Abstract
Oculocutaneous albinism (OCA) is associated with a wide range of clinical presentations and has been categorized with syndromic and non-syndromic features. The most common causative genes in non-syndromic OCA are TYR and OCA2 and HSP1 is in the syndromic albinism. The objective of this study was to identify pathogenic variants in congenital OCA families from Pakistan. Eight consanguineous families were recruited, and clinical and ophthalmological examination was carried out to diagnose the disease. Whole blood was collected from the participating individuals, and genomic DNA was extracted for sequencing analysis. TruSight one-panel sequencing was carried out on one affected individual of each family, and termination Sanger sequencing was carried out to establish the co-segregation of the causative gene or genes. In silico analysis was conducted to predict the causative pathogenic variants. Two families were found to have novel genetic pathogenic variants, and six families harbored previously reported variants. One novel compound heterozygous pathogenic variant in the TYR gene, c.1002delA; p.Ala335LeufsTer20, a novel frameshift deletion pathogenic variant and c.832C>T; and p.Arg278Ter (a known pathogenic variant) were found in one family, whereas HPS1; c.437G>A; and p.Trp146Ter were detected in another family. The identification of new and previous pathogenic variants in TYR, OCA2, and HPS1 genes are causative of congenital OCA, and these findings are expanding the heterogeneity of OCA.
1. Introduction
Albinism is a rare genetic disorder arising from the impairment of melanin biosynthesis. It is categorized into ocular albinism, which influences only the eyes; and oculocutaneous albinism, which involves the skin, eyes, and hair. Oculocutaneous albinism (OCA) is a rare genetic anomaly consequence due to the complete melanin deficiency or partial or reduced synthesis of pigment in eyes, hair, and skin. OCA-affected individuals have relatively fair complexions compared to their unaffected relatives [1]. The clinical presentations of OCA are variable in each subtype. These features include nystagmus, photophobia, hypopigmentation of skin, hair and eyes, mild to moderate reduced visual acuity, and iridis trans-illumination [2,3,4]. OCA is further divided into syndromic and non-syndromic OCA (nsOCA) types. As of now, eighteen OCA-associated genes have been reported in syndromic and non-syndromic phenotypes. Out of these genes, seven genes/loci are linked to autosomal recessive nsOCA. These genes include TYR, OCA2, TYRP1, SLC45A2, SLC24A5, C10orf11, and DCT [1,2,3,4]. These genes are subsequently related to type OCA1-8, except the OCA5 locus, which has been described in OCA families of Pakistani origin. The causative gene has not yet been detected [5]. The recently identified type OCA8 is linked to DCT, as reported in Turkish and French populations [3,4]. Worldwide, more than 400 pathogenic variants have been identified in TYR, and more than 300 pathogenic variants are associated with OCA2. The most commonly reported subtype is OCA1 in the Caucasian population, and about 50% of cases are documented worldwide [6,7]. In Pakistani families, OCA1 is the most common OCA type, followed by OCA2 [1,2,8,9].
On the other hand, Hermansky–Pudlak syndrome (HPS) is one of the syndromes associated with albinism. It is a multisystem autosomal recessive syndrome characterized by OCA, excessive bleeding, and, in some individuals, fatal pulmonary fibrosis, granulomatous colitis, and immunodeficiency. The associated ocular abnormalities include nystagmus and foveal hypoplasia with a substantial reduction in visual acuity [10]. Eleven different HPS types have been reported (HPS1–HPS-11) with common visual acuity and variable bleeding manifestations [11,12,13]. HPS-1 affects individuals of different ethnicities, including those from Western Europe, Japan, the Middle East, the Indian Subcontinent, and non-Puerto Rican Hispanic people [14,15,16]. HPS affects 1/1,500,000–1,000,000 individuals globally, although the prevalence is 1/1800 in individuals of Puerto Rican origin [17]. The group of BLOC-2 including HPS-3 and HPS-5&6 contains milder and quite rare subtypes [18]. The group of BLOC-1 including HPS-7, HPS-8, and HPS-9 contains extremely rare subtypes [19].
In the present study, we have identified the consanguineous OCA families’ pathogenic variant of the TYR, OCA2, and HPS-1 genes in the Pakistani population.
