Circum-Saharan Prehistory through the Lens of mtDNA Diversity
Abstract
1. Introduction
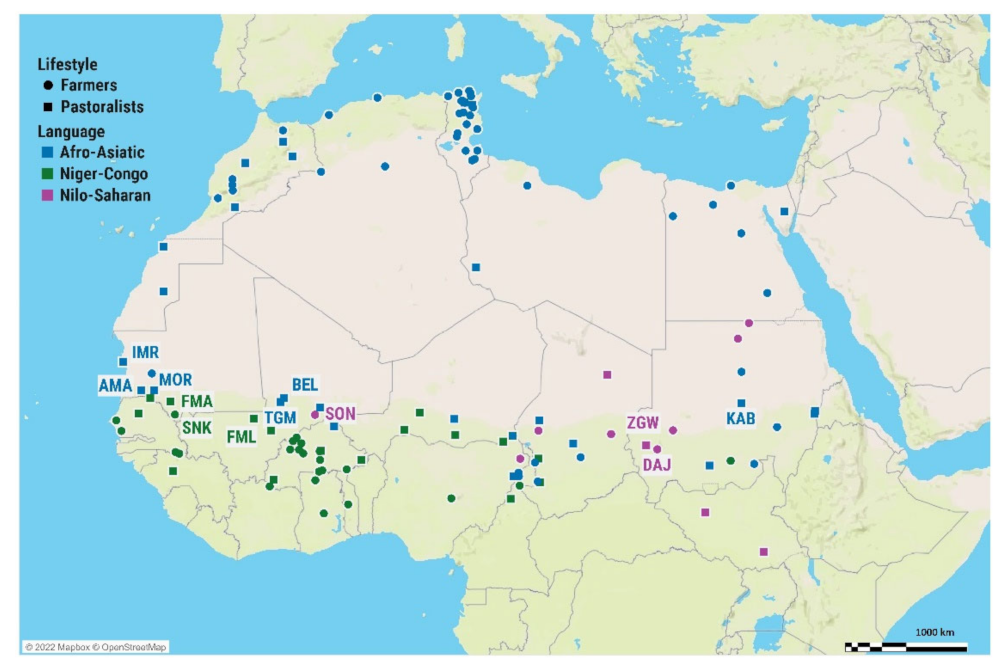
2. Materials and Methods
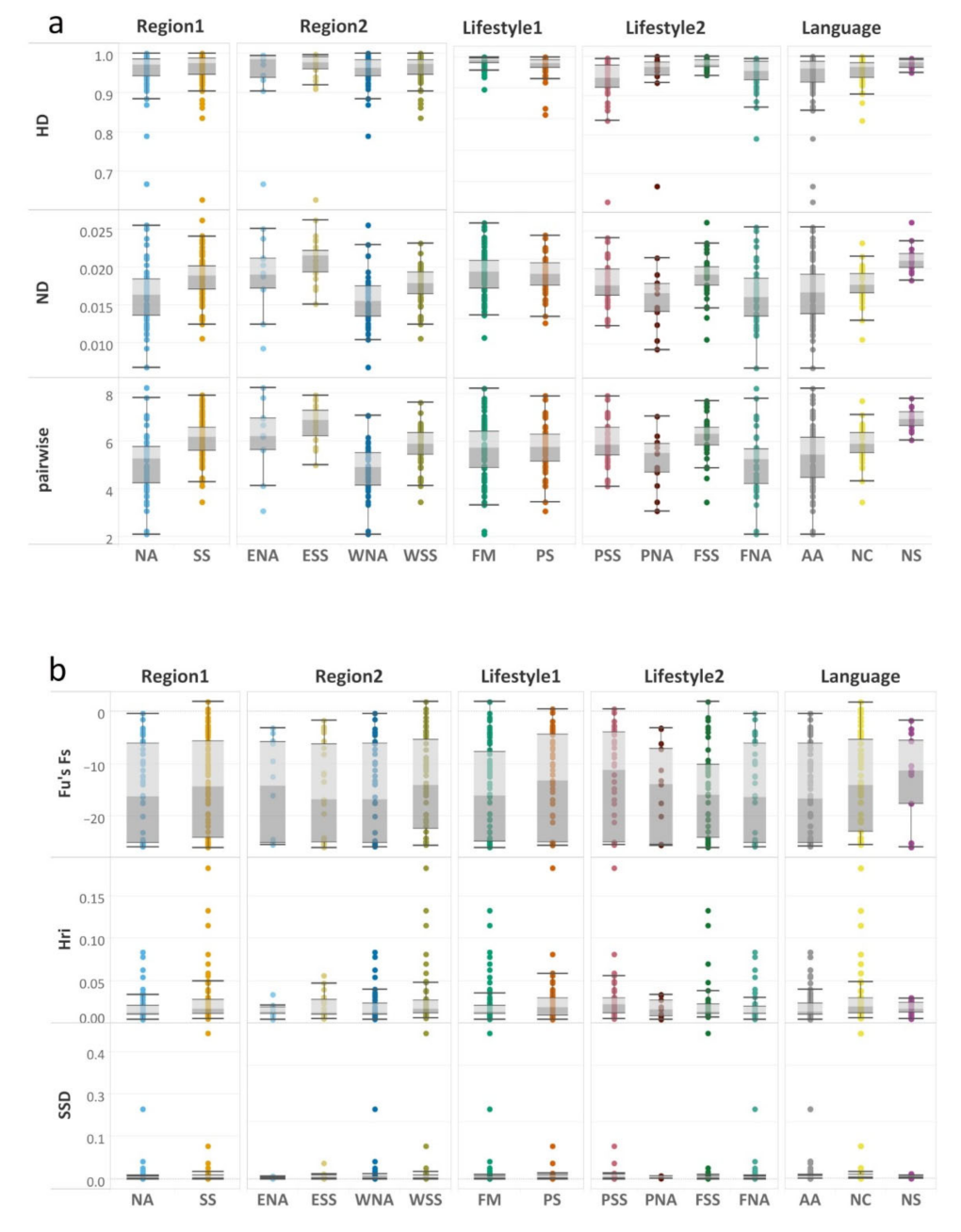
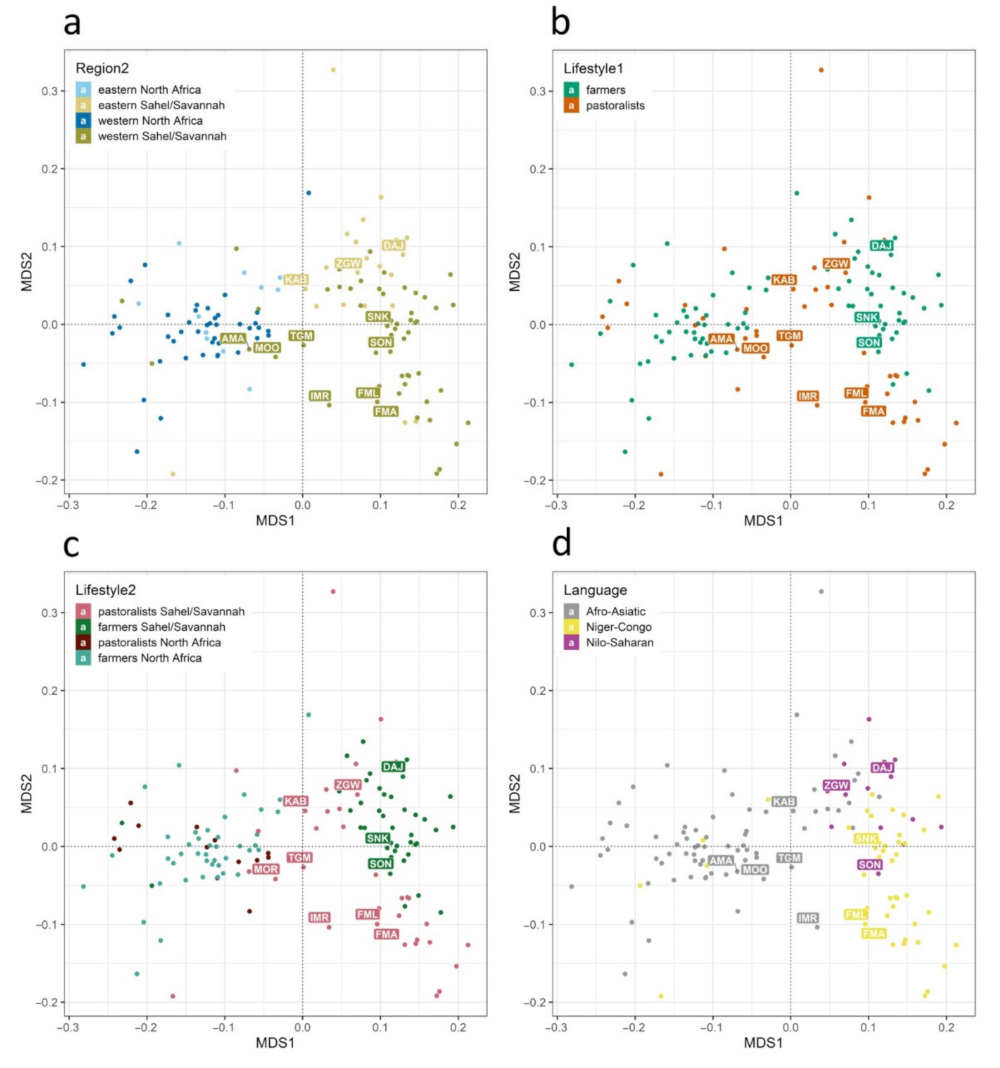
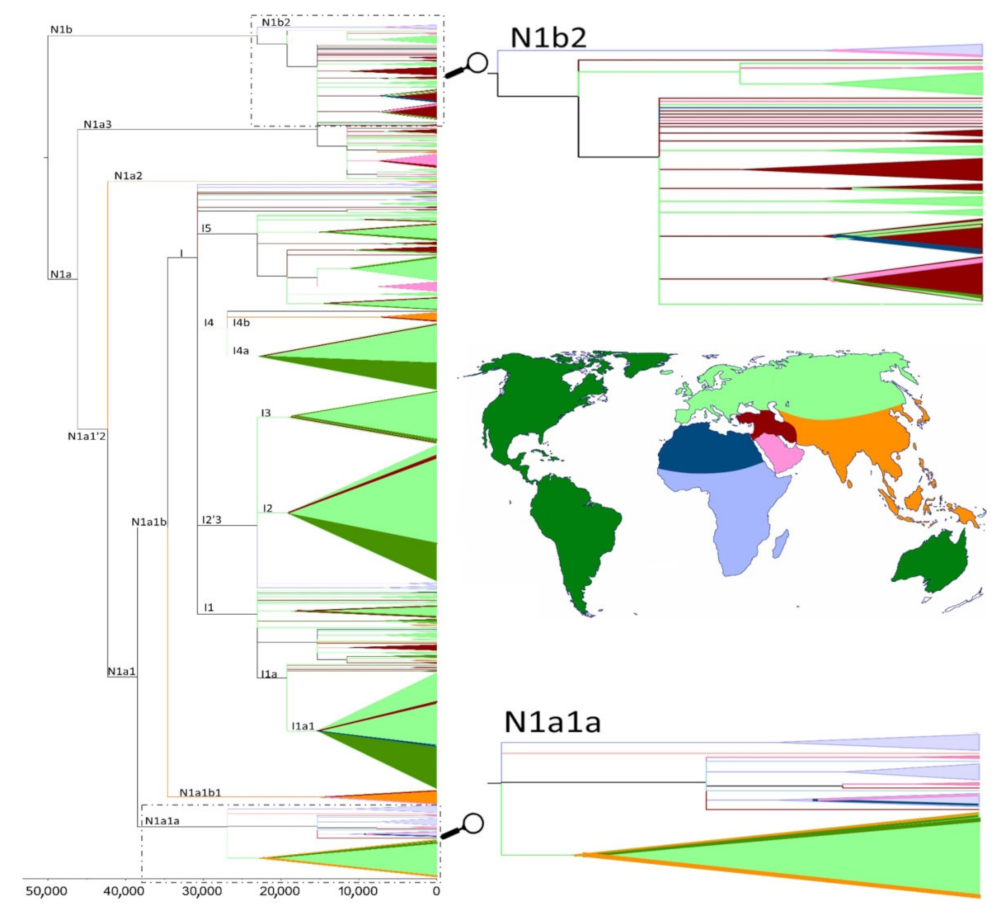
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bae, C.J.; Douka, K.; Petraglia, M.D. On the Origin of Modern Humans: Asian Perspectives. Science 2017, 358, eaai9067. [Google Scholar] [CrossRef] [PubMed]
- Harvati, K.; Röding, C.; Bosman, A.M.; Karakostis, F.A.; Grün, R.; Stringer, C.; Karkanas, P.; Thompson, N.C.; Koutoulidis, V.; Moulopoulos, L.A.; et al. Apidima Cave Fossils Provide Earliest Evidence of Homo Sapiens in Eurasia. Nature 2019, 571, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Malaspinas, A.-S.; Westaway, M.C.; Muller, C.; Sousa, V.C.; Lao, O.; Alves, I.; Bergström, A.; Athanasiadis, G.; Cheng, J.Y.; Crawford, J.E.; et al. A Genomic History of Aboriginal Australia. Nature 2016, 538, 207–214. [Google Scholar] [CrossRef]
- Fernandes, V.; Alshamali, F.; Alves, M.; Costa, M.D.; Pereira, J.B.; Silva, N.M.; Cherni, L.; Harich, N.; Cerny, V.; Soares, P.; et al. The Arabian Cradle: Mitochondrial Relicts of the First Steps along the Southern Route out of Africa. Am. J. Hum. Genet. 2012, 90, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Behar, D.M.; Metspalu, E.; Kivisild, T.; Rosset, S.; Tzur, S.; Hadid, Y.; Yudkovsky, G.; Rosengarten, D.; Pereira, L.; Amorim, A.; et al. Counting the Founders: The Matrilineal Genetic Ancestry of the Jewish Diaspora. PLoS ONE 2008, 3, e2062. [Google Scholar] [CrossRef] [PubMed]
- Kivisild, T. Maternal Ancestry and Population History from Whole Mitochondrial Genomes. Investig. Genet. 2015, 6, 3. [Google Scholar] [CrossRef]
- Černý, V.; Pereira, L.; Musilová, E.; Kujanová, M.; Vašíková, A.; Blasi, P.; Garofalo, L.; Soares, P.; Diallo, I.; Brdička, R.; et al. Genetic Structure of Pastoral and Farmer Populations in the African Sahel. Mol. Biol. Evol. 2011, 28, 2491–2500. [Google Scholar] [CrossRef]
- Čížková, M.; Munclinger, P.; Diallo, M.Y.; Kulichová, I.; Mokhtar, M.G.; Dème, A.; Pereira, L.; Černý, V. Genetic Structure of the Western and Eastern African Sahel/Savannah Belt and the Role of Nomadic Pastoralists as Inferred from the Variation of D-Loop Mitochondrial DNA Sequences. Hum. Biol. 2017, 89, 281–302. [Google Scholar] [CrossRef] [PubMed]
- Nováčková, J.; Čížková, M.; Mokhtar, M.G.; Duda, P.; Stenzl, V.; Tříska, P.; Hofmanová, Z.; Černý, V. Subsistence Strategy Was the Main Factor Driving Population Differentiation in the Bidirectional Corridor of the African Sahel. Am. J. Phys. Anthr. 2020, 171, 496–508. [Google Scholar] [CrossRef]
