; de Vries, I.; Dietrich, R.; Farmer, A.D.; et al. Combating a global threat to a clonal crop: Banana black Sigatoka pathogen Pseudocercospora fijiensis (synonym Mycosphaerella fijiensis) genomes reveal clues for disease control. PLoS Genet. 2016, 12, e1005876. [Google Scholar] [CrossRef] [PubMed]
- Maryani, N.; Lombard, L.; Poerba, Y.S.; Subandiyah, S.; Crous, P.W.; Kema, G.H.J. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud. Mycol. 2019, 92, 155–194. [Google Scholar] [CrossRef] [PubMed]
- Pegg, K.G.; Coates, L.M.; O’Neill, W.T.; Turner, D.W. The epidemiology of Fusarium wilt of banana. Front. Plant Sci. 2019, 10, 1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Esse, H.P.; Reuber, T.L.; van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Churchill, A.C.L. Mycosphaerella Fijiensis, the black leaf streak pathogen of banana: Progress towards understanding pathogen biology and detection, disease development, and the challenges of control. Mol. Plant Pathol. 2011, 12, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Li, S.; Jiang, Y.; Zhang, Y.; Wang, Z.; Jia, S. Isolation, characterization and phylogenetic analysis of the resistance gene analogues (RGAs) in banana (Musa spp.). Plant Sci. 2007, 172, 1166–1174. [Google Scholar] [CrossRef]
- Peraza-Echeverria, S.; Dale, J.L.; Harding, R.M.; Smith, M.K.; Collet, C. Characterization of disease resistance gene candidates of the nucleotide binding site (NBS) type from banana and correlation of a transcriptional polymorphism with resistance to Fusarium oxysporum f. sp. cubense race 4. Mol. Breed. 2008, 22, 565–579. [Google Scholar] [CrossRef]
- Peraza-Echeverria, S.; Dale, J.L.; Harding, R.M.; Collet, C. Molecular cloning and in silico analysis of potential Fusarium resistance genes in banana. Mol. Breed. 2009, 23, 431–443. [Google Scholar] [CrossRef]
- Miller, R.N.G.; Bertioli, D.J.; Baurens, F.C.; Santos, C.M.R.; Alves, P.C.; Martins, N.F.; Togawa, R.C.; Souza, M.T., Jr.; Pappas, G.J., Jr. Analysis of non-TIR NBS-LRR resistance gene analogs in Musa acuminata Colla: Isolation, RFLP marker development, and physical mapping. BMC Plant Biol. 2008, 8, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dale, J.; James, A.; Paul, J.Y.; Khanna, H.; Smith, M.; Peraza-Echeverria, S.; Garcia-Bastidas, F.; Kema, G.; Waterhouse, P.; Mengersen, K.; et al. Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 2017, 8, 1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Rouard, M.; Droc, G.; Martin, G.; Sardos, J.; Hueber, Y.; Guignon, V.; Cenci, A.; Geigle, B.; Hibbins, M.S.; Yahiaoui, N.; et al. Three new genome assemblies support a rapid radiation in Musa acuminata (wild banana). Genome Biol. Evol. 2018, 10, 3129–3140. [Google Scholar] [CrossRef] [Green Version]
- Chang, W.; Li, H.; Chen, H.; Qiao, F.; Zeng, H. NBS-LRR gene family in banana (Musa acuminata): Genome-wide identification and responses to Fusarium oxysporum f. sp. cubense race 1 and tropical race 4. Eur. J. Plant Pathol. 2020, 157, 549–563. [Google Scholar] [CrossRef]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanyuka, K.; Rudd, J.J. Cell surface immune receptors: The guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 2019, 50, 1–8. [Google Scholar] [CrossRef]