2. Patients and Methods
2.1. Detail of Families
Ethical approval for this research was obtained from the Ethical Review Committee (ERC) of University of Health Sciences Lahore, and the human subjects were included according to the Helsinki guidelines (2008). This study was carried out on eight OCA Pakistani families. All the congenital families originated and were recruited from the province of the Punjab with distinct ethnic backgrounds; 1—Mayo (Lahore), 2—Cheema (Sargodha), 3—Rajpoot (Rawalpindi), 4—Arain (Sheikhupora), 5—Dogar (Burewala), 6—Dogar (Kasur), 7—Bhatti Rajpoot (Lahore), 8—Bukhari Sayyed (Rahim Yar Khan). To establish the diagnosis of a pro-band in each family, familial history and clinical presentation were assisted by clinicians/ophthalmologists from the regional teaching hospitals. The phenotype was examined by physical inspection of skin, hair, and eye color. Clinical images/photos of the individuals affected with OCA were taken to confirm the disease pattern. Ophthalmic examinations were carried out to assess the visual acuity and color vision testing, and direct ophthalmoscopy was used for fundoscopic examination. These findings were documented on data forms.
2.2. Molecular Genetic Analysis
An ethylenediaminetetraacetic acid (EDTA) blood sample was collected from each participating individual, and genomic DNA extraction was carried out as previously described [8]. One affected individual of every family underwent TruSight gene panel sequencing, which contained all the syndromic and non-syndromic variants associated with albinism. Then, Sanger sequencing carried out the co-segregation of pathogenic variants in putative albinism genes (TYR, OCA2, and HPS1) in respective families. All the data was aligned to the human genome reference sequence (hg19) to observe sequence variants using BioEdit 7.0 software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html, accessed on 27 August 2019), Chromas Lite (http://technelysium.com.au/wp/chromas/, accessed on 27 August 2019) software, and CLC sequence viewer (https://www.qiagenbioinformatics.com/products/clc-sequence-viewer/, accessed on 27 August 2019).
The in silico tools applied to predict the pathogenicity were PolyPhen2, SIFT, and Pathogenic variant Taster [20]. For the missense pathogenic variants, multiple sequence alignment (MSA) determined the evolutionary conservation of mutated amino acid residues in TYR and OCA2 orthologs by using MEGA-X software, Pennsylvania State University (https://www.megasoftware.net/, accessed on 21 December 2020). In the absence of an experimentally determined crystal protein structure, the SWISS-MODEL server (https://swissmodel.expasy.org/, accessed on 21 December 2020) was used for homology protein modelling of wild-type TYR protein. Subsequently, 3D protein modelling analysis and visualization of the TYR p.Arg278Ter and p.Ala335LeufsTer20 pathogenic variants were performed using PyMOL software (http://www.pymol.org, accessed on 17 November 2021).
3. Results
3.1. Clinical Presentation of OCA Cases
A total of eight OCA families were recruited and investigated for pathogenic variant analysis. Inherent and pedigree analysis established an autosomal recessive mode of inheritance in all families, and each affected individual of the family presented cardinal features consistent with OCA. These characteristics included pale to reddish-white skin, white to golden blonde hair, photophobia, nystagmus, reduced visual acuity of variable degree, and strabismus. In OCA families, the ophthalmological examinations showed the classical characteristics of albinism, such as nystagmus, foveal hypoplasia (moderate degree), hypo-pigmented fundus, and strabismus. A summary of clinical findings observed in OCA families is represented in Table 1.

Table 1.
Physical and clinical features observed in OCA families.
3.2. Genetic Pathogenic Variants
3.2.1. Novel and Known Pathogenic Variants in TYR Gene
In albinism family number 01 (AL01), the combination of sequencing techniques determined a novel compound heterozygous pathogenic variant in the TYR gene in which one allele c.832C>T; p.Arg278Ter was a previously known pathogenic variant, whereas another pathogenic variant c.1002delA; p.Ala335LeufsTer20 was a novel frameshift deletion. This is the native compound heterozygous pathogenic variant associated with the OCA phenotype. In co-segregation analysis, the father was a homozygous wild type (WT) for c.1002delA and a heterozygous carrier for allele c.832C>T, whereas the mother was homozygous WT for c.832C>T and heterozygous for c.1002delA variant. Both the affected members showed heterozygous alleles, one c.832C>T contributed by the father and another allele c.1002delA received from the mother to produce the OCA phenotype, as seen in Figure 1A–C.
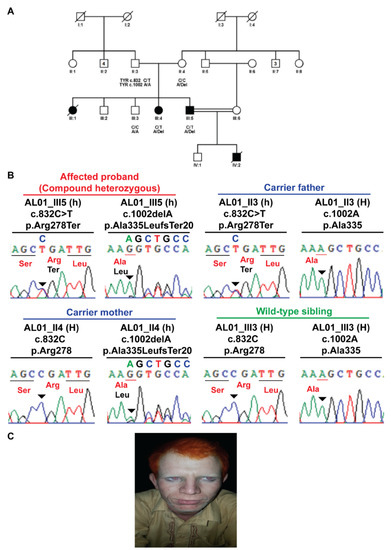
Figure 1.
