Ovarian Transcriptomic Analysis of Ninghai Indigenous Chickens at Different Egg-Laying Periods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal and Sample Collection
2.2. Ethical Statement
2.3. RNA Sequencing (RNA-seq)
2.4. Transcriptomic Bioinformatics Analyses
2.5. Differential Expression Analysis
2.6. GO and KEGG Enrichment Analysis of DEGs
2.7. Gene Expression Analysis by qPCR
3. Results
3.1. Sequencing Data and Read Mapping
3.2. Differential Expression Genes
3.3. GO and KEGG Analysis
4. Discussion
4.1. Analysis of DEGs
4.2. GO and KEGG Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zaheer, K. An updated review on chicken eggs: Production, consumption, management aspects and nutritional benefits to human health. Food Sci. Nutr. 2015, 6, 1208. [Google Scholar] [CrossRef] [Green Version]
- Iannotti, L.L.; Lutter, C.K.; Bunn, D.A.; Stewart, C.P. Eggs: The uncracked potential for improving maternal and young child nutrition among the world’s poor. Nutr. Rev. 2014, 72, 355–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Chen, X.; Zhao, J.; Ma, C.; Yan, C.; Liswaniso, S.; Xu, R.; Qin, N. Transcriptome comparative analysis of ovarian follicles reveals the key genes and signaling pathways implicated in hen egg production. BMC Genom. 2021, 22, 899. [Google Scholar] [CrossRef] [PubMed]
- Webb, R.; Garnsworthy, P.C.; Gong, J.G.; Armstrong, D.G. Control of follicular growth: Local interactions and nutritional influences. J. Anim. Sci. 2004, 82 E-Suppl, E63–E74. [Google Scholar] [CrossRef]
- Mishra, S.K.; Chen, B.; Zhu, Q.; Xu, Z.; Ning, C.; Yin, H.; Wang, Y.; Zhao, X.; Fan, X.; Yang, M. Transcriptome analysis reveals differentially expressed genes associated with high rates of egg production in chicken hypothalamic-pituitary-ovarian axis. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Woodruff, T.K.; Shea, L.D.J.R.S. The role of the extracellular matrix in ovarian follicle development. Reprod. Sci. 2007, 14, 6–10. [Google Scholar] [CrossRef] [Green Version]
- Tao, Z.; Song, W.; Zhu, C.; Xu, W.; Liu, H.; Zhang, S.; Huifang, L. Comparative transcriptomic analysis of high and low egg-producing duck ovaries. Poult. Sci. 2017, 96, 4378–4388. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, L.; Wu, T.; Feng, Y.; Ding, Y.; Ye, P.; Yin, Z. Transcriptomic analysis of ovaries from pigs with high and low litter size. PLoS ONE 2015, 10, e0139514. [Google Scholar] [CrossRef] [Green Version]
- Lan, D.; Xiong, X.; Huang, C.; Mipam, T.D.; Li, J. Toward understanding the genetic basis of yak ovary reproduction: A characterization and comparative analyses of estrus ovary transcriptiome in yak and cattle. PLoS ONE 2016, 11, e0152675. [Google Scholar] [CrossRef]