- Van der Burgh, A.M.; Joosten, M.H.A.J. Plant immunity: Thinking outside and inside the box. Trends Plant Sci. 2019, 24, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Han, L.; Yang, L.; Zeng, H.; Fan, D.; Zhu, Y.; Feng, Y.; Wang, G.; Peng, C.; Jiang, X.; et al. Genome and transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. cubense causing banana vascular wilt disease. PLoS ONE 2014, 9, e95543. [Google Scholar] [CrossRef] [PubMed]
- Simons, G.; Groenendijk, J.; Wijbrandi, J.; Reijans, M.; Groenen, J.; Diergaarde, P.; Van der Lee, T.; Bleeker, M.; Onstenk, J.; de Both, M.; et al. Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell 1998, 10, 1055–1068. [Google Scholar] [CrossRef] [Green Version]
- Catanzariti, A.M.; Lim, G.T.T.; Jones, D.A. The tomato I-3 gene: A novel gene for resistance to fusarium wilt disease. New Phytol. 2015, 207, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Cendales, Y.; Catanzariti, A.M.; Baker, B.; Mcgrath, D.J.; Jones, D.A. Identification of I-7 expands the repertoire of genes for resistance to fusarium wilt in tomato to three resistance gene classes. Mol. Plant Pathol. 2016, 17, 448–463. [Google Scholar] [CrossRef] [PubMed]
- Catanzariti, A.M.; Do, H.T.T.; Bru, P.; de Sain, M.; Thatcher, L.F.; Rep, M.; Jones, D.A. The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 2017, 89, 1195–1209. [Google Scholar] [CrossRef] [Green Version]
- Stergiopoulos, I.; Van Den Burg, H.A.; Ökmen, B.; Beenen, H.G.; Van Liere, S.; Kema, G.H.J.; De Wit, P.J.G.M. Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA 2010, 107, 7610–7615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutrot, F.; Zipfel, C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef]
- Martin, G.; Baurens, F.C.; Droc, G.; Rouard, M.; Cenci, A.; Kilian, A.; Hastie, A.; Doležel, J.; Aury, J.M.; Alberti, A.; et al. Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genom. 2016, 17, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, W.H.; Yeom, S.I. Genome-wide identification, classification, and expression analysis of the receptor-like protein family in tomato. Plant Pathol. J. 2018, 34, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Mulder, N.; Apweiler, R. InterPro and InterProScan: Tools for protein sequence classification and comparison. Methods Mol. Biol. 2007, 396, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Gasteriger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–603. ISBN 978-1-58829-343-5. [Google Scholar]
- Armenteros, J.J.A.; Sønderby, C.K.; Sønderby, S.K.; Nielsen, H.; Winther, O. Sequence analysis Deeploc: Prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Jupe, F.; Pritchard, L.; Etherington, G.J.; MacKenzie, K.; Cock, P.J.A.; Wright, F.; Sharma, S.K.; Bolser, D.; Bryan, G.J.; Jones, J.D.G.; et al. Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genom. 2012, 13, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef] [Green Version]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz-Laylin, L.K.; Krishnamurthy, N.; Tor, M.; Sjolander, K.V.; Jones, J.D.G. Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 2005, 138, 611–623. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Jamieson, P.A.; Shan, L.; He, P. Plant cell surface molecular cypher: Receptor-like proteins and their roles in immunity and development. Plant Sci. 2018, 274, 242–251. [Google Scholar] [CrossRef]