Compound heterozygous TYR p.Arg278Ter and p.Ala335LeufsTer20 variants in AL01 family manifesting albinism. (A) Pedigree structure of AL01 family. (B) Sanger sequencing electropherograms reflecting familial segregation of TYR p.Arg278Ter and p.Ala335LeufsTer20 pathogenic variants in AL01 family. (C) Clinical image depicting clinical features of albinism in pro-band III-5 of family AL01.
An in silico analysis was carried out to explore the relationship between the mutant TYR and the wild-type protein (Figure 2A). It was predicted that a nonsense mutant allele of p.Arg278Ter may result in early termination and causes premature truncation in the TYR protein of 251 amino acids only (Figure 2B). For the mutant p.Ala335LeufsTer20 allele, the expasy translation analysis proposed a 335 amino acid reading frameshift due to a premature stop codon, which resulted in a truncated TYR protein instead of 529 residues in normal protein (Figure 2C). The comparison between WT and mutant protein was made directly, which demonstrated that the truncated TYR protein for the p.Arg278Ter mutant showed loss of significant secondary structures and also showed conformational changes, which impacted the altered quaternary structures of the protein. Altogether, the reported pathogenic mutants (compound heterozygous) consequently suggested the defective synthesis of tyrosinase protein with a reduced enzymatic function that leads to the OCA1 phenotype (Figure 2A–C).
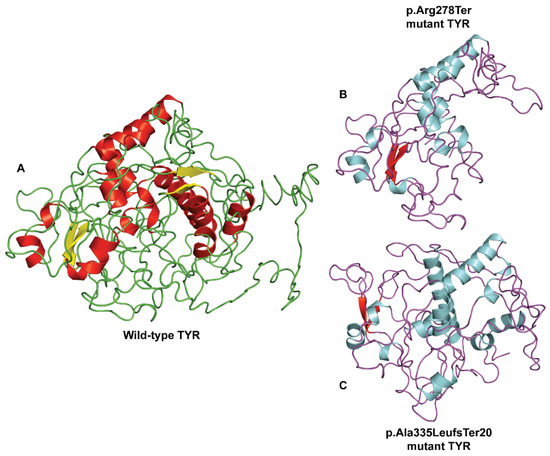
Figure 2.
The 3D protein modelling of p.Arg278Ter and p.Ala335LeufsTer20 pathogenic variants in the predicted crystal structure of human TYR protein. (A) Wild-type protein structure of TYR protein as predicted using SWISS-MODEL. Comparison of the wild-type and mutant TYR protein structures reveals loss of secondary structure elements and changes in the overall 3D protein structure for truncating (B) p.Arg278Ter and (C) p.Ala335LeufsTer20 pathogenic variants.
In families AL02, AL03, and AL04, previously reported pathogenic variants including a splice-site error (c.1037-7T>A) and a missense pathogenic variant (c.1255G>A; p.Gly419Arg) in exon 4 and another non-sense pathogenic variant (c.832C>T; p.Arg278Ter) in exon 2 were identified, respectively, in the TYR gene (Scheme 1 and Table 2).
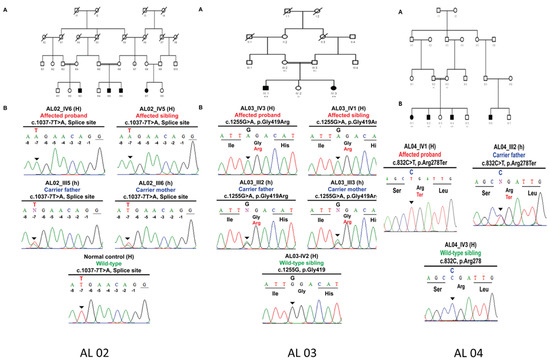
Scheme 1.
Homozygous splice site TYR c.1037-7T>A variant in AL02 family manifesting albinism. (A) Pedigree structure of AL02 family. (B) Sanger sequencing electropherograms reflecting familial segregation of TYR c.1037-7T>A pathogenic variant in AL02 family. Homozygous missense p.Gly419Arg variant in AL03 family manifesting albinism. (A) Pedigree structure of AL03 family. (B) Sanger sequencing electropherograms reflecting familial segregation of TYR p.Gly419Arg pathogenic variant in AL03 family. Homozygous nonsense TYR p.Arg278Ter variant in AL04 family manifesting albinism. (A) Pedigree structure of AL04 family. (B) Sanger sequencing electropherograms reflecting familial segregation of TYR p.Arg278Ter pathogenic variants in AL04 family.

Table 2.