- Černý, V.; Salas, A.; Hájek, M.; Zaloudková, M.; Brdicka, R. A Bidirectional Corridor in the Sahel-Sudan Belt and the Distinctive Features of the Chad Basin Populations: A History Revealed by the Mitochondrial DNA Genome. Ann. Hum. Genet. 2007, 71, 433–452. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Černý, V.; Cerezo, M.; Silva, N.M.; Hájek, M.; Vasíková, A.; Kujanová, M.; Brdicka, R.; Salas, A. Linking the Sub-Saharan and West Eurasian Gene Pools: Maternal and Paternal Heritage of the Tuareg Nomads from the African Sahel. Eur. J. Hum. Genet. 2010, 18, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Podgorná, E.; Soares, P.; Pereira, L.; Černý, V. The Genetic Impact of the Lake Chad Basin Population in North Africa as Documented by Mitochondrial Diversity and Internal Variation of the L3e5 Haplogroup. Ann. Hum. Genet. 2013, 77, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Blench, R. The Westward Wanderings of Cushitic Pastoralists: Explorations in the Prehistory of Cental Africa. In L’Homme et l’animal dans le bassin du lac Tchad; Editions IRD: Paris, France, 1999; pp. 39–80. [Google Scholar]
- Černý, V.; Fernandes, V.; Costa, M.D.; Hájek, M.; Mulligan, C.J.; Pereira, L. Migration of Chadic Speaking Pastoralists within Africa Based on Population Structure of Chad Basin and Phylogeography of Mitochondrial L3f Haplogroup. BMC Evol. Biol. 2009, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Cerezo, M.; Černý, V.; Carracedo, Á.; Salas, A. New Insights into the Lake Chad Basin Population Structure Revealed by High-Throughput Genotyping of Mitochondrial DNA Coding SNPs. PLoS ONE 2011, 6, e18682. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Rengo, C.; Battaglia, V.; Pala, M.; Olivieri, A.; Fornarino, S.; Magri, C.; Scozzari, R.; Babudri, N.; Santachiara-Benerecetti, A.S.; et al. Saami and Berbers--an Unexpected Mitochondrial DNA Link. Am. J. Hum. Genet. 2005, 76, 883–886. [Google Scholar] [CrossRef] [PubMed]
- Kulichová, I.; Fernandes, V.; Deme, A.; Nováčková, J.; Stenzl, V.; Novelletto, A.; Pereira, L.; Černý, V. Internal Diversification of Non-Sub-Saharan Haplogroups in Sahelian Populations and the Spread of Pastoralism beyond the Sahara. Am. J. Phys. Anthropol. 2017, 164, 424–434. [Google Scholar] [CrossRef]
- Gifford-Gonzalez, D.; Hanotte, O. Domesticating Animals in Africa: Implications of Genetic and Archaeological Findings. J. World Prehistory 2011, 24, 1–23. [Google Scholar] [CrossRef]
- Ozainne, S. Un Néolithique Ouest-Africain: Cadre Chrono-Culturel, Économique et Environnemental de l’Holocène Récent En Pays Dogon (Mali); Africa Magna Verlag: Frankfurt am Main, Germany, 2013; Volume 8, ISBN 3-937248-33-1. [Google Scholar]
- Ozainne, S.; Lespez, L.; Garnier, A.; Ballouche, A.; Neumann, K.; Pays, O.; Huysecom, E. A Question of Timing: Spatio-Temporal Structure and Mechanisms of Early Agriculture Expansion in West Africa. J. Archaeol. Sci. 2014, 50, 359–368. [Google Scholar] [CrossRef]
