Are Roma People Descended from the Punjab Region of Pakistan: A Y-Chromosomal Perspective
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Laboratory Procedures
2.3. Statistical Analysis
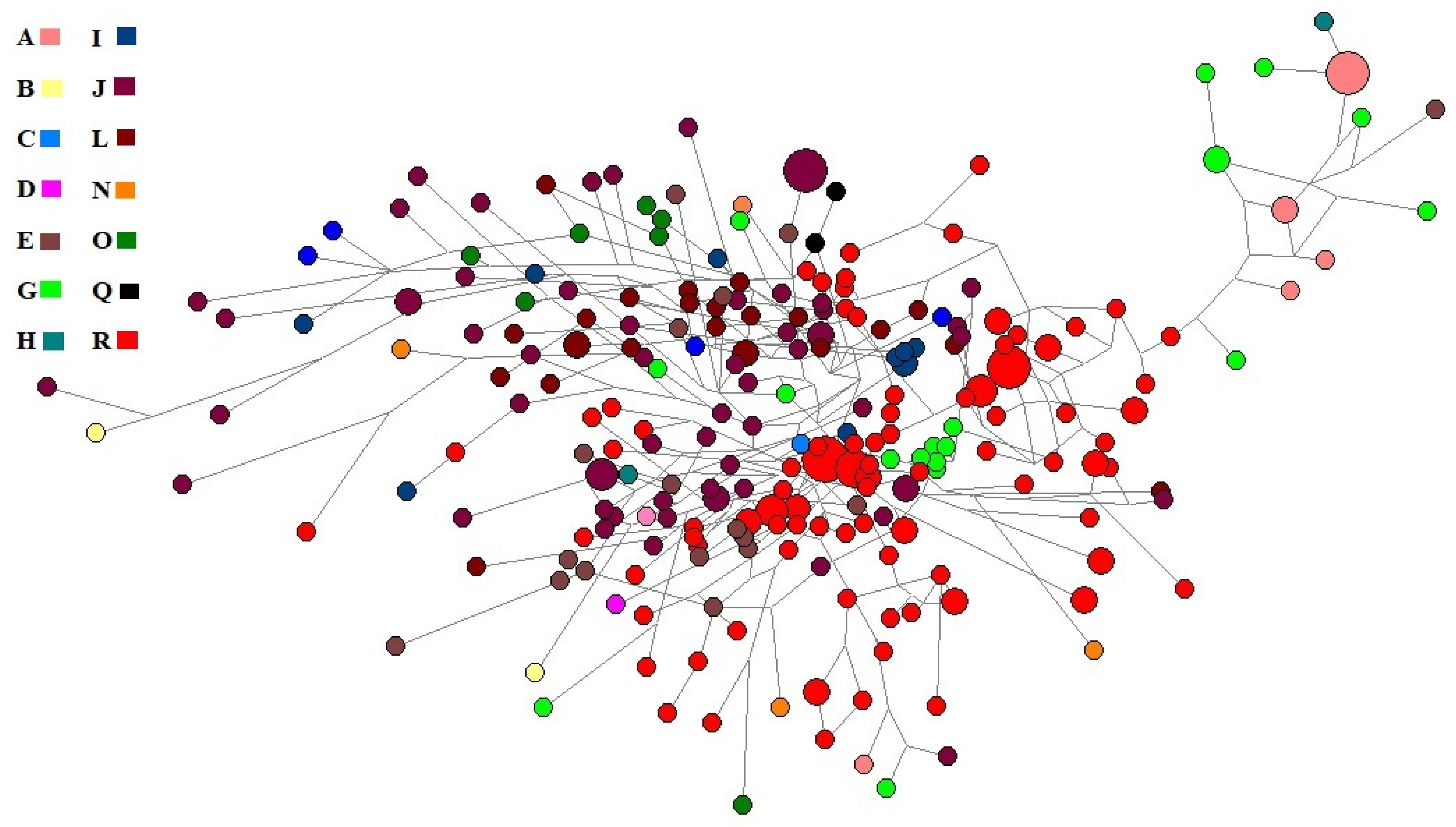
2.3.1. The Median-Joining Network
2.3.2. Assessment of Gene Flow and Migration in Roma Population
3. Results and Discussion
3.1. Genetic Diversity
3.2. Population Pairwise Rst and Fst Distance Comparisons
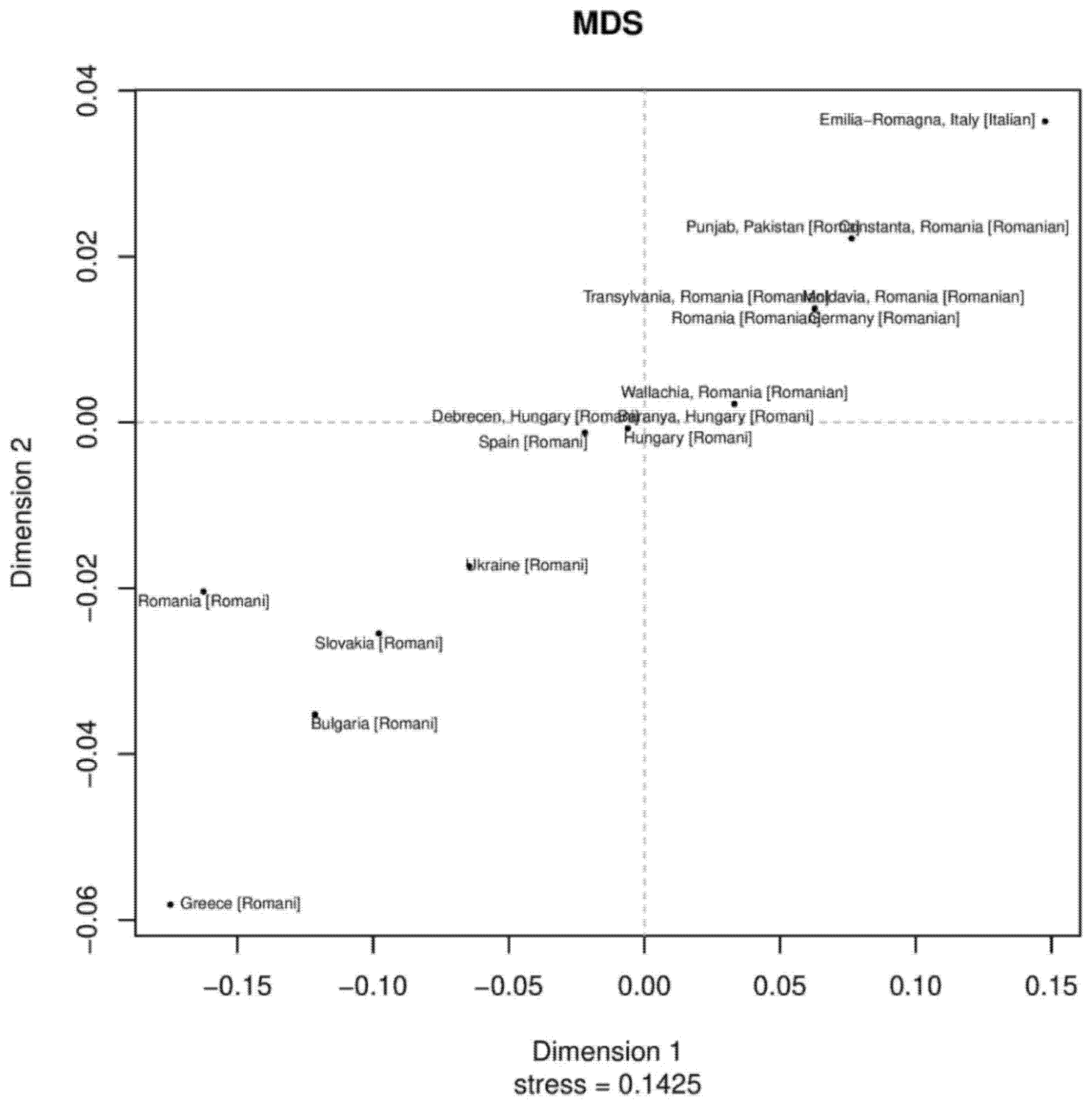
3.3. MDS and Neighbor-Joining Tree Analyses
3.4. AMOVA Analyses
3.5. Y-Chromosome Haplogroups
3.6. Estimation of Gene Flow and Migration Rate in Pakistani Roma Population
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Compliance with Ethical Standards
References
- Fraser, A.M. The Gypsies, 2nd ed.; The Peoples of Europe; Blackwell: Oxford, UK; Cambridge, MA, USA, 1995; ISBN 978-0-631-19605-1. [Google Scholar]
- Hancock, I.F. A Handbook of Vlax Romani; Slavica: Columbus, OH, USA, 1995; ISBN 978-0-89357-258-7. [Google Scholar]
- Grierson, G.A. Linguistic Survey of India; Office of the Superintendent of Government Printing: Calcutta, India, 1903. [Google Scholar]
- Radu, P.I.; Schurr, T.G. Reconstructing the Origins and Migrations of Diasporic Populations: The Case of the European Gypsies. Wiley 2004, 106, 267–281. [Google Scholar]
- Trail, G. Statistical Sketch of Kamaon. 1828. Available online: https://archive.org/stream/in.ernet.dli.2015.53201/2015.53201.Statistical-Sketch-Of-Kumaon_djvu.txt (accessed on 27 January 2022).