Novel and reported TYR, OCA2 and HPS-1 variants identified in albinism families in this study.
3.2.2. Previous Pathogenic Variants in OCA2
Family AL05 and AL06 were identified for known homozygous c.1456G>T; p.Asp486Tyr pathogenic variant in exon 13 of the OCA2 gene. The homozygous pattern of mutants carries T/T allele at position c.1456G>T, whereas the heterozygous carrier showed a T/G allele pattern at the same position Scheme 2 and Table 2.
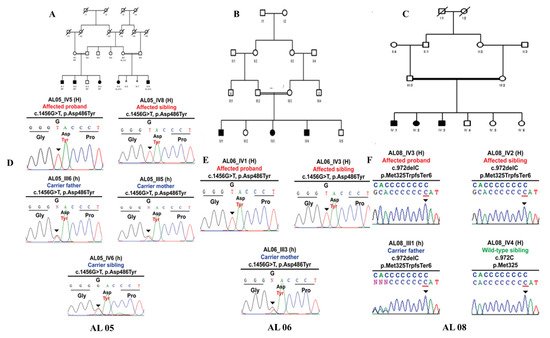
Scheme 2.
Homozygous Missense OCA2 p.Asp486Tyr variant in AL05 family manifesting albinism (A–F). (A) Pedigree structure of AL05 family. (D) Sanger sequencing electropherograms reflecting familial segregation of OCA2 p.Asp486Tyr pathogenic variant in AL05 family. Homozygous Missense OCA2 p.Asp486Tyr variant in AL06 family manifesting albinism. (E) Pedigree structure of AL06 family. (B) Sanger sequencing electropherograms reflecting familial segregation of OCA2 p.Asp486Tyr pathogenic variant in AL06 family. Homozygous frameshift HPS-1 p.Met325TrpfsTer6 variant in AL08 family manifesting albinism. (C) Pedigree structure of AL08 family. (F) Sanger sequencing electropherograms reflecting familial segregation of HPS-1 p.Met325TrpfsTer6 pathogenic variant in AL01 family.
3.2.3. Novel and Known HPS-1 Pathogenic Variants
In AL07, both affected individuals presented with all the cardinal features of nsOCA except eye color, but this family was not linked to any known gene of nsOCA. TruSight exome sequencing of one affected child revealed that it was linked to the Hermansky–Pudlak Syndrome 1 (HPS1) gene. Co-segregation analysis identified a novel nonsense pathogenic variant in family AL07; HPS-1 c.437G>A; p.Trp146Ter. The heterozygous carrier parents had a G/A change, ehereas homozygous affected individuals had A/A instead of WT G/G, which resulted in the formation of a termination codon causing the premature termination of the protein (Figure 3 and Table 2).
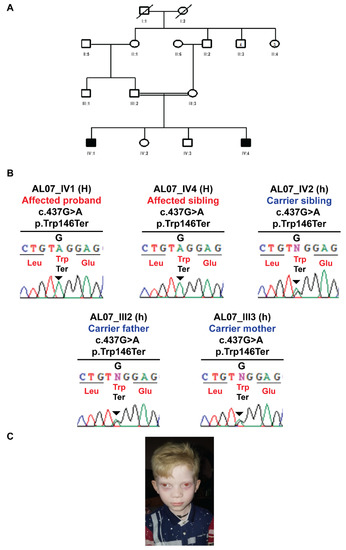
Figure 3.
Homozygous nonsense HPS1 p.Trp146Ter variant in AL07 family manifesting albinism. (A) Pedigree structure of AL07 family. (B) Sanger sequencing electropherograms reflecting the familial segregation of HPS1 p.Trp146Ter novel pathogenic variant in AL07 family. (C) Clinical image depicting clinical features of albinism in pro-band IV-1 of family AL07.
In family AL08, the combination of TruSight exome sequencing and Sanger sequencing found the known frame-shift pathogenic variant in HPS-1; c.972delC; p.Met325TrpfsTer6 due to deletion of one base. The heterozygous carrier mother had a C/del change, whereas homozygous-affected individuals had del/del instead of WT C/C. This sequence change creates a premature translational stop signal (p.Met325TrpfsTer6) in the HPS-1 gene (Scheme 2 and Table 2). It is expected to result in an absent or disrupted protein product. This variant is present in normal population databases (rs281865082, gnomAD 0.0001%) but not reported in a homozygous form.