- Sikora, M.; Carpenter, M.L.; Moreno-Estrada, A.; Henn, B.M.; Underhill, P.A.; Sánchez-Quinto, F.; Zara, I.; Pitzalis, M.; Sidore, C.; Busonero, F.; et al. Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe. PLoS Genet. 2014, 10, e1004353. [Google Scholar] [CrossRef]
- Chiang, C.W.K.; Marcus, J.H.; Sidore, C.; Biddanda, A.; Al-Asadi, H.; Zoledziewska, M.; Pitzalis, M.; Busonero, F.; Maschio, A.; Pistis, G.; et al. Genomic History of the Sardinian Population. Nat. Genet. 2018, 50, 1426–1434. [Google Scholar] [CrossRef] [PubMed]
- Vicente, M.; Priehodová, E.; Diallo, I.; Podgorná, E.; Poloni, E.S.; Černý, V.; Schlebusch, C.M. Population History and Genetic Adaptation of the Fulani Nomads: Inferences from Genome-Wide Data and the Lactase Persistence Trait. BMC Genom. 2019, 20, 915. [Google Scholar] [CrossRef] [PubMed]
- Priehodová, E.; Austerlitz, F.; Čížková, M.; Nováčková, J.; Ricaut, F.-X.; Hofmanová, Z.; Schlebusch, C.M.; Černý, V. Sahelian Pastoralism from the Perspective of Variants Associated with Lactase Persistence. Am. J. Phys. Anthr. 2020, 173, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Černý, V.; Fortes-Lima, C.; Tříska, P. Demographic History and Admixture Dynamics in African Sahelian Populations. Hum. Mol. Genet. 2021, 30, R29–R36. [Google Scholar] [CrossRef] [PubMed]
- Triska, P.; Soares, P.; Patin, E.; Fernandes, V.; Cerny, V.; Pereira, L. Extensive Admixture and Selective Pressure Across the Sahel Belt. Genome Biol. Evol. 2015, 7, 3484–3495. [Google Scholar] [CrossRef] [PubMed]
- Priehodová, E.; Austerlitz, F.; Čížková, M.; Mokhtar, M.G.; Poloni, E.S.; Černý, V. The Historical Spread of Arabian Pastoralists to the Eastern African Sahel Evidenced by the Lactase Persistence −13,915*G Allele and Mitochondrial DNA. Am. J. Hum. Biol. 2017, 29, e22950. [Google Scholar] [CrossRef] [PubMed]
- Levy, T.E.; Holl, A.F.C. Migrations, Ethnogenesis, and Settlement Dynamics: Israelites in Iron Age Canaan and Shuwa-Arabs in the Chad Basin. J. Anthropol. Archaeol. 2002, 21, 83–118. [Google Scholar] [CrossRef]
- Pereira, L.; Mutesa, L.; Tindana, P.; Ramsay, M. African Genetic Diversity and Adaptation Inform a Precision Medicine Agenda. Nat. Rev. Genet. 2021, 22, 284–306. [Google Scholar] [CrossRef]
- Mulindwa, J.; Noyes, H.; Ilboudo, H.; Pagani, L.; Nyangiri, O.; Kimuda, M.P.; Ahouty, B.; Asina, O.F.; Ofon, E.; Kamoto, K.; et al. High Levels of Genetic Diversity within Nilo-Saharan Populations: Implications for Human Adaptation. Am. J. Hum. Genet. 2020, 107, 473–486. [Google Scholar] [CrossRef]
- Choudhury, A.; Aron, S.; Botigué, L.R.; Sengupta, D.; Botha, G.; Bensellak, T.; Wells, G.; Kumuthini, J.; Shriner, D.; Fakim, Y.J.; et al. High-Depth African Genomes Inform Human Migration and Health. Nature 2020, 586, 741–748. [Google Scholar] [CrossRef]