- Wang, G.; Ellendorff, U.; Kemp, B.; Mansfield, J.W.; Forsyth, A.; Mitchell, K.; Bastas, K.; Liu, C.M.; Woods-Tör, A.; Zipfel, C.; et al. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 2008, 147, 503–517. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Shao, J.; Wang, Y.; Li, W.; Guo, D.; Yan, B.; Xia, Y.; Peng, M. Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp. cubense. BMC Genom. 2013, 14, 851. [Google Scholar] [CrossRef] [Green Version]
- Van den Berg, N.; Berger, D.K.; Hein, I.; Birch, P.R.J.; Wingfield, M.J.; Viljoen, A. Tolerance in banana to Fusarium wilt is associated with early up-regulation of cell wall-strengthening genes in the roots. Mol. Plant Pathol. 2007, 8, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Ahmad, F.; Martawi, N.M.; Poerba, Y.S.; de Jong, H.; Schouten, H.; Kema, G.H.J. Genetic mapping of Fusarium wilt resistance in a wild banana Musa acuminata ssp. malaccensis accession. Theor. Appl. Genet. 2020, 133, 3409–3418. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Diener, A.C. Arabidopsis thaliana resistance to Fusarium oxysporum 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet. 2013, 9, e1003525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.M.; Chai, J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013, 342, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, Z.; Zhang, Z.; Lv, Y.; Yang, N.; Zhang, G.; Wu, M.; Lv, S.; Pan, L.; Joosten, M.H.A.J.; et al. Transcriptional regulation of receptor-like protein genes by environmental stresses and hormones and their overexpression activities in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 3339–3351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steidele, C.E.; Stam, R. Multi-Omics approach highlights differences between RLP classes in Arabidopsis thaliana. BMC Genom. 2021, 22, 557. [Google Scholar] [CrossRef]
- Petre, B.; Hacquard, S.; Duplessis, S.; Rouhier, N. Genome analysis of poplar LRR-RLP gene clusters reveals RISP, a defense-related gene coding a candidate endogenous peptide elicitor. Front. Plant Sci. 2014, 5, 111. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.Y.; Huang, J.Q.; Li, N.Y.; Ma, X.F.; Wang, J.L.; Liu, C.; Liu, Y.F.; Liang, Y.; Bao, Y.M.; Dai, X.F. Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to verticillium wilt. BMC Plant Biol. 2015, 15, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Restrepo-Montoya, D.; Brueggeman, R.; McClean, P.E.; Osorno, J.M. Computational identification of receptor-like kinases “RLK” and receptor-like proteins “RLP” in legumes. BMC Genom. 2020, 21, 459. [Google Scholar] [CrossRef]