4. Discussion
In this study, pathogenic variants of TYR, OCA2, and HPS1 genes were detected in OCA families of Pakistani origin. Molecular genetic studies in eight albinism families detected novel and previously known pathogenic variants in different OCA genes. TYR pathogenic variants reported in the present study have also been investigated widely in various ethnic/language groups. In our study, known pathogenic variants c.1255G>A, c.832C>T, and 1037-7T>A of TYR, were found in three OCA families. To date, more than 600 pathogenic variants are linked to the TYR gene, and some pathogenic variants are very common, with high frequency in different populations (HGMD Professional, 2020). Pakistan is one of the leading countries in reported genetic disorders, and more than 200 OCA families have been reported from different research groups.
The most common pathogenic variant of the TYR gene reported in Pakistani OCA families, with a frequency of about 23%, is c.1255G>A; p.Gly419Arg and is predominantly found in the Punjabi-speaking language group. On the other hand, OCA families of other language groups, including Saraiki, Sindhi, Balochi, and Kashmiri, are reported for this common pathogenic variant [1,2,8,24,25,26,27]. The c.832C>T; p.Arg278Ter pathogenic variant is the second most frequent pathogenic variant, representing about 21% of total reported pathogenic variants, and it is also documented in the current study [23,25]. In addition, a previous splice-site pathogenic variant c.1037-7T>A has been reported in heterogeneous groups of Pakistan ancestry [2,24,25,27], and in the current study.
In Pakistan, the overall prevalence of pathogenic variant associated with TYR is nearly 44%, compared to the rate in the European population, where it is 46%. The reports of TYR pathogenic variants from other Asian groups are variable, but it is about 70% in a regional group of the Chinese population, showing remarkable heterogeneity [23,28]. A report from an Indian ethnic group showed a 60% prevalence of TYR pathogenic variants in patients with OCA features [29,30]. In addition, in other populations, including American, Korean, Japanese, and Italian people, the higher rate of TYR pathogenic variants is the main cause of the OCA phenotype [6].
In the Pakistani population, OCA2 is the second causative gene of OCA, and 24% of diverse and frequently recurrent pathogenic variants are reported in this gene. In the present study, we identified one known c.1456G>T; p.Asp486Tyr pathogenic variant in two OCA families. Previously, this pathogenic variant was identified in five OCA families of the Pakistan region. Its frequency is almost 22.4%, making it one of the common alterations of the OCA2 gene in the current study and in [2,8,23]. Interestingly, in the present study, an OCA2 pathogenic variant was detected in two families belonging to the Dogar caste; one residing in Burewala, and the other was in Kasur. They were not distant relatives but might have had some common ancestors who harbored this particular pathogenic variant among the generations. Different in silico tools predicted this pathogenic variant as pathogenic. The residue was found in the conserved region when compared in different species orthologs of OCA2. It was anticipated that the biological action of the subsequent protein would be abnormal.
HPS subtype identification is important for accurate diagnosis, treatment options, clinical management, and prognosis. For example, BLOC-2 deficiency causes a milder type of phenotype with no life-threatening traits, and AP-3 defects result in immunodeficiency, whereas BLOC-3 and AP-3 pathogenic variants cause severe pulmonary fibrosis.
In addition, the current study investigated the involvement of the syndromic gene or genes in OCA families. In this study, one novel pathogenic variant, HPS-1 c.437G>A; p.Trp146Ter, and one known pathogenic variant, p.Met325TrpfsTer6, have been identified in the HPS-1 gene. HPS-1, the most common pathogenic variant of HPS, has been identified in individuals of different ethnicities all over the world. HPS-1 and HPS-4 manifest the most severe phenotypes. HPS-1-affected individuals are presented with more severe ocular findings than in other subtypes, and cutaneous albinism is more prominent with increased hypopigmentation of hair and skin [10]. The known frameshift pathogenic variant in exon 11 of HPS-1, p.Met325TrpfsTer6, present in one family, has also been described in different ethnicities, for example, in Japanese, Chinese, African American, Mexican, and Northern European populations. More than 76 HPS-1 pathogenic variants including 15 (20%) missense, 11 (14.5%) nonsense, 11 (14.5%) splice site, 20 (26%) frameshift, and 18 (24%) insertions and/or deletions pathogenic variants have been described to date [10,31,32,33,34,35].
In a previous study, it was noted that HPS-1 pathogenic variants in mice appeared to have only a slight effect on bleeding and pigmentation for unknown reasons [36]. The same was observed in two families in this study with novel and known HPS-1 pathogenic variants, as they did not show any bleeding tendency. According to Huizing et al., 2020, some HPS-1 affected individuals develop granulomatous colitis, most of them develop pulmonary fibrosis by middle age, and most of the affected females have menorrhagia [10], whereas HPS-1 affected cases in this study did not show any such symptoms. Phenotypically, we cannot distinguish between syndromic and non-syndromic OCA, so genetic testing is the gold standard for the exact diagnosis of such conditions.