- Miao, X.; Luo, Q. Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing. Reproduction 2013, 145, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Miao, X.; Luo, Q.; Qin, X. Genome-wide transcriptome analysis in the ovaries of two goats identifies differentially expressed genes related to fecundity. Gene 2016, 582, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.; Asiamah, C.A.; Lu, L.-L.; Liu, Y.; Pan, Y.; Chen, T.; Zhao, Z.; Su, Y. Ovarian transcriptomic analysis and follicular development of Leizhou black duck. Poult. Sci. 2020, 99, 6173–6187. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Liu, J.; Cao, J.; Zhang, H.; Liu, X. Ovarian transcriptomic analysis of black Muscovy duck at the early, peak and late egg-laying stages. Gene 2021, 777, 145449. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. In Babraham Bioinformatics; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357-U121. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Garber, M.; Grabherr, M.G.; Guttman, M.; Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 2011, 8, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate - a Practical and Powerful Approach To Multiple Testing. J R Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.Y.; Song, Y.P.; Li, T.; Zhang, S.S.; Huang, L.H.; Zhang, S.Y.; Cao, J.T.; Liu, X.L.; Zhang, J.Q. Comparative Transcriptome Profiling of Ovary Tissue between Black Muscovy Duck and White Muscovy Duck with High- and Low-Egg Production. Genes 2021, 12, 57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Wang, P.F.; Cong, G.L.; Liu, M.H.; Shi, S.R.; Shao, D.; Tan, B.J. Comparative transcriptomic analysis of ovaries from high and low egg-laying Lingyun black-bone chickens. Vet. Med. Sci. 2021, 7, 1867–1880. [Google Scholar] [CrossRef]
- Akhavan, S.R.; Falahatkar, B.; McCormick, S.P.; Lokman, P.M.; Physiology, C. Changes in lipid biology during ovarian development in farmed beluga sturgeon, Huso huso L. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2020, 319, R376–R386. [Google Scholar] [CrossRef]
- Olson, G.E.; Winfrey, V.P.; NagDas, S.K.; Hill, K.E.; Burk, R.F. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J. Biol. Chem. 2007, 282, 12290–12297. [Google Scholar] [CrossRef] [Green Version]
- Andersen, O.M.; Yeung, C.-H.; Vorum, H.; Wellner, M.; Andreassen, T.K.; Erdmann, B.; Mueller, E.-C.; Herz, J.; Otto, A.; Cooper, T.G. Essential role of the apolipoprotein E receptor-2 in sperm development. J. Biol. Chem. 2003, 278, 23989–23995. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Chen, Z.; Xu, G.; Wang, X.; Ning, Z.; Zheng, J.; Qu, L.; Yang, N. Low-density lipoprotein receptor-related protein 8 gene association with egg traits in dwarf chickens. Poult. Sci. 2010, 89, 883–886. [Google Scholar] [CrossRef]