- Van Driem, G. Languages of the Himalayas: An Ethnolinguistic Handbook of the Greater Himalayan Region: Containing an Introduction to the Symbiotic Theory of Language; Handbook of Oriental Studies. Section Two, India, Handbuch der Orientalistik. Indien; Brill: Leiden, The Netherlands; New York, NY, USA, 2001; ISBN 978-90-04-10390-0. [Google Scholar]
- Ramkumar, N. The Caste Culture in India: Contemporary Approach; Global Vision Pub. House: New Delhi, India, 2011; ISBN 978-81-8220-389-1. [Google Scholar]
- von dem Knesebeck, J. (Ed.) The Roma Struggle for Compensation in Post-War Germany; University of Hertfordshire Press: Hatfield, UK, 2011; ISBN 978-1-907396-11-3. [Google Scholar]
- Turner, R.L. The Position of Romani in Indo-Aryan; Monographs (Gypsy Lore Society), No. 4; B. Quaritch: London, UK, 1927. [Google Scholar]
- Berger, H. Die Burušaski-Lehnwörter in der Zigeunersprache. Indo-Iran. J. 1959, 3, 17–43. [Google Scholar] [CrossRef] [Green Version]
- Marushiakova, E.; Popov, V. Gypsies (Roma) in Bulgaria; Studien zur Tsiganologie und Folkloristik; P. Lang: Frankfurt am Main, Germany; New York, NY, USA, 1997; ISBN 978-0-8204-3216-8. [Google Scholar]
- Liégeois, J.-P.; Acton, T.A. Roma, Gypsies, Travellers; Collection Education; Council of Europe: Strasbourg, France; Sales Agent, Manhattan Pub. Co.: Croton-on-Hudson, NY, USA, 1994; ISBN 978-92-871-2349-7. [Google Scholar]
- Mendizabal, I.; Valente, C.; Gusmão, A.; Alves, C.; Gomes, V.; Goios, A.; Parson, W.; Calafell, F.; Alvarez, L.; Amorim, A.; et al. Reconstructing the Indian Origin and Dispersal of the European Roma: A Maternal Genetic Perspective. PLoS ONE 2011, 6, e15988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Font-Porterias, N.; Arauna, L. European Roma groups show complex West Eurasian admixture footprints and a common South Asian genetic origin. PLoS Gen. 2019, 15, e1008417. [Google Scholar] [CrossRef]
- Pamjav, H.; Zalán, A.; Béres, J.; Nagy, M.; Chang, Y.M. Genetic Structure of the Paternal Lineage of the Roma People. Am. J. Phys. Anthropol. 2011, 145, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Regueiro, M.; Rivera, L.; Chennakrishnaiah, S.; Popovic, B.; Andjus, S.; Milasin, J.; Herrera, R.J. Ancestral Modal Y-STR Haplotype Shared among Romani and South Indian Populations. Gene 2012, 504, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cruz, B.; Mendizabal, I. Origins, admixture and founder lineages in European Roma. Eur. J. Hum. Genet. 2016, 24, 937–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendizabal, I.; Lao, O.; Marigorta, U.M.; Wollstein, A.; Gusmão, L.; Ferak, V.; Ioana, M.; Jordanova, A.; Kaneva, R.; Kouvatsi, A.; et al. Reconstructing the Population History of European Romani from Genome-Wide Data. Curr. Biol. 2012, 22, 2342–2349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Slatkin, M. A Measure of Population Subdivision Based on Microsatellite Allele Frequencies. Genetics 1995, 139, 457–462. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Adnan, A.; Rakha, A.; Nazir, S.; Alghafri, R.; Hassan, Q.; Wang, C.-C.; Lu, J. Forensic Features and Genetic Legacy of the Baloch Population of Pakistan and the Hazara Population across Durand Line Revealed by Y-Chromosomal STRs. Int. J. Legal Med. 2021, 135, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- MIGRATE. 2021. Available online: https://peterbeerli.com/migrate-html5/index.html (accessed on 27 January 2022).