Our study identified the pathogenic variants in TYR, OCA2, and HSP-1 associated with the OCA phenotype. The results showed two novel pathogenic variants in TYR and HSP-1 in two OCA families. In the other six families, the previously known pathogenic variants were identified in TYR, OCA2, and HSP-1. It is suggested that exploring the causative genes for OCA may expand the understanding of the molecular mechanism of the disease in specific ethnic groups, helping the community in genetic counseling. It may also be helpful in evolving the gene panels for accurate, cost-effective diagnosis of the most common OCA pathogenic variants present in the community that may further pave the pathway towards the treatment of such inherited diseases in future.
Author Contributions
Conceptualization, H.K. and M.A.Q.; Data curation, N.M.A. and M.Z.L.; Formal analysis, M.S., A.A. (Abida Akbar), A.A. (Ali Amar) and M.A.Q.; Investigation, I.H., M.Z.L. and M.A.Q.; Methodology, A.A. (Abida Akbar), N.M.A. and M.I.U.; Project administration, S.M.; Software, A.A. (Ali Amar); Supervision, S.M.; Validation, S.M.; Writing—original draft, M.S., M.I.U. and M.A.; Writing—review and editing, H.K. and S.M. All authors have read and agreed to the published version of the manuscript.
Funding
Pakistan Science Foundation (PSF) provided funding for this research vide reference No. PSF/Res/P-UHS/Med (492).
Informed Consent Statement
Ethical permission was obtained to conduct this study and consent of congenital OCA families was obtained according to the Helsinki guidelines (2008). Approval of this study was obtained by the ethical approval committee/institutional review board of the University of Health Sciences, Lahore, Pakistan.
Data Availability Statement
Data available within the article or its Supplementary Materials. The authors confirm that the data supporting the findings of this study are available within the article [and/or] its Supplementary Materials.
Acknowledgments
We acknowledge the families who participated in this study. I am also thankful to University of Health Sciences for providing the equipment facilities and thankful to Pakistan Science Foundation (PSF) for providing research funds. We are also thankful to Andrew H Crosby and Ema L Baple, University of Exeter Medical School, RILD Building, RD&E Hospital Wonford, Barrack Road, Exeter, EX2 5DW, UK for their support in Genetic analyses.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Shakil, M.; Harlalka, G.V.; Ali, S.; Lin, S.; D’Atri, I.; Hussain, S.; Nasir, A.; Shahzad, M.A.; Ullah, M.I.; Self, J.E.; et al. Tyrosinase (TYR) gene sequencing and literature review reveals recurrent pathogenic variants and multiple population founder gene pathogenic variants as causative of oculocutaneous albinism (OCA) in Pakistani families. Eye 2019, 33, 1339–1346. [Google Scholar] [CrossRef]
- Shahzad, M.; Yousaf, S.; Waryah, Y.M.; Gul, H.; Kausar, T.; Tariq, N.; Mahmood, U.; Ali, M.; Khan, M.A.; Waryah, A.M.; et al. Molecular outcomes, clinical consequences, and genetic diagnosis of Oculocutaneous Albinism in Pakistani population. Sci. Rep. 2017, 7, 44185. [Google Scholar] [CrossRef] [PubMed]
- Pennamen, P.; Tingaud-Sequeira, A.; Gazova, I.; Keighren, M.; McKie, L.; Marlin, S.; Gherbi Halem, S.; Kaplan, J.; Delevoye, C.; Lacombe, D.; et al. Dopachrome tautomerase variants in patients with oculocutaneous albinism. Genet. Med. 2021, 23, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Volk, A.E.; Hedergott, A.; Preising, M.; Rading, S.; Fricke, J.; Herkenrath, P.; Nürnberg, P.; Altmüller, J.; von Ameln, S.; Lorenz, B.; et al. Biallelic pathogenic variants in L-dopachrome tautomerase (DCT) cause infantile nystagmus and oculocutaneous albinism. Hum. Genet. 2021, 140, 1157–1168. [Google Scholar] [CrossRef] [PubMed]