- Wang, C.; Li, S.; Li, C.; Yu, G.; Feng, Y.; Peng, X.; Gong, Y.J.B.P.S. Molecular cloning, expression and association study with reproductive traits of the duck LRP8 gene. Br. Poult. Sci. 2013, 54, 567–574. [Google Scholar] [CrossRef]
- Dias, F.; Khan, M.; Sirard, M.; Adams, G.; Singh, J. Transcriptome analysis of granulosa cells after conventional vs long FSH-induced superstimulation in cattle. BMC Genom. 2018, 19, 258. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.-M.; Qiao, J.; Leung, P.C. Oocyte–somatic cell interactions in the human ovary—novel role of bone morphogenetic proteins and growth differentiation factors. Hum. Reprod. Update 2017, 23, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, G.; Guo, B.; Pan, D.; Mu, Y.; Feng, S. Expression of bone morphogenetic proteins and receptors in porcine cumulus–oocyte complexes during in vitro maturation. Anim. Reprod. Sci. 2008, 104, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Frota, I.M.; Leitão, C.C.; Costa, J.J.; van den Hurk, R.; Saraiva, M.V.; Figueiredo, J.R.; Silva, J.R. Levels of BMP-6 mRNA in goat ovarian follicles and in vitro effects of BMP-6 on secondary follicle development. Zygote 2013, 21, 270–278. [Google Scholar] [CrossRef]
- De Conto, E.; Matte, U.; Cunha-Filho, J.S. BMP-6 and SMAD4 gene expression is altered in cumulus cells from women with endometriosis-associated infertility. Acta Obstet. Gynecol. Scand. 2021, 100, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, K.; Johnson, A.L. Follicle dynamics and granulosa cell differentiation in the turkey hen ovary. Poult. Sci. 2018, 97, 3755–3761. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Rico, M.; Jimenez-Movilla, M.; Llop, E.; Perez-Oliva, A.; Ballesta, J.; Gutierrez-Gallego, R.; Jimenez-Cervantes, C.; Aviles, M. Hamster zona pellucida is formed by four glycoproteins: ZP1, ZP2, ZP3, and ZP4. Proteome Res. 2009, 8, 926–941. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ni, C.X.; Wu, L.; Chen, B.B.; Xu, Y.; Zhang, Z.H.; Mu, J.; Li, B.; Yan, Z.; Fu, J.; et al. Novel mutations in ZP1, ZP2, and ZP3 cause female infertility due to abnormal zona pellucida formation. Hum. Genet. 2019, 138, 327–337. [Google Scholar] [CrossRef]
- Serizawa, M.; Kinoshita, M.; Rodler, D.; Tsukada, A.; Ono, H.; Yoshimura, T.; Kansaku, N.; Sasanami, T. Oocytic expression of zona pellucida protein ZP4 in Japanese quail (Coturnix japonica). Anim. Sci. J. 2011, 82, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Russell, D.L.; Ochsner, S.A.; Hsieh, M.; Mulders, S.; Richards, J.S. Hormone-regulated expression and localization of versican in the rodent ovary. Endocrinology 2003, 144, 1020–1031. [Google Scholar] [CrossRef]