- Mizuno, H.; Katagiri, S.; Kanamori, H.; Mukai, Y.; Sasaki, T.; Matsumoto, T.; Wu, J. Evolutionary dynamics and impacts of chromosome regions carrying R-gene clusters in rice. Sci. Rep. 2020, 10, 872. [Google Scholar] [CrossRef] [PubMed]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004, 20, 113–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houterman, P.M.; Cornelissen, B.J.C.; Rep, M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 2008, 4, e1000061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Does, H.C.; Constantin, M.E.; Houterman, P.M.; Takken, F.L.W.; Cornelissen, B.J.C.; Haring, M.A.; van den Burg, H.A.; Rep, M. Fusarium oxysporum colonizes the stem of resistant tomato plants, the extent varying with the R-gene present. Eur. J. Plant Pathol. 2019, 154, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Paul, J.Y.; Becker, D.K.; Dickman, M.B.; Harding, R.M.; Khanna, H.K.; Dale, J.L. Apoptosis-related genes confer resistance to fusarium wilt in transgenic “Lady finger” bananas. Plant Biotechnol. J. 2011, 9, 1141–1148. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, K.H.; Gouy, M.; Yang, Y.W.; Sharp, P.M.; Li, W.H. Date of the Monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 1989, 86, 6201–6205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.-T.; Zhou, G.C.; Feng, X.Y.; Zeng, Z.; Liu, Y.; Shao, Z.Q. Frequent gene duplication/loss shapes distinct evolutionary patterns of NLR genes in Arecaceae species. Horticulturae 2021, 7, 539. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Heal, R.; Wyler, M.; Schmid, M.W.; Jones, J.D.G. Concerted expansion and contraction of immune receptor gene repertoires in plant genomes. bioRxiv 2022. [Google Scholar] [CrossRef]

| Gene Name | Sequence ID | Domains a | Protein Length | kDa | pI | Subcellular Location |
|---|---|---|---|---|---|---|
| MaLRR-RLP01 | Ma01_p05590.1 | SP-LRR-TM | 976 | 104.65 | 6.27 | Cell membrane |
| MaLRR-RLP02 | Ma01_p07780.1 | SP-LRR-TM | 280 | 29.80 | 7.60 | Cell membrane |
| MaLRR-RLP03 | Ma02_p10510.1 | SP-LRR-TM | 280 | 29.77 | 6.71 | Cell membrane |
| MaLRR-RLP04 | Ma02_p11370.1 | SP-LRR-TM | 465 | 49.35 | 9.33 | Cell membrane |
| MaLRR-RLP05 | Ma02_p12110.1 | SP-LRR-TM | 1098 | 123.51 | 5.75 | Cell membrane |
| MaLRR-RLP06 | Ma02_p12120.1 | LRR-TM | 684 | 76.37 | 5.00 | Cell membrane |
| MaLRR-RLP07 | Ma02_p12140.1 | LRR-TM | 732 | 82.10 | 6.08 | Cell membrane |
| MaLRR-RLP08 | Ma02_p12150.1 | SP-LRR-TM | 903 | 102.55 | 5.98 | Cell membrane |
| MaLRR-RLP09 | Ma02_p12160.1 | LRR-TM | 1096 | 123.30 | 5.30 | Cell membrane |
| MaLRR-RLP10 | Ma02_p12190.1 | LRR-TM | 584 | 65.46 | 5.73 | Cell membrane |
| MaLRR-RLP11 | Ma02_p12580.1 | SP-LRR-TM | 976 | 108.33 | 5.75 | Cell membrane |
| MaLRR-RLP12 | Ma02_p12610.1 | SP-LRR-TM | 795 | 89.78 | 5.55 | Cell membrane |
| MaLRR-RLP13 | Ma02_p12630.1 | LRR-TM | 998 | 113.02 | 5.96 | Cell membrane |
| MaLRR-RLP14 | Ma02_p12660.1 | SP-LRR-TM | 853 | 96.55 | 5.71 | Cell membrane |
| MaLRR-RLP15 | Ma02_p12670.1 | SP-LRR-TM | 1076 | 120.27 | 6.48 | Cell membrane |
| MaLRR-RLP16 | Ma03_p02400.1 | SP-LRR-TM | 973 | 106.68 | 5.15 | Cell membrane |
| MaLRR-RLP17 | Ma03_p19590.1 | SP-LRR-TM | 772 | 84.27 | 6.44 | Cell membrane |
| MaLRR-RLP18 | Ma03_p27010.1 | SP-LRR-TM | 281 | 29.90 | 5.88 | Cell membrane |
| MaLRR-RLP19 | Ma03_p28990.1 | SP-LRR-TM | 1219 | 134.93 | 5.40 | Cell membrane |
| MaLRR-RLP20 | Ma03_p29020.1 | SP-LRR-TM | 851 | 95.65 | 5.71 | Cell membrane |
| MaLRR-RLP21 | Ma03_p29780.1 | SP-LRR-TM | 709 | 78.56 | 5.25 | Cell membrane |
| MaLRR-RLP22 | Ma03_p29790.1 | SP-LRR-TM | 1073 | 119.30 | 5.36 | Cell membrane |