- Vai, S.; Sarno, S.; Lari, M.; Luiselli, D.; Manzi, G.; Gallinaro, M.; Mataich, S.; Hübner, A.; Modi, A.; Pilli, E.; et al. Ancestral Mitochondrial N Lineage from the Neolithic ‘Green’ Sahara. Sci. Rep. 2019, 9, 3530. [Google Scholar] [CrossRef] [PubMed]
- Van Oven, M.; Kayser, M. Updated Comprehensive Phylogenetic Tree of Global Human Mitochondrial DNA Variation. Hum. Mutat. 2009, 30, E386–E394. [Google Scholar] [CrossRef] [PubMed]
- Gonder, M.K.; Mortensen, H.M.; Reed, F.A.; de Sousa, A.; Tishkoff, S.A. Whole-MtDNA Genome Sequence Analysis of Ancient African Lineages. Mol. Biol. Evol. 2007, 24, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Bendall, K.E.; Sykes, B.C. Length Heteroplasmy in the First Hypervariable Segment of the Human MtDNA Control Region. Am. J. Hum. Genet. 1995, 57, 248. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical Tests of Neutrality of Mutations against Population Growth, Hitchhiking and Background Selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Ramos-Onsins, S.E.; Rozas, J. Statistical Properties of New Neutrality Tests against Population Growth. Mol. Biol. Evol. 2002, 19, 2092–2100. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population Growth Makes Waves in the Distribution of Pairwise Genetic Differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [CrossRef]
- Reynolds, J.; Weir, B.S.; Cockerham, C.C. Estimation of the Coancestry Coefficient: Basis for a Short-Term Genetic Distance. Genetics 1983, 105, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. A Correction to the Exact Test Based on the Ewens Sampling Distribution. Genet. Res. 1996, 68, 259–260. [Google Scholar] [CrossRef]
- Poloni, E.S.; Naciri, Y.; Bucho, R.; Niba, R.; Kervaire, B.; Excoffier, L.; Langaney, A.; Sanchez-Mazas, A. Genetic Evidence for Complexity in Ethnic Differentiation and History in East Africa. Ann. Hum. Genet. 2009, 73, 582–600. [Google Scholar] [CrossRef]
- Jari Oksanen, F.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Peter, R.; Minchin, R.B.; O’Hara, G.; Simpson, L.; et al. Vegan: Community Ecology Package. R Package Version; R Package: Vienna, Austria, 2020; pp. 2–5. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2016. [Google Scholar]
- Beerli, P. How to Use MIGRATE or Why Are Markov Chain Monte Carlo Programs Difficult to Use? In Population Genetics for Animal Conservation; Conservation Biology; Rizzoli, A., Vernesi, C., Bertorelle, G., Hauffe, H.C., Bruford, M.W., Eds.; Cambridge University Press: Cambridge, UK, 2009; pp. 42–79. ISBN 978-0-521-86630-9. [Google Scholar]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and Enhances Circular Visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Henn, B.M.; Botigué, L.R.; Gravel, S.; Wang, W.; Brisbin, A.; Byrnes, J.K.; Fadhlaoui-Zid, K.; Zalloua, P.A.; Moreno-Estrada, A.; Bertranpetit, J.; et al. Genomic Ancestry of North Africans Supports Back-to-Africa Migrations. PLoS Genet. 2012, 8, e1002397. [Google Scholar] [CrossRef] [PubMed]