- Dunning, K.R.; Lane, M.; Brown, H.M.; Yeo, C.; Robker, R.L.; Russell, D.L. Altered composition of the cumulus-oocyte complex matrix during in vitro maturation of oocytes. Hum. Reprod. 2007, 22, 2842–2850. [Google Scholar] [CrossRef] [Green Version]
- Papler, T.B.; Bokal, E.V.; Maver, A.; Lovrečić, L. Specific gene expression differences in cumulus cells as potential biomarkers of pregnancy. Reprod. Biomed. Online 2015, 30, 426–433. [Google Scholar] [CrossRef] [Green Version]
- Shen, Q.; Chen, M.; Zhao, X.; Liu, Y.; Ren, X.; Zhang, L. Versican expression level in cumulus cells is associated with human oocyte developmental competence. Syst. Biol. Reprod. Med. 2020, 66, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Terauchi, K.J.; Miyagawa, S.; Iguchi, T.; Sato, T. Hedgehog signaling regulates the basement membrane remodeling during folliculogenesis in the neonatal mouse ovary. Cell Tissue Res. 2020, 381, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Franchi, F.F.; Hernandes, M.P.; Ferreira, A.L.C.; de Lima, V.A.V.; de Oliveira Mendes, L.; de Aquino, A.M.; Scarano, W.R.; de Souza Castilho, A.C. Fractal analysis and histomolecular phenotyping provides insights into extracellular matrix remodeling in the developing bovine fetal ovary. Biochem. Biophys. Res. Commun. 2020, 523, 823–828. [Google Scholar] [CrossRef]
- Tang, J.; Hu, W.; Chen, S.; Di, R.; Liu, Q.; Wang, X.; He, X.; Gan, S.; Zhang, X.; Zhang, J. The genetic mechanism of high prolificacy in small tail han sheep by comparative proteomics of ovaries in the follicular and luteal stages. J. Proteom. 2019, 204, 103394. [Google Scholar] [CrossRef]
- Bao, Y.; Yao, X.; Li, X.; Ei-Samahy, M.; Yang, H.; Liang, Y.; Liu, Z.; Wang, F. INHBA transfection regulates proliferation, apoptosis and hormone synthesis in sheep granulosa cells. Theriogenology 2021, 175, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Pangas, S.A.; Jorgez, C.J.; Tran, M.; Agno, J.; Li, X.H.; Brown, C.W.; Kumar, T.R.; Matzuk, M.M. Intraovarian activins are required for female fertility. Mol. Endocrinol. 2007, 21, 2458–2471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, S.Z.; Gao, L.; Wu, C.J.; Qin, F.; Yuan, J.H. Role of the lysyl oxidase family in organ development (Review). Exp. Ther. Med. 2020, 20, 163–172. [Google Scholar] [CrossRef]
- Laczko, R.; Csiszar, K.J.B. Lysyl oxidase (LOX): Functional contributions to signaling pathways. Biomolecules 2020, 10, 1093. [Google Scholar] [CrossRef]