- Beerli, P. Comparison of Bayesian and Maximum-Likelihood Inference of Population Genetic Parameters. Bioinformatics 2006, 22, 341–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beerli, P. Estimation of the Population Scaled Mutation Rate from Microsatellite Data. Genetics 2007, 177, 1967–1968. [Google Scholar] [CrossRef] [Green Version]
- Beerli, P.; Palczewski, M. Unified Framework to Evaluate Panmixia and Migration Direction Among Multiple Sampling Locations. Genetics 2010, 185, 313–326. [Google Scholar] [CrossRef] [Green Version]
- Lischer, H.E.L.; Excoffier, L. PGDSpider: An Automated Data Conversion Tool for Connecting Population Genetics and Genomics Programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef] [Green Version]
- Kopelman, N.M.; Stone, L.; Gascuel, O.; Rosenberg, N.A. The Behavior of Admixed Populations in Neighbor-Joining Inference of Population Trees. Pac. Symp. Biocomput. 2013, 2013, 273–284. [Google Scholar]
- Bianco, E.; Laval, G. Recent Common Origin, Reduced Population Size, and Marked Admixture Have Shaped European Roma Genomes. Mol. Biol. Evol. 2020, 37, 3175–3187. [Google Scholar] [CrossRef]
- Underhill, P.A.; Passarino, G.; Lin, A.A.; Shen, P.; Mirazon Lahr, M.; Foley, R.A.; Oefner, P.J.; Cavalli-Sforza, L.L. The Phylogeography of Y Chromosome Binary Haplotypes and the Origins of Modern Human Populations. Ann. Hum. Genet. 2001, 65, 43–62. [Google Scholar] [CrossRef]
- Chandrasekar, A.; Saheb, S.Y.; Gangopadyaya, P.; Gangopadyaya, S.; Mukherjee, A.; Basu, D.; Lakshmi, G.R.; Sahani, A.K.; Das, B.; Battacharya, S.; et al. YAP Insertion Signature in South Asia. Ann. Hum. Biol. 2007, 34, 582–586. [Google Scholar] [CrossRef] [PubMed]
- Haber, M.; Jones, A.L.; Connell, B.A.; Asan; Arciero, E.; Yang, H.; Thomas, M.G.; Xue, Y.; Tyler-Smith, C. A Rare Deep-Rooting D0 African Y-Chromosomal Haplogroup and Its Implications for the Expansion of Modern Humans Out of Africa. Genetics 2019, 212, 1421–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gresham, D.; Morar, B.; Underhill, P.A.; Passarino, G.; Lin, A.A.; Wise, C.; Angelicheva, D.; Calafell, F.; Oefner, P.J.; Shen, P.; et al. Origins and Divergence of the Roma (Gypsies). Am. J. Hum. Genet. 2001, 69, 1314–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peričić, M.; Klarić, I.M.; Lauc, L.B.; Janićijević, B.; Đorđević, D.; Efremovska, L.; Rudan, P. Population Genetics of 8 Y Chromosome STR Loci in Macedonians and Macedonian Romani (Gypsy). Forensic Sci. Int. 2005, 154, 257–261. [Google Scholar] [CrossRef]
- Peričić, M.; Lauc, L.B.; Klarić, I.M.; Rootsi, S.; Janićijević, B.; Rudan, I.; Terzić, R.; Čolak, I.; Kvesić, A.; Popović, D.; et al. High-Resolution Phylogenetic Analysis of Southeastern Europe Traces Major Episodes of Paternal Gene Flow Among Slavic Populations. Mol. Biol. Evol. 2005, 22, 1964–1975. [Google Scholar] [CrossRef]
- Poznik, G.D.; Xue, Y.; Mendez, F.L.; Willems, T.F.; Massaia, A.; Wilson Sayres, M.A.; Ayub, Q.; McCarthy, S.A.; Narechania, A.; Kashin, S.; et al. Punctuated Bursts in Human Male Demography Inferred from 1244 Worldwide Y-Chromosome Sequences. Nat. Genet. 2016, 48, 593–599. [Google Scholar] [CrossRef] [Green Version]
- Gusmão, A.; Gusmão, L.; Gomes, V.; Alves, C.; Calafell, F.; Amorim, A.; Prata, M.J. A Perspective on the History of the Iberian Gypsies Provided by Phylogeographic Analysis of Y-Chromosome Lineages. Ann. Hum. Genet. 2008, 72, 215–227. [Google Scholar] [CrossRef]