- Andrews, R.M.; Kubacka, I.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Reanalysis and Revision of the Cambridge Reference Sequence for Human Mitochondrial DNA. Nat. Genet. 1999, 23, 147. [Google Scholar] [CrossRef] [PubMed]
- Forster, P.; Harding, R.; Torroni, A.; Bandelt, H.J. Origin and Evolution of Native American MtDNA Variation: A Reappraisal. Am. J. Hum. Genet. 1996, 59, 935–945. [Google Scholar]
- Soares, P.; Ermini, L.; Thomson, N.; Mormina, M.; Rito, T.; Röhl, A.; Salas, A.; Oppenheimer, S.; Macaulay, V.; Richards, M.B. Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock. Am. J. Hum. Genet. 2009, 84, 740–759. [Google Scholar] [CrossRef]
- Saillard, J.; Forster, P.; Lynnerup, N.; Bandelt, H.-J.; Nørby, S. MtDNA Variation among Greenland Eskimos: The Edge of the Beringian Expansion. Am. J. Hum. Genet. 2000, 67, 718–726. [Google Scholar] [CrossRef]
- Yang, Z. PAML: A Program Package for Phylogenetic Analysis by Maximum Likelihood. Comput. Appl. Biosci. 1997, 13, 555–556. [Google Scholar] [CrossRef]
- Černý, V.; Hájek, M.; Bromová, M.; Čmejla, R.; Diallo, I.; Brdička, R. MtDNA of Fulani nomads and their genetic relationships to neighboring sedentary populations. Hum. Biol. 2006, 78, 9–27. [Google Scholar] [CrossRef]
- van de Loosdrecht, M.; Bouzouggar, A.; Humphrey, L.; Posth, C.; Barton, N.; Aximu-Petri, A.; Nickel, B.; Nagel, S.; Talbi, E.H.; El Hajraoui, M.A.; et al. Pleistocene North African Genomes Link Near Eastern and Sub-Saharan African Human Populations. Science 2018, 360, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.C.; Alshamali, F.; Montinaro, F.; Cavadas, B.; Torroni, A.; Pereira, L.; Raveane, A.; Fernandes, V. Projecting Ancient Ancestry in Modern-Day Arabians and Iranians: A Key Role of the Past Exposed Arabo-Persian Gulf on Human Migrations. Genome Biol. Evol. 2021, 13, evab194. [Google Scholar] [CrossRef]
- Arredi, B.; Poloni, E.S.; Paracchini, S.; Zerjal, T.; Fathallah, D.M.; Makrelouf, M.; Pascali, V.L.; Novelletto, A.; Tyler-Smith, C. A Predominantly Neolithic Origin for Y-Chromosomal DNA Variation in North Africa. Am. J. Hum. Genet. 2004, 75, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Kujanová, M.; Pereira, L.; Fernandes, V.; Pereira, J.B.; Cerný, V. Near Eastern Neolithic Genetic Input in a Small Oasis of the Egyptian Western Desert. Am. J. Phys. Anthr. 2009, 140, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Černý, V.; Čížková, M.; Poloni, E.S.; Al-Meeri, A.; Mulligan, C.J. Comprehensive View of the Population History of Arabia as Inferred by MtDNA Variation. Am. J. Phys. Anthr. 2016, 159, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Currat, M.; Excoffier, L.; Penny, D. Modern Humans Did Not Admix with Neanderthals during Their Range Expansion into Europe. PLoS Biol. 2004, 2, e421. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, J.; Lopes, A.M.; Comas, D.; Amorim, A.; Arenas, M. Evaluating the Neolithic Expansion at Both Shores of the Mediterranean Sea. Mol. Biol. Evol. 2017, 34, 3232–3242. [Google Scholar] [CrossRef] [PubMed]