- Liu, C.; Zhu, P.; Wang, W.; Li, W.; Shu, Q.; Chen, Z.-J.; Myatt, L.; Sun, K. Inhibition of lysyl oxidase by prostaglandin E2 via EP2/EP4 receptors in human amnion fibroblasts: Implications for parturition. Mol. Cell. Endocrinol. 2016, 424, 118–127. [Google Scholar] [CrossRef]
- Harlow, C.R.; Rae, M.; Davidson, L.; Trackman, P.C.; Hillier, S.G. Lysyl oxidase gene expression and enzyme activity in the rat ovary: Regulation by follicle-stimulating hormone, androgen, and transforming growth factor-β superfamily members in vitro. Endocrinology 2003, 144, 154–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irez, T.; Sahin, Y.; Malik, E.; Guralp, O. The Predictive Value of the Pentraxin 3 Concentration in Cumulus Cell Culture Media for the Embryo Implantation. J. Gynecol. Obstet. Hum. Reprod. 2022, 1–7. [Google Scholar] [CrossRef]
- Zhang, X.; Jafari, N.; Barnes, R.B.; Confino, E.; Milad, M.; Kazer, R.R. Studies of gene expression in human cumulus cells indicate pentraxin 3 as a possible marker for oocyte quality. Fertil. Steril. 2005, 83, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, H.; Sheng, Y.; Wang, Z. MicroRNA-224 delays oocyte maturation through targeting Ptx3 in cumulus cells. Mech. Dev. 2017, 143, 20–25. [Google Scholar] [CrossRef]
- Hatzirodos, N.; Irving-Rodgers, H.F.; Hummitzsch, K.; Harland, M.L.; Morris, S.E.; Rodgers, R.J. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genom. 2014, 15, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Peng, J.; Matzuk, M.M.; Yao, H.H.-C. Lineage specification of ovarian theca cells requires multicellular interactions via oocyte and granulosa cells. Nat. Commun. 2015, 6, 6934. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, H.; Zhao, X.; Das, S.K.; Hogan, B.L.; Dey, S.K. Indian hedgehog as a progesterone-responsive factor mediating epithelial–mesenchymal interactions in the mouse uterus. Dev. Biol. 2002, 245, 280–290. [Google Scholar] [CrossRef]
- Bhurke, A.S.; Bagchi, I.C.; Bagchi, M.K. Progesterone-Regulated Endometrial Factors Controlling Implantation. Am. J. Reprod. Immunol. 2016, 75, 237–245. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Jeong, J.; Kwak, I.; Yu, C.-T.; Lanske, B.; Soegiarto, D.W.; Toftgard, R.; Tsai, M.-J.; Tsai, S.; Lydon, J.P. Indian hedgehog is a major mediator of progesterone signaling in the mouse uterus. Nature Genet. 2006, 38, 1204–1209. [Google Scholar] [CrossRef]
- Mulayim, N.; Palter, S.F.; Kayisli, U.A.; Senturk, L.; Arici, A. Chemokine receptor expression in human endometrium. Biol. Reprod. 2003, 68, 1491–1495. [Google Scholar] [CrossRef]