- Semino, O.; Magri, C.; Benuzzi, G.; Lin, A.A.; Al-Zahery, N.; Battaglia, V.; Maccioni, L.; Triantaphyllidis, C.; Shen, P.; Oefner, P.J.; et al. Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area. Am. J. Hum. Genet. 2004, 74, 1023–1034. [Google Scholar] [CrossRef] [Green Version]
- Mahal, D.G.; Matsoukas, I.G. The Geographic Origins of Ethnic Groups in the Indian Subcontinent: Exploring Ancient Footprints with Y-DNA Haplogroups. Front. Genet. 2018, 9, 4. [Google Scholar] [CrossRef] [Green Version]
- Wells, S. Deep Ancestry: Inside the Genographic Project; National Geographic: Washington, DC, USA, 2007; ISBN 978-1-4262-0118-9. [Google Scholar]
- Smolenyak, M.; Turner, A. Trace Your Roots with DNA: Using Genetic Tests to Explore Your Family Tree; Rodale: Kutztown, PA, USA, 2004; ISBN 978-1-59486-006-5. [Google Scholar]
| Groups | Fst |
|---|---|
| Pakistani Roma vs. Croatian Roma | 0.35647 |
| Pakistani Roma vs. Serbian Roma | 0.32266 |
| Pakistani Roma vs. Portuguese Roma | 0.29913 |
| Pakistani Roma vs. Turkish population | 0.09929 |
| Pakistani Roma vs. Bulgarian Roma | 0.09198 |
| Pakistani Roma vs. Slovenian Roma | 0.07805 |
| Pakistani Roma vs. Romanian Roma | 0.06276 |
| Pakistani Roma vs. Hungarian Roma | 0.0462 |
| Migration Route | Model | Log(mL) | LBF | Model Probability |
|---|---|---|---|---|
| India + Pakistan | 4 | −33,811.1 | −31,982.5 | 0 |
| India→Pakistan | 1 | −28,444.7 | −26,616 | 0 |
| Pakistan→India | 1 | −26,025.5 | −24,196.9 | 0 |
| India→Pakistan | 3 | −2016.59 | −187.92 | 0 |
| Pakistan→India | 3 | −1863.45 | −34.78 | 0 |
| India→Pakistan | 2 | −1836.47 | −7.8 | 0.0004 |
| Pakistan→India | 2 | −1828.67 | 0 | 0.9996 |
| Route | Model | Log(mL) | LBF | Model Probability |
|---|---|---|---|---|
| Pakistan→Turkey→eastern Europe | 1 | −2900.02 | −370.62 | 0 |
| Eastern Europe→Turkey→Pakistan | 1 | −2851.56 | −322.16 | 0 |
| Pakistan→Turkey→eastern Europe | 3 | −2823.05 | −293.65 | 0 |
| Eastern Europe→Turkey→Pakistan | 3 | −2758.07 | −228.67 | 0 |
| Pakistan→Turkey→eastern Europe | 2 | −2533.65 | −4.25 | 0.0141 |
| Eastern Europe→Turkey→Pakistan | 2 | −2529.4 | 0 | 0.9859 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adnan, A.; Rakha, A.; Lazim, H.; Nazir, S.; Al-Qahtani, W.S.; Abdullah Alwaili, M.; Hadi, S.; Wang, C.-C. Are Roma People Descended from the Punjab Region of Pakistan: A Y-Chromosomal Perspective. Genes 2022, 13, 532. https://doi.org/10.3390/genes13030532
Adnan A, Rakha A, Lazim H, Nazir S, Al-Qahtani WS, Abdullah Alwaili M, Hadi S, Wang C-C. Are Roma People Descended from the Punjab Region of Pakistan: A Y-Chromosomal Perspective. Genes. 2022; 13(3):532. https://doi.org/10.3390/genes13030532
Chicago/Turabian StyleAdnan, Atif, Allah Rakha, Hayder Lazim, Shahid Nazir, Wedad Saeed Al-Qahtani, Maha Abdullah Alwaili, Sibte Hadi, and Chuan-Chao Wang. 2022. "Are Roma People Descended from the Punjab Region of Pakistan: A Y-Chromosomal Perspective" Genes 13, no. 3: 532. https://doi.org/10.3390/genes13030532
APA StyleAdnan, A., Rakha, A., Lazim, H., Nazir, S., Al-Qahtani, W. S., Abdullah Alwaili, M., Hadi, S., & Wang, C.-C. (2022). Are Roma People Descended from the Punjab Region of Pakistan: A Y-Chromosomal Perspective. Genes, 13(3), 532. https://doi.org/10.3390/genes13030532