- Fregel, R.; Méndez, F.L.; Bokbot, Y.; Martín-Socas, D.; Camalich-Massieu, M.D.; Santana, J.; Morales, J.; Ávila-Arcos, M.C.; Underhill, P.A.; Shapiro, B.; et al. Ancient Genomes from North Africa Evidence Prehistoric Migrations to the Maghreb from Both the Levant and Europe. Proc. Natl. Acad. Sci. USA 2018, 115, 6774–6779. [Google Scholar] [CrossRef]
- Hernández, C.L.; Pita, G.; Cavadas, B.; López, S.; Sánchez-Martínez, L.J.; Dugoujon, J.-M.; Novelletto, A.; Cuesta, P.; Pereira, L.; Calderón, R. Human Genomic Diversity Where the Mediterranean Joins the Atlantic. Mol. Biol. Evol. 2020, 37, 1041–1055. [Google Scholar] [CrossRef]
- Bekada, A. Caractéristaion Anthropogénétique d’un Échantillon de la Population Algérienne: Analyse des Marqueurs Parentaux; Université d’Oran 1: Es Senia, Algeria, 2015. [Google Scholar]
- Adams, W.Y. Nubia: Corridor to Africa; Princeton University Press: Princeton, NJ, USA, 1977; ISBN 978-0-691-09370-3. [Google Scholar]
- Fox, C.L. MtDNA Analysis in Ancient Nubians Supports the Existence of Gene Flow between Sub-Sahara and North Africa in the Nile Valley. Ann. Hum. Biol. 1997, 24, 217–227. [Google Scholar] [CrossRef]
- Krings, M.; Salem, A.E.; Bauer, K.; Geisert, H.; Malek, A.K.; Chaix, L.; Simon, C.; Welsby, D.; Di Rienzo, A.; Utermann, G.; et al. MtDNA Analysis of Nile River Valley Populations: A Genetic Corridor or a Barrier to Migration? Am. J. Hum. Genet. 1999, 64, 1166–1176. [Google Scholar] [CrossRef]
- Segal, R. Islam’s Black Slaves: The History of Africa’s Other Black Diaspora; Atlantic Books: London, UK, 2002; ISBN 978-1-903809-81-5. [Google Scholar]
- Turner, M.D.; Schlecht, E. Livestock Mobility in Sub-Saharan Africa: A Critical Review. Pastoralism 2019, 9, 13. [Google Scholar] [CrossRef]
- Jesse, F.; Keding, B.; Lenssen-Erz, T.; Pöllath, N. I Hope Your Cattle Are Well’: Archaeological Evidence for Early Cattle-Centred Behaviour in the Eastern Sahara of Sudan and Chad. In Pastoralism in Africa: Past, Present and Future; Berghahn Books: New York, NY, USA, 2013; pp. 66–103. [Google Scholar]
- Linseele, V. From First Stock Keepers to Specialised Pastoralists in the West African Savannah. In Pastoralism in Africa: Past, Present and Future; Bollig, M., Schnegg, M., Wotzka, H.-P., Eds.; Berghan Books: New York, NY, USA, 2013; pp. 145–170. [Google Scholar]
- Ozainne, S.; Mayor, A.; Huysecom, E. Chronology of Human Occupation during the Holocene in West Africa. The Dogon country radiocarbon record. In Winds of change; Rudolf Habelt Verlag: Bonn, Germany, 2017; pp. 211–227. [Google Scholar]