- Zhang, J.; Lathbury, L.J.; Salamonsen, L.A. Expression of the chemokine eotaxin and its receptor, CCR3, in human endometrium. Biol. Reprod. 2000, 62, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.G.A.; Pereira, L.A.A.C.; Viana, J.H.M.; Russo, R.C.; Campos-Junior, P.H.A. The CC-chemokine receptor 2 is involved in the control of ovarian folliculogenesis and fertility lifespan in mice. J. Reprod. Immunol. 2020, 141, 103174. [Google Scholar] [CrossRef] [PubMed]
- Walma, D.A.C.; Yamada, K.M. The extracellular matrix in development. Development 2020, 147, dev175596. [Google Scholar] [CrossRef] [PubMed]
- Kulus, J.; Kulus, M.; Kranc, W.; Jopek, K.; Zdun, M.; Józkowiak, M.; Jaśkowski, J.M.; Piotrowska-Kempisty, H.; Bukowska, D.; Antosik, P. Transcriptomic Profile of New Gene Markers Encoding Proteins Responsible for Structure of Porcine Ovarian Granulosa Cells. Biol.-Basel 2021, 10, 1214. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, L.; Yoneda, M.; Zhao, M.; Yingsung, W.; Yoshida, N.; Kitagawa, Y.; Kawamura, K.; Suzuki, T.; Kimata, K.J.J.O.B.C. Defect in SHAP-hyaluronan complex causes severe female infertility: A study by inactivation of the bikunin gene in mice. J. Biol. Chem. 2001, 276, 7693–7696. [Google Scholar] [CrossRef] [Green Version]
- Bello, S.F.; Xu, H.; Guo, L.; Li, K.; Zheng, M.; Xu, Y.; Zhang, S.; Bekele, E.J.; Bahareldin, A.A.; Zhu, W.; et al. Hypothalamic and ovarian transcriptome profiling reveals potential candidate genes in low and high egg production of white Muscovy ducks (Cairina moschata). Poult. Sci. 2021, 100, 101310. [Google Scholar] [CrossRef]
- Li, S.; Ao, L.; Yan, Y.; Jiang, J.; Chen, B.; Duan, Y.; Shen, F.; Chen, J.; Inglis, B.; Ni, R. Differential motility parameters and identification of proteomic profiles of human sperm cryopreserved with cryostraw and cryovial. Clin. Proteom. 2019, 16, 24. [Google Scholar] [CrossRef]
- Heffner, K.; Hizal, D.B.; Majewska, N.I.; Kumar, S.; Dhara, V.G.; Zhu, J.; Bowen, M.; Hatton, D.; Yerganian, G.; Yerganian, A.J.S.R. Expanded Chinese hamster organ and cell line proteomics profiling reveals tissue-specific functionalities. Sci. Rep. 2020, 10, 15841. [Google Scholar] [CrossRef]
- Park, Y.-J.; Yoo, S.-A.; Kim, M.; Kim, W.-U. The Role of Calcium–Calcineurin–NFAT Signaling Pathway in Health and Autoimmune Diseases. Front. Immunol. 2020, 11, 195. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, X.; Chen, L.; Wang, J.; Duan, Y.; Li, H.; Lu, L. Transcriptomic analyses of the hypothalamic-pituitary-gonadal axis identify candidate genes related to egg production in Xinjiang Yili geese. Animals 2020, 10, 90. [Google Scholar] [CrossRef] [Green Version]