| MaLRR-RLP23 | Ma03_p29840.1 | SP-LRR-TM | 1065 | 117.41 | 4.94 | Cell membrane |
| MaLRR-RLP24 | Ma03_p29850.1 | LRR-TM | 704 | 78.89 | 5.81 | Cell membrane |
| MaLRR-RLP25 | Ma03_p29910.1 | LRR-TM | 717 | 80.29 | 5.91 | Cell membrane |
| MaLRR-RLP26 | Ma03_p29970.1 | SP-LRR-TM | 1728 | 191.53 | 5.29 | Cell membrane |
| MaLRR-RLP27 | Ma03_p30010.1 | LRR-TM | 716 | 79.78 | 5.50 | Cell membrane |
| MaLRR-RLP28 | Ma03_p31870.1 | SP-LRR-TM | 287 | 30.02 | 5.40 | Cell membrane |
| MaLRR-RLP29 | Ma04_p06430.1 | SP-LRR-TM | 1092 | 118.88 | 5.18 | Cell membrane |
| MaLRR-RLP30 | Ma04_p07610.1 | SP-LRR-TM | 721 | 77.83 | 8.41 | Cell membrane |
| MaLRR-RLP31 | Ma04_p20720.1 | SP-LRR-TM | 841 | 92.49 | 5.50 | Cell membrane |
| MaLRR-RLP32 | Ma04_p22770.1 | SP-LRR-TM | 728 | 81.22 | 5.01 | Cell membrane |
| MaLRR-RLP33 | Ma04_p22780.1 | SP-LRR-TM | 943 | 104.62 | 5.64 | Cell membrane |
| MaLRR-RLP34 | Ma04_p29910.1 | SP-LRR-TM | 984 | 107.17 | 5.60 | Cell membrane |
| MaLRR-RLP35 | Ma04_p32290.1 | SP-LRR-TM | 599 | 66.07 | 7.07 | Cell membrane |
| MaLRR-RLP36 | Ma04_p32300.1 | SP-LRR-TM | 597 | 65.97 | 5.97 | Cell membrane |
| MaLRR-RLP37 | Ma04_p32760.1 | SP-LRR-TM | 974 | 108.41 | 5.11 | Cell membrane |
| MaLRR-RLP38 | Ma04_p32770.1 | LRR-TM | 1009 | 111.79 | 4.82 | Cell membrane |
| MaLRR-RLP39 | Ma04_p32780.1 | LRR-TM | 871 | 96.61 | 4.64 | Cell membrane |
| MaLRR-RLP40 | Ma04_p32810.1 | SP-LRR-TM | 993 | 110.20 | 4.74 | Cell membrane |
| MaLRR-RLP41 | Ma04_p33500.1 | SP-LRR-TM | 338 | 36.02 | 9.24 | Cell membrane |
| MaLRR-RLP42 | Ma04_p35850.1 | SP-LRR-TM | 961 | 103.93 | 5.12 | Cell membrane |
| MaLRR-RLP43 | Ma04_p36340.1 | SP-LRR-TM | 980 | 110.29 | 6.02 | Cell membrane |
| MaLRR-RLP44 | Ma05_p12870.1 | SP-LRR-TM | 854 | 94.06 | 8.66 | Cell membrane |
| MaLRR-RLP45 | Ma05_p16530.1 | SP-LRR-TM | 462 | 50.00 | 8.33 | Cell membrane |
| MaLRR-RLP46 | Ma05_p19050.1 | LRR-TM | 481 | 52.82 | 4.94 | Cell membrane |
| MaLRR-RLP47 | Ma06_p10770.1 | SP-LRR-TM | 982 | 107.99 | 6.20 | Cell membrane |
| MaLRR-RLP48 | Ma06_p10790.1 | SP-LRR-TM | 925 | 103.14 | 5.72 | Cell membrane |
| MaLRR-RLP49 | Ma06_p10800.1 | SP-LRR-TM | 1667 | 183.83 | 5.41 | Cell membrane |
| MaLRR-RLP50 | Ma06_p10810.1 | LRR-TM | 974 | 107.05 | 5.93 | Cell membrane |
| MaLRR-RLP51 | Ma06_p10820.1 | SP-LRR-TM | 990 | 109.57 | 5.94 | Cell membrane |
| MaLRR-RLP52 | Ma06_p10830.1 | SP-LRR-TM | 972 | 107.84 | 5.78 | Cell membrane |
| MaLRR-RLP53 | Ma06_p10880.1 | SP-LRR-TM | 475 | 51.97 | 5.45 | Cell membrane |
| MaLRR-RLP54 | Ma06_p26220.1 | SP-LRR-TM | 275 | 29.17 | 8.51 | Cell membrane |
| MaLRR-RLP55 | Ma06_p33510.1 | SP-LRR-TM | 427 | 45.03 | 8.64 | Cell membrane |
| MaLRR-RLP56 | Ma07_p04260.1 | SP-LRR-TM | 733 | 79.43 | 6.71 | Cell membrane |
| MaLRR-RLP57 | Ma07_p18320.1 | LRR-TM | 891 | 97.53 | 5.57 | Cell membrane |
| MaLRR-RLP58 | Ma07_p19540.1 | SP-LRR-TM | 1115 | 122.28 | 5.73 | Cell membrane |
| MaLRR-RLP59 | Ma07_p26290.1 | SP-LRR-TM | 465 | 49.96 | 5.06 | Cell membrane |
| MaLRR-RLP60 | Ma07_p28580.1 | SP-LRR-TM | 285 | 29.63 | 5.16 | Cell membrane |
| MaLRR-RLP61 | Ma08_p16720.1 | SP-LRR-TM | 484 | 52.98 | 5.32 | Cell membrane |