- Wright, D.K. Humans as Agents in the Termination of the African Humid Period. Front. Earth Sci. 2017, 5, 4. [Google Scholar] [CrossRef]
- Manning, K.; Pelling, R.; Higham, T.; Schwenniger, J.-L.; Fuller, D.Q. 4500-Year Old Domesticated Pearl Millet (Pennisetum Glaucum) from the Tilemsi Valley, Mali: New Insights into an Alternative Cereal Domestication Pathway. J. Archaeol. Sci. 2011, 38, 312–322. [Google Scholar] [CrossRef]
- Burgarella, C.; Cubry, P.; Kane, N.A.; Varshney, R.K.; Mariac, C.; Liu, X.; Shi, C.; Thudi, M.; Couderc, M.; Xu, X.; et al. A Western Sahara Centre of Domestication Inferred from Pearl Millet Genomes. Nat. Ecol. Evol. 2018, 2, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Cubry, P.; Tranchant-Dubreuil, C.; Thuillet, A.-C.; Monat, C.; Ndjiondjop, M.-N.; Labadie, K.; Cruaud, C.; Engelen, S.; Scarcelli, N.; Rhoné, B.; et al. The Rise and Fall of African Rice Cultivation Revealed by Analysis of 246 New Genomes. Curr. Biol. 2018, 28, 2274–2282.e6. [Google Scholar] [CrossRef]
- Scarcelli, N.; Cubry, P.; Akakpo, R.; Thuillet, A.-C.; Obidiegwu, J.; Baco, M.N.; Otoo, E.; Sonké, B.; Dansi, A.; Djedatin, G.; et al. Yam Genomics Supports West Africa as a Major Cradle of Crop Domestication. Sci. Adv. 2019, 5, eaaw1947. [Google Scholar] [CrossRef]
- Winchell, F.; Stevens, C.J.; Murphy, C.; Champion, L.; Fuller, D.Q. Evidence for Sorghum Domestication in Fourth Millennium BC Eastern Sudan: Spikelet Morphology from Ceramic Impressions of the Butana Group. Curr. Anthropol. 2017, 58, 673–683. [Google Scholar] [CrossRef]
- Cervigni, R.; Morris, M. Confronting Drought in Africa’s Drylands: Opportunities for Enhancing Resilience; World Bank Publications: Herndon, VA, USA, 2016; ISBN 978-1-4648-0818-0. [Google Scholar]
- Kleisner, K.; Pokorný, Š.; Čížková, M.; Froment, A.; Černý, V. Nomadic Pastoralists and Sedentary Farmers of the Sahel/Savannah Belt of Africa in the Light of Geometric Morphometrics Based on Facial Portraits. Am. J. Phys. Anthropol. 2019, 169, 632–645. [Google Scholar] [CrossRef]
- Naino Jika, A.K.; Dussert, Y.; Raimond, C.; Garine, E.; Luxereau, A.; Takvorian, N.; Djermakoye, R.S.; Adam, T.; Robert, T. Unexpected Pattern of Pearl Millet Genetic Diversity among Ethno-Linguistic Groups in the Lake Chad Basin. Heredity 2017, 118, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Musilová, E.; Fernandes, V.; Silva, N.M.; Soares, P.; Alshamali, F.; Harich, N.; Cherni, L.; Gaaied, A.B.; Al-Meeri, A.; Pereira, L.; et al. Population history of the Red Sea—Genetic exchanges between the Arabian Peninsula and East Africa signaled in the mitochondrial DNA HV1 haplogroup. Am. J. Phys. Anthropol. 2011, 145, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, F.; Trombetta, B.; Sellitto, D.; Massaia, A.; Destro-Bisol, G.; Watson, E.; Beraud Colomb, E.; Dugoujon, J.