- Ye, P.; Ge, K.; Li, M.; Yang, L.; Jin, S.; Zhang, C.; Chen, X.; Geng, Z. Egg-laying and brooding stage-specific hormonal response and transcriptional regulation in pituitary of Muscovy duck (Cairina moschata). Poult. Sci. 2019, 98, 5287–5296. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, X.; Chimbaka, I.M.; Qin, N.; Xu, X.; Liswaniso, S.; Xu, R.; Gonzalez, J.M. Transcriptome analysis of ovarian follicles reveals potential pivotal genes associated with increased and decreased rates of chicken egg production. Front. Genet. 2021, 12, 250. [Google Scholar] [CrossRef] [PubMed]
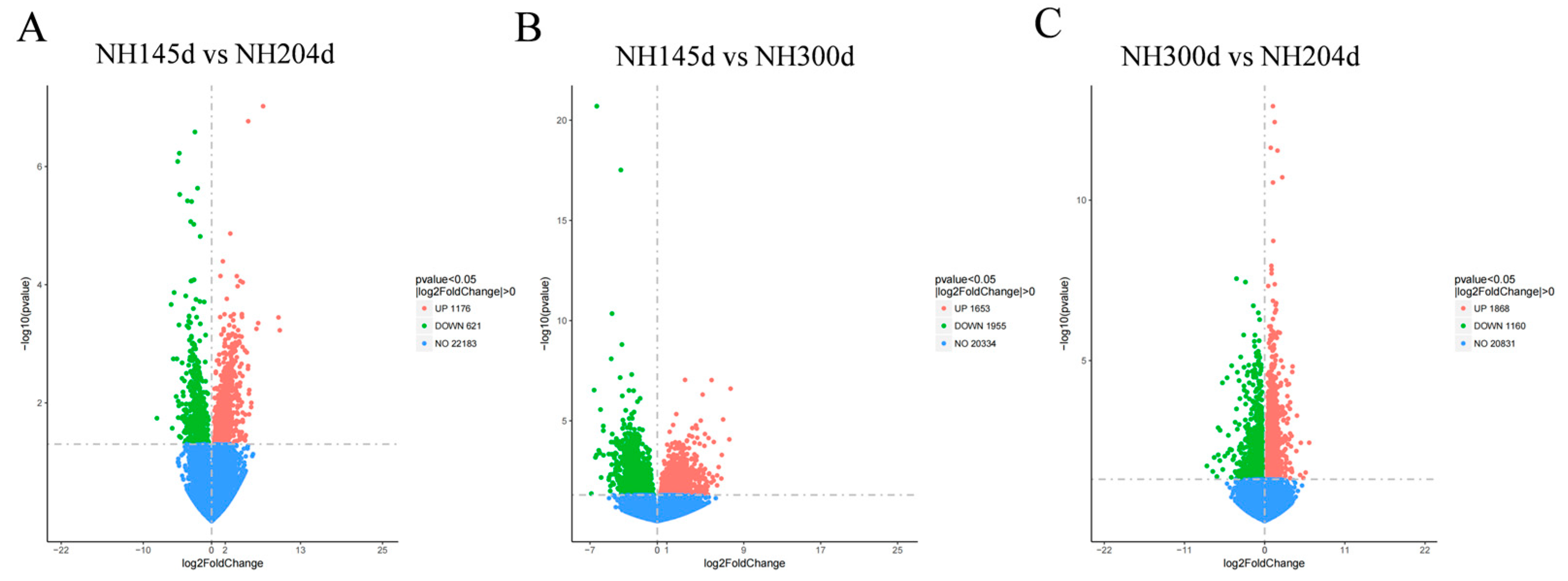
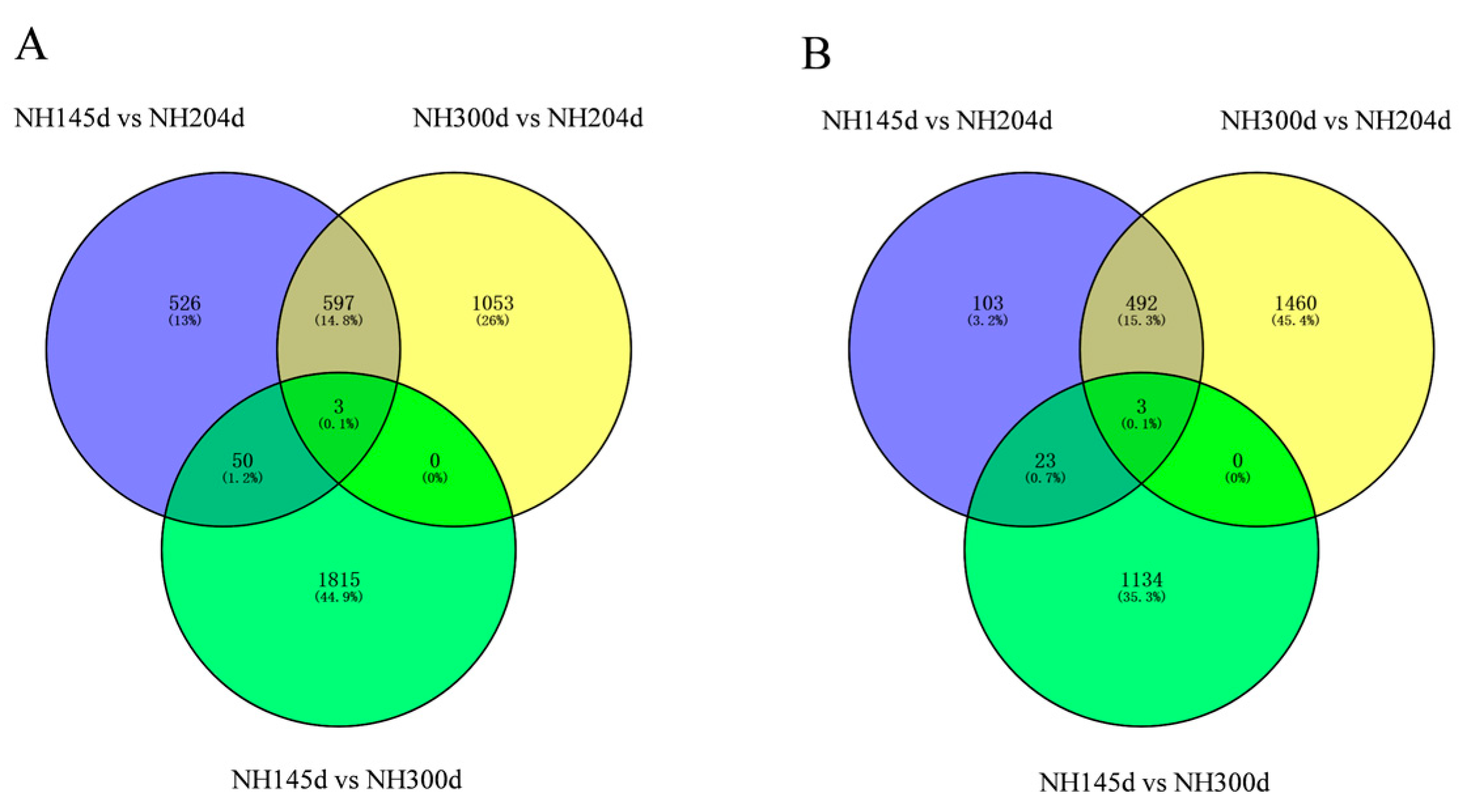
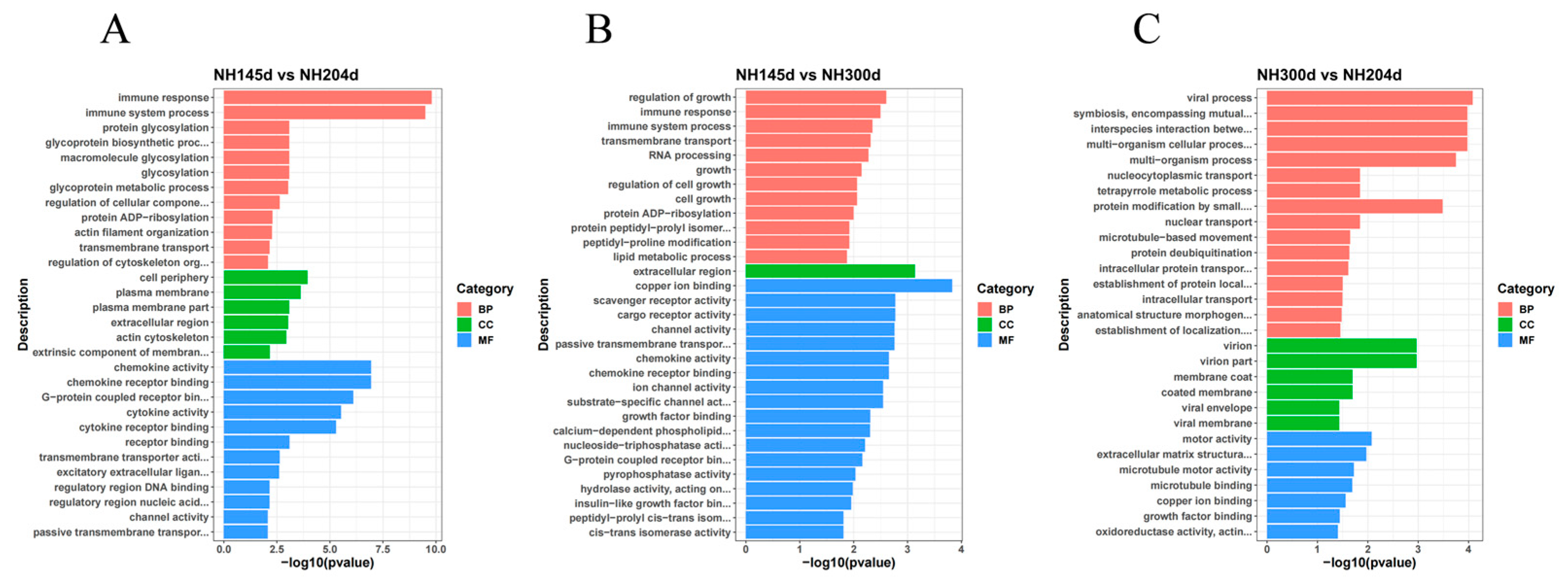
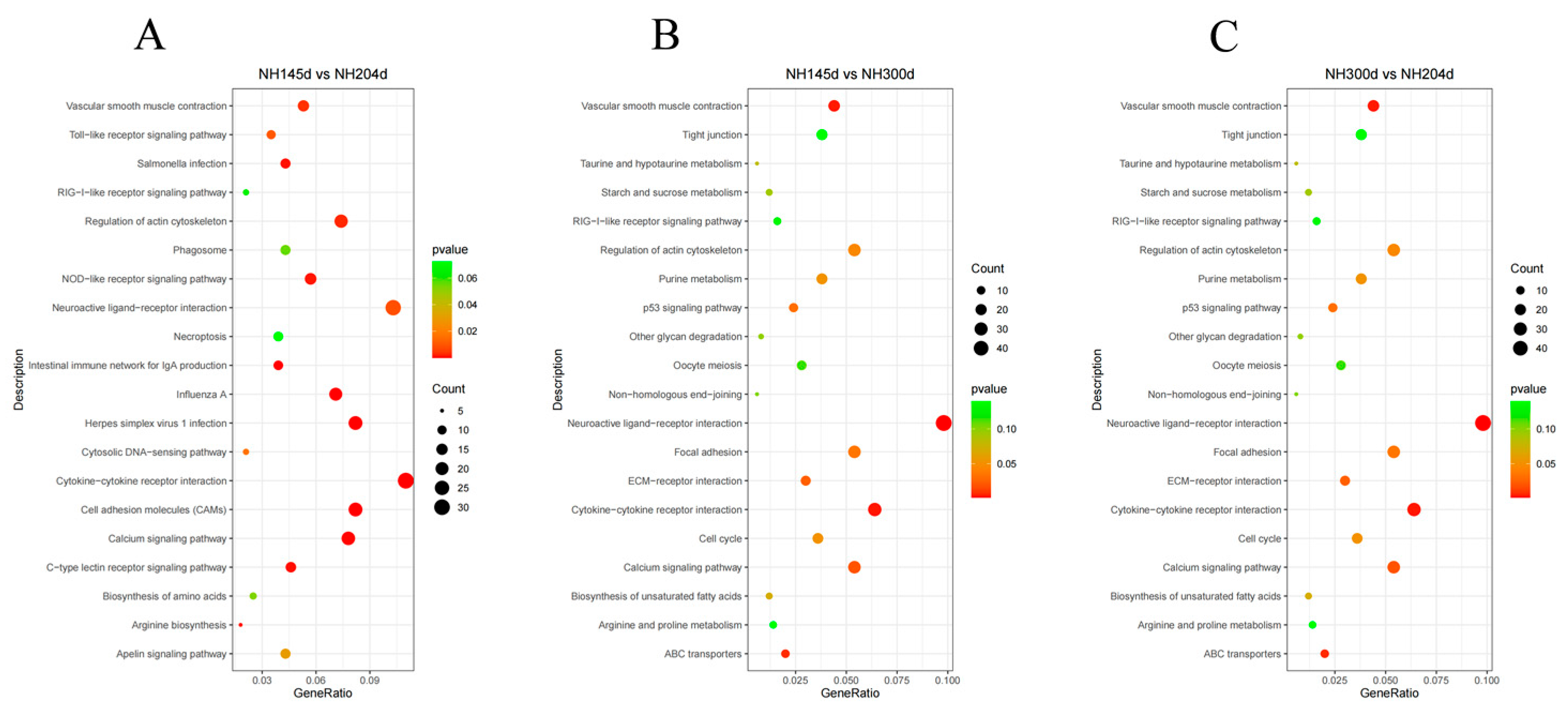

| Sample | Raw Reads | Clean Reads | Total Map | Q20 | Q30 | GC Pct |
|---|---|---|---|---|---|---|
| NH300d_1 | 66888644 | 66121522 | 60514494 (91.52%) | 97.88% | 94.22% | 50.69% |
| NH300d_2 | 63552028 | 62595376 | 55486806 (88.64%) | 97.66% | 93.86% | 51.78% |
| NH300d_3 | 58809960 | 58226874 | 54031796 (92.8%) | 98.01% | 94.49% | 50.84% |
| NH300d_4 | 52437050 | 51709354 | 47297226 (91.47%) | 98% | 94.52% | 50.4% |
| NH204d_1 | 59434470 | 58593266 | 53253424 (90.89%) | 97.86% | 94.22% | 50.91% |
| NH204d_2 | 53435540 | 52503528 | 47500358 (90.47%) | 97.85% | 94.22% | 50.53% |
| NH204d_3 | 62255842 | 61476260 | 56020220 (91.12%) | 97.82% | 94.17% | 51.38% |
| NH204d_4 | 53250580 | 52498894 | 46560294 (88.69%) | 97.85% | 94.17% | 51.69% |
| NH145d_1 | 73572594 | 72533526 | 64007420 (88.25%) | 97.82% | 94.2% | 51.65% |
| NH145d_2 | 57911030 | 56397798 | 47287950 (83.85%) | 96.71% | 92.32% | 50.4% |
| NH145d_3 | 57453868 | 56745306 | 51098056 (90.05%) | 97.95% | 94.46% | 51.52% |
| NH145d_4 | 52929654 | 52233058 | 48179156 (92.24%) | 97.81% | 94.02% | 50.81% |
| Group | Pathway | GeneRatio | BgRatio | p | Q | KEGG ID |
|---|---|---|---|---|---|---|
| NH145dvsNH204d | Cell adhesion molecules (CAMs) | 23/282 | 107/4684 | 5.1 × 10−8 | 6.0 × 10−6 | gga04514 |
| Cytokine–cytokine receptor interaction | 31/282 | 187/4684 | 1.4 × 10−7 | 8.5 × 10−6 | gga04060 | |
| Herpes simplex virus 1 infection | 23/282 | 132/4684 | 2.7 × 10−6 | 1.1 × 10−4 | gga05168 | |
| Intestinal immune network for IgA production | 11/282 | 36/4684 | 4.8 × 10−6 | 1.4 ×10−4 | gga04672 | |
| Influenza A | 20/282 | 112/4684 | 8.4 × 10−6 | 2.0 ×10−4 | gga05164 | |
| Calcium signaling pathway | 22/282 | 168/4684 | 4.0 × 10−4 | 7.8 ×10−3 | gga04020 | |
| C–type lectin receptor signaling pathway | 13/282 | 80/4684 | 8.5 × 10−4 | 1.3 ×10−2 | gga04625 | |
| Salmonella infection | 12/282 | 71/4684 | 9.3 × 10−4 | 1.3 ×10−2 | gga05132 | |
| Arginine biosynthesis | 5/282 | 14/4684 | 9.7 × 10−4 | 1.3 ×10−2 | gga00220 | |
| NOD–like receptor signaling pathway | 16/282 | 115/4684 | 1.3 × 10−3 | 1.5 ×10−2 | gga04621 | |