| MaLRR-RLP62 | Ma09_p00750.1 | SP-LRR-TM | 478 | 52.30 | 6.09 | Cell membrane |
| MaLRR-RLP63 | Ma09_p05420.1 | SP-LRR-TM | 424 | 44.62 | 8.48 | Cell membrane |
| MaLRR-RLP64 | Ma09_p22650.1 | SP-LRR-TM | 1013 | 112.09 | 5.28 | Cell membrane |
| MaLRR-RLP65 | Ma09_p22680.1 | LRR-TM | 1040 | 114.95 | 5.03 | Cell membrane |
| MaLRR-RLP66 | Ma09_p22690.1 | LRR-TM | 1082 | 119.59 | 5.07 | Cell membrane |
| MaLRR-RLP67 | Ma09_p22710.1 | SP-LRR-TM | 1336 | 148.51 | 5.11 | Cell membrane |
| MaLRR-RLP68 | Ma09_p22720.1 | LRR-TM | 587 | 65.49 | 5.64 | Cell membrane |
| MaLRR-RLP69 | Ma10_p00450.1 | SP-LRR-TM | 624 | 70.87 | 5.51 | Cell membrane |
| MaLRR-RLP70 | Ma10_p00470.1 | LRR-TM | 585 | 65.92 | 6.66 | Cell membrane |
| MaLRR-RLP71 | Ma10_p00480.1 | LRR-TM | 631 | 71.10 | 5.38 | Cell membrane |
| MaLRR-RLP72 | Ma10_p00490.1 | SP-LRR-TM | 677 | 76.94 | 6.39 | Cell membrane |
| MaLRR-RLP73 | Ma10_p00500.1 | SP-LRR-TM | 676 | 75.97 | 5.14 | Cell membrane |
| MaLRR-RLP74 | Ma10_p00550.1 | SP-LRR-TM | 902 | 100.82 | 5.43 | Cell membrane |
| MaLRR-RLP75 | Ma10_p00570.1 | SP-LRR-TM | 917 | 102.26 | 5.22 | Cell membrane |
| MaLRR-RLP76 | Ma10_p16560.1 | SP-LRR-TM | 722 | 77.99 | 4.99 | Cell membrane |
| MaLRR-RLP77 | Ma10_p20250.1 | LRR-TM | 449 | 49.50 | 5.28 | Cell membrane |
| MaLRR-RLP78 | Ma10_p28400.1 | SP-LRR-TM | 1034 | 112.83 | 5.72 | Cell membrane |
| # | MaLRR-RLP Gene | Sequence ID | Domains a | Protein Length | kDa | pI | Subcellular Location |
|---|---|---|---|---|---|---|---|
| 1 | Ma10_p00400.1 | Kinase | 179 | 19.80 | 5.29 | Nucleus | |
| 2 | Ma10_p00410.1 | Malectin | 397 | 44.11 | 8.62 | Extracellular | |
| 3 | Ma10_p00420.1 | SP-LRR | 377 | 42.13 | 6.13 | Extracellular | |
| 4 | Ma10_p00430.1 | Kinase | 128 | 14.25 | 6.28 | Cytoplasm | |
| 5 | Ma10_p00440.1 | LRR | 209 | 23.29 | 5.94 | Nucleus | |
| 6 | MaLRR-RLP69 | Ma10_p00450.1 | SP-LRR-TM | 624 | 70.87 | 5.51 | Cell membrane |
| 7 | Ma10_p00460.1 | Kinase | 138 | 15.77 | 5.91 | Mitochondrion | |
| 8 | MaLRR-RLP70 | Ma10_p00470.1 | LRR-TM | 585 | 65.92 | 6.66 | Cell membrane |
| 9 | MaLRR-RLP71 | Ma10_p00480.1 | LRR-TM | 631 | 71.10 | 5.38 | Cell membrane |
| 10 | MaLRR-RLP72 | Ma10_p00490.1 | SP-LRR-TM | 677 | 76.94 | 6.39 | Cell membrane |
| 11 | MaLRR-RLP73 | Ma10_p00500.1 | SP-LRR-TM | 676 | 75.97 | 5.14 | Cell membrane |
| 12 | Ma10_p00510.1 | LRR | 480 | 53.69 | 5.79 | Cytoplasm | |
| 13 | Ma10_p00520.1 | Malectin | 241 | 26.28 | 5.26 | Cytoplasm | |
| 14 | Ma10_p00530.1 | SP-LRR | 166 | 18.92 | 8.58 | Extracellular | |
| 15 | MaLRR-RLP74 | Ma10_p00550.1 | SP-LRR-TM | 902 | 100.82 | 5.43 | Cell membrane |
| 16 | Ma10_p00560.1 | SP-LRR | 348 | 38.59 | 5.84 | Extracellular | |
| 17 | MaLRR-RLP75 | Ma10_p00570.1 | SP-LRR-TM | 917 | 102.26 | 5.22 | Cell membrane |
| 18 | Ma10_p00650.1 | LRR | 641 | 71.47 | 5.62 | Cytoplasm | |
| 19 | Ma10_p00660.1 | LRR-TM | 143 | 16.84 | 5.47 | Endoplasmic reticulum membrane | |
| I (Solyc11g011180) b | SP-LRR-TM | 994 | 111.79 | 5.56 | Cell membrane | ||
| I-7 (AKR80573) c | SP-LRR-TM | 966 | 108.15 | 5.72 | Cell membrane |
| Clade Name | LRR-RLP Gene | Role in Plant Immunity |
|---|---|---|
| MaLRR-RLP58 | Resistance gene candidate | |
| I-7 | Cf-2 | Resistance to fungus Cladosporium fulvum |