- Kausar, T.; Bhatti, M.A.; Ali, M.; Shaikh, R.S.; Ahmed, Z.M. OCA5, a novel locus for non-syndromic oculocutaneous albinism, maps to chromosome 4q24. Clin. Genet. 2013, 84, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Hutton, S.M.; Spritz, R.A. Comprehensive analysis of oculocutaneous albinism among non-Hispanic caucasians shows that OCA1 is the most prevalent OCA type. J. Investig. Dermatol. 2008, 128, 2442–2450. [Google Scholar] [CrossRef] [PubMed]
- Rooryck, C.; Roudaut, C.; Robine, E.; Müsebeck, J.; Arveiler, B. Oculocutaneous albinism with TYRP1 gene pathogenic variants in a Caucasian patient. Pigment. Cell Res. 2006, 19, 239–242. [Google Scholar] [CrossRef]
- Arshad, M.W.; Harlalka, G.V.; Lin, S.; D’Atri, I.; Mehmood, S.; Shakil, M.; Hassan, M.J.; Chioza, B.A.; Self, J.E.; Ennis, S.; et al. Pathogenic variants in TYR and OCA2 associated with oculocutaneous albinism in Pakistani families. Meta Gene 2018, 17, 48–55. [Google Scholar] [CrossRef]
- Chuan, Z.; Yan, Y.; Hao, S.; Zhang, Q.; Zhou, B.; Feng, X.; Wang, X.; Liu, F.; Zheng, L.; Cao, Z.; et al. Pathogenic variant Analysis of 63 Northwest Chinese Probands with Oculocutaneous Albinism. Curr. Eye Res. 2021, 46, 140–143. [Google Scholar] [CrossRef]
- Huizing, M.; Malicdan, M.C.V.; Wang, J.A.; Pri-Chen, H.; Hess, R.A.; Fischer, R.; O’Brien, K.J.; Merideth, M.A.; Gahl, W.A.; Gochuico, B.R. Hermansky-Pudlak syndrome: Pathogenic variant update. Hum. Mutat. 2020, 41, 543–580. [Google Scholar] [CrossRef]
- Gochuico, B.R.; Huizing, M.; Golas, G.A.; Scher, C.D.; Tsokos, M.; Denver, S.D.; Frei-Jones, M.J.; Gahl, W.A. Interstitial lung disease and pulmonary fibrosis in Hermansky-Pudlak syndrome type 2, an adaptor protein-3 complex disease. Mol. Med. 2012, 18, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Ammann, S.; Schulz, A.; Krägeloh-Mann, I.; Dieckmann, N.M.; Niethammer, K.; Fuchs, S.; Eckl, K.M.; Plank, R.; Werner, R.; Altmüller, J.; et al. Pathogenic variants in AP3D1 associated with immunodeficiency and seizures define a new type of Hermansky-Pudlak syndrome. Blood 2016, 127, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Cullinane, A.R.; Curry, J.A.; Golas, G.; Pan, J.; Carmona-Rivera, C.; Hess, R.A.; White, J.G.; Huizing, M.; Gahl, W.A. A BLOC-1 pathogenic variant screen reveals a novel BLOC1S3 pathogenic variant in Hermansky-Pudlak Syndrome type 8. Pigment. Cell Melanoma Res. 2012, 25, 584–591. [Google Scholar] [CrossRef]
- Jones, M.L.; Murden, S.L.; Brooks, C.; Maloney, V.; Manning, R.A.; Gilmour, K.C.; Bharadwaj, V.; de la Fuente, J.; Chakravorty, S.; Mumford, A.D. Disruption of AP3B1 by a chromosome 5 inversion: A new disease mechanism in Hermansky-Pudlak syndrome type 2. BMC Med. Genet. 2013, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Garrido, G.; Fernández, A.; Montoliu, L. HPS11 and OCA8: Two new types of albinism associated with pathogenic variants in BLOC1S5 and DCT genes. Pigment. Cell Melanoma Res. 2021, 34, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Kanazu, M.; Arai, T.; Sugimoto, C.; Kitaichi, M.; Akira, M.; Abe, Y.; Hozumi, Y.; Suzuki, T.; Inoue, Y. An intractable case of Hermansky-Pudlak syndrome. Intern. Med. 2014, 53, 2629–2634. [Google Scholar] [CrossRef] [PubMed]
- De Jesus Rojas, W.; Young, L.R. Hermansky-Pudlak Syndrome. Semin. Respir Crit. Care Med. 2020, 41, 238–246. [Google Scholar] [CrossRef]
- Anikster, Y.; Huizing, M.; White, J.; Shevchenko, Y.O.; Fitzpatrick, D.L.; Touchman, J.W.; Compton, J.G.; Bale, S.J.; Swank, R.T.; Gahl, W.A.; et al. Pathogenic variant of a new gene causes a unique form of Hermansky-Pudlak syndrome in a genetic isolate of central Puerto Rico. Nat. Genet. 2001, 28, 376–380. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Q.; Oiso, N.; Novak, E.K.; Gautam, R.; O’Brien, E.P.; Tinsley, C.L.; Blake, D.J.; Spritz, R.A.; Copeland, N.G.; et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1). Nat. Genet. 2003, 35, 84–89. [Google Scholar] [CrossRef]