| Regulation of actin cytoskeleton | 21/282 | 182/4684 | 2.7 × 10−3 | 3.0 ×10−2 | gga04810 | |
| Vascular smooth muscle contraction | 15/282 | 116/4684 | 3.7 × 10−3 | 3.7 ×10−2 | gga04270 | |
| Neuroactive ligand–receptor interaction | 29/282 | 305/4684 | 8.5 × 10−3 | 7.6 ×10−2 | gga04080 | |
| Toll-like receptor signaling pathway | 10/282 | 72/4684 | 1.0 × 10−2 | 8.7 ×10−2 | gga04620 | |
| Cytosolic DNA-sensing pathway | 6/282 | 35/4684 | 1.7 × 10−2 | 1.3 ×10−1 | gga04623 | |
| Apelin signaling pathway | 12/282 | 109/4684 | 3.0 × 10−2 | 2.2 ×10−1 | gga04371 | |
| NH145dvsNH300d | Neuroactive ligand–receptor interaction | 49/499 | 305/4685 | 1.7 × 10−3 | 2.3 ×10−1 | gga04080 |
| Cytokine–cytokine receptor interaction | 32/499 | 187/4685 | 4.1 × 10−3 | 2.3 ×10−1 | gga04060 | |
| Vascular smooth muscle contraction | 22/499 | 116/4685 | 4.7 × 10−3 | 2.3 ×10−1 | gga04270 | |
| ABC transporters | 10/499 | 40/4685 | 7.6 × 10−3 | 2.7 ×10−1 | gga02010 | |
| Calcium signaling pathway | 27/499 | 169/4685 | 1.9 × 10−2 | 5.5 ×10−1 | gga04020 | |
| ECM–receptor interaction | 15/499 | 82/4685 | 2.4 × 10−2 | 5.8 ×10−1 | gga04512 | |
| p53 signaling pathway | 12/499 | 63/4685 | 3.1 × 10−2 | 6.3 ×10−1 | gga04115 | |
| Focal adhesion | 27/499 | 178/4685 | 3.5 × 10−2 | 6.3 ×10−1 | gga04510 | |
| Regulation of actin cytoskeleton | 27/499 | 182/4685 | 4.5 × 10−2 | 6.8 ×10−1 | gga04810 | |
| NH300dvsNH204d | Progesterone–mediated oocyte maturation | 21/611 | 76/4684 | 5.0 × 10−4 | 7.3 ×10−2 | gga04914 |
| RNA degradation | 18/611 | 67/4684 | 1.7 × 10−3 | 1.3 ×10−1 | gga03018 | |
| Oocyte meiosis | 19/611 | 92/4684 | 2.6 × 10−2 | 1.0 | gga04114 | |
| Homologous recombination | 9/611 | 37/4684 | 4.4 × 10−2 | 1.0 | gga03440 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Zhou, W.; Cao, H.; Zhang, H.; Xiang, X.; Yin, Z. Ovarian Transcriptomic Analysis of Ninghai Indigenous Chickens at Different Egg-Laying Periods. Genes 2022, 13, 595. https://doi.org/10.3390/genes13040595
Huang X, Zhou W, Cao H, Zhang H, Xiang X, Yin Z. Ovarian Transcriptomic Analysis of Ninghai Indigenous Chickens at Different Egg-Laying Periods. Genes. 2022; 13(4):595. https://doi.org/10.3390/genes13040595
Chicago/Turabian StyleHuang, Xuan, Wei Zhou, Haiyue Cao, Haiyang Zhang, Xin Xiang, and Zhaozheng Yin. 2022. "Ovarian Transcriptomic Analysis of Ninghai Indigenous Chickens at Different Egg-Laying Periods" Genes 13, no. 4: 595. https://doi.org/10.3390/genes13040595
APA StyleHuang, X., Zhou, W., Cao, H., Zhang, H., Xiang, X., & Yin, Z. (2022). Ovarian Transcriptomic Analysis of Ninghai Indigenous Chickens at Different Egg-Laying Periods. Genes, 13(4), 595. https://doi.org/10.3390/genes13040595