| Cf-4 | Resistance to fungus Cladosporium fulvum | |
| Cf-5 | Resistance to fungus Cladosporium fulvum | |
| Cf-9 | Resistance to fungus Cladosporium fulvum | |
| Hcr9-4E | Resistance to fungus Cladosporium fulvum | |
| ELR | Resistance to oomycete Phytophthora infestans | |
| I-7 | Resistance to fungus Fusarium oxysporum | |
| Ve1 | Resistance to fungi Verticillium dahliae and V. albo-atrum | |
| RLP23 | Resistance to fungus Botrytis cinerea | |
| RLP30 | Resistance to fungus Sclerotinia sclerotiorum | |
| RBPG1 | Resistance to fungus Hyaloperonospora arabidopsidis | |
| RLM2 | Resistance to fungus Leptosphaeria maculans | |
| MaLRR-RLP78 | Resistance gene candidate | |
| I | I | Resistance to fungus Fusarium oxysporum |
| CuRe1 | Resistance to parasitic plant Cuscuta reflexa | |
| ReMAX | Perception of the Xanthomonas protein eMAX | |
| Foc race 1 RGCs | MaLRR-RLP69 | Resistance gene candidate |
| MaLRR-RLP70 | Resistance gene candidate | |
| MaLRR-RLP71 | Resistance gene candidate | |
| MaLRR-RLP72 | Resistance gene candidate | |
| MaLRR-RLP73 | Resistance gene candidate | |
| MaLRR-RLP74 | Resistance gene candidate | |
| MaLRR-RLP75 | Resistance gene candidate | |
| HcrVf2 | MaLRR-RLP33 | Resistance gene candidate |
| HcrVf2 | Resistance to fungus Venturia inaequalis | |
| EIX2 | Perception of a fungal xylanase (EIX) | |
| RFO2 | MaLRR-RLP35 | Resistance gene candidate |
| MaLRR-RLP36 | Resistance gene candidate | |
| MaLRR-RLP46 | Resistance gene candidate | |
| MaLRR-RLP56 | Resistance gene candidate | |
| RFO2 | Resistance to fungus Fusarium oxysporum |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez-López, D.; Herrera-Valencia, V.A.; Góngora-Castillo, E.; García-Laynes, S.; Puch-Hau, C.; López-Ochoa, L.A.; Lizama-Uc, G.; Peraza-Echeverria, S. Genome-Wide Analysis of the LRR-RLP Gene Family in a Wild Banana (Musa acuminata ssp. malaccensis) Uncovers Multiple Fusarium Wilt Resistance Gene Candidates. Genes 2022, 13, 638. https://doi.org/10.3390/genes13040638
Álvarez-López D, Herrera-Valencia VA, Góngora-Castillo E, García-Laynes S, Puch-Hau C, López-Ochoa LA, Lizama-Uc G, Peraza-Echeverria S. Genome-Wide Analysis of the LRR-RLP Gene Family in a Wild Banana (Musa acuminata ssp. malaccensis) Uncovers Multiple Fusarium Wilt Resistance Gene Candidates. Genes. 2022; 13(4):638. https://doi.org/10.3390/genes13040638
Chicago/Turabian StyleÁlvarez-López, Dulce, Virginia Aurora Herrera-Valencia, Elsa Góngora-Castillo, Sergio García-Laynes, Carlos Puch-Hau, Luisa Alhucema López-Ochoa, Gabriel Lizama-Uc, and Santy Peraza-Echeverria. 2022. "Genome-Wide Analysis of the LRR-RLP Gene Family in a Wild Banana (Musa acuminata ssp. malaccensis) Uncovers Multiple Fusarium Wilt Resistance Gene Candidates" Genes 13, no. 4: 638. https://doi.org/10.3390/genes13040638
APA StyleÁlvarez-López, D., Herrera-Valencia, V. A., Góngora-Castillo, E., García-Laynes, S., Puch-Hau, C., López-Ochoa, L. A., Lizama-Uc, G., & Peraza-Echeverria, S. (2022). Genome-Wide Analysis of the LRR-RLP Gene Family in a Wild Banana (Musa acuminata ssp. malaccensis) Uncovers Multiple Fusarium Wilt Resistance Gene Candidates. Genes, 13(4), 638. https://doi.org/10.3390/genes13040638