-M.; Moral, P.; Scozzari, R. Human Y Chromosome Haplogroup R-V88: A Paternal Genetic Record of Early Mid Holocene Trans-Saharan Connections and the Spread of Chadic Languages. Eur. J. Hum. Genet. 2010, 18, 800–807. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shriner, D.; Rotimi, C.N. Genetic History of Chad. Am. J. Phys. Anthropol. 2018, 167, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, J.K.; Patterson, N.; Loh, P.-R.; Lipson, M.; Berger, B.; Stoneking, M.; Pakendorf, B.; Reich, D. Ancient West Eurasian Ancestry in Southern and Eastern Africa. Proc. Natl. Acad. Sci. USA 2014, 111, 2632–2637. [Google Scholar] [CrossRef] [PubMed]


| Kruskal–Wallis Test (p-Values) | ||||||
|---|---|---|---|---|---|---|
| Haplotype Diversity | Nucleotide Diversity | Mean Number of Pairwise Differences | Fu’s Fs (p) | Hri (p) | SSD (p) | |
| NA vs. SS | 0.380 | 0.000 | 0.000 | 0.563 | 0.068 | 0.208 |
| FM vs. PS | 0.004 | 0.856 | 0.630 | 0.133 | 0.271 | 0.249 |
| AA vs. NC vs. NS | 0.293 | 0.011 | 0.001 | 0.682 | 0.033 | 0.147 |
| eNA vs. eSS vs. wNA vs. wSS | 0.453 | 0.000 | 0.000 | 0.940 | 0.266 | 0.231 |
| NAFM vs. NAPS vs. SSFM vs. SSPS | 0.000 | 0.000 | 0.000 | 0.507 | 0.482 | 0.874 |
| Maximum Parsimony | Maximum Likelihood | |||||
|---|---|---|---|---|---|---|
| Total Clock | Synonymous Clock | All SNPs | ||||
| Haplogroup | Age | 95% CI | Age | 95% CI | Age | 95% CI |
| N1a1a | 24,005 | 16,482–31,778 | 20,727 | 10,873–30,580 | 21,754 | 21,304–22,182 |
| N1a1a3 | 13,900 | 10,071–17,808 | 14,266 | 7442–21,090 | 17,948 | 17,676–18,211 |
| N1a3a+195C! | 3188 | 1266–5135 | 1819 | −838–4477 | 3610 | 3577–3643 |
| N1b2+9325C | 6816 | 553–13,327 | 14,191 | −3 562–31,944 | 6908 | 6762–7048 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diallo, M.Y.; Čížková, M.; Kulichová, I.; Podgorná, E.; Priehodová, E.; Nováčková, J.; Fernandes, V.; Pereira, L.; Černý, V. Circum-Saharan Prehistory through the Lens of mtDNA Diversity. Genes 2022, 13, 533. https://doi.org/10.3390/genes13030533
Diallo MY, Čížková M, Kulichová I, Podgorná E, Priehodová E, Nováčková J, Fernandes V, Pereira L, Černý V. Circum-Saharan Prehistory through the Lens of mtDNA Diversity. Genes. 2022; 13(3):533. https://doi.org/10.3390/genes13030533
Chicago/Turabian StyleDiallo, Mame Yoro, Martina Čížková, Iva Kulichová, Eliška Podgorná, Edita Priehodová, Jana Nováčková, Veronica Fernandes, Luísa Pereira, and Viktor Černý. 2022. "Circum-Saharan Prehistory through the Lens of mtDNA Diversity" Genes 13, no. 3: 533. https://doi.org/10.3390/genes13030533
APA StyleDiallo, M. Y., Čížková, M., Kulichová, I., Podgorná, E., Priehodová, E., Nováčková, J., Fernandes, V., Pereira, L., & Černý, V. (2022). Circum-Saharan Prehistory through the Lens of mtDNA Diversity. Genes, 13(3), 533. https://doi.org/10.3390/genes13030533