- Norman, C.S.; O’Gorman, L.; Gibson, J.; Pengelly, R.J.; Baralle, D.; Ratnayaka, J.A.; Griffiths, H.; Rose-Zerilli, M.; Ranger, M.; Bunyan, D.; et al. Identification of a functionally significant tri-allelic genotype in the Tyrosinase gene (TYR) causing hypomorphic oculocutaneous albinism (OCA1B). Sci. Rep. 2017, 7, 4415. [Google Scholar] [CrossRef]
- Chaki, M.; Sengupta, M.; Mondal, M.; Bhattacharya, A.; Mallick, S.; Bhadra, R.; Indian Genome Variation Consortium; Ray, K. Molecular and functional studies of tyrosinase variants among Indian oculocutaneous albinism type 1 patients. J. Investig. Dermatol. 2011, 131, 260–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, Y.; Shen, N.; Peng, J.; Wang, C.; Liu, H.; Lu, Y. Pathogenic variant analysis of a Chinese family with oculocutaneous albinism. Oncotarget 2016, 7, 84981–84988. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jaworek, T.J.; Kausar, T.; Bell, S.M.; Tariq, N.; Maqsood, M.I.; Sohail, A.; Ali, M.; Iqbal, F.; Rasool, S.; Riazuddin, S.; et al. Molecular genetic studies and delineation of the oculocutaneous albinism phenotype in the Pakistani population. Orphanet. J. Rare Dis. 2012, 7, 44. [Google Scholar] [CrossRef]
- Gul, H.; Shah, A.H.; Harripaul, R.; Mikhailov, A.; Prajapati, K.; Khan, E.; Ullah, F.; Zubair, M.; Ali, M.Z.; Shah, A.H.; et al. Genetic studies of multiple consanguineous Pakistani families segregating oculocutaneous albinism identified novel and reported pathogenic variants. Ann. Hum. Genet. 2019, 83, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Forshew, T.; Khaliq, S.; Tee, L.; Smith, U.; Johnson, C.A.; Mehdi, S.Q.; Maher, E.R. Identification of novel TYR and TYRP1 pathogenic variants in oculocutaneous albinism. Clin. Genet. 2005, 68, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.K.; Strunk, K.M.; Giebel, L.B.; Weleber, R.G.; Spritz, R.A. Tyrosinase gene pathogenic variants in type I (tyrosinase-deficient) oculocutaneous albinism define two clusters of missense substitutions. Am. J. Med. Genet. 1992, 43, 865–871. [Google Scholar] [CrossRef]
- Gul, H.; Ali, M.Z.; Khan, E.; Zubair, M.; Badar, M.; Khan, S.; Shah, A.H.; Khan, M.A. Ophthalmo-genetic analysis of Pakistani patients with nonsyndromic oculocutaneous albinism through whole exome sequencing. J. Pak. Med. Assoc. 2017, 67, 790–792. [Google Scholar]
- Lin, Y.; Chen, X.; Yang, Y.; Che, F.; Zhang, S.; Yuan, L.; Wu, Y. Pathogenic variantal Analysis of TYR, OCA2, and SLC45A2 Genes in Chinese Families with Oculocutaneous Albinism. Mol. Genet. Genomic Med. 2019, 7, e00687. [Google Scholar] [CrossRef]
- Sengupta, M.; Mondal, M.; Jaiswal, P.; Sinha, S.; Chaki, M.; Samanta, S.; Ray, K. Comprehensive analysis of the molecular basis of oculocutaneous albinism in Indian patients lacking a pathogenic variant in the tyrosinase gene. Br. J. Dermatol. 2010, 163, 487–494. [Google Scholar] [CrossRef]
- Okamura, K.; Hayashi, M.; Abe, Y.; Kono, M.; Nakajima, K.; Aoyama, Y.; Nishigori, C.; Ishimoto, H.; Ishimatsu, Y.; Nakajima, M.; et al. NGS-based targeted resequencing identified rare subtypes of albinism: Providing accurate molecular diagnosis for Japanese patients with albinism. Pigment. Cell Melanoma Res. 2019, 32, 848–853. [Google Scholar] [CrossRef]
- Merideth, M.A.; Vincent, L.M.; Sparks, S.E.; Hess, R.A.; Manoli, I.; O’Brien, K.J.; Tsilou, E.; White, J.G.; Huizing, M.; Gahl, W.A. Hermansky-Pudlak syndrome in two African-American brothers. Am. J. Med. Genet. A 2009, 149A, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Rivera, C.; Hess, R.A.; O’Brien, K.; Golas, G.; Tsilou, E.; White, J.G.; Gahl, W.A.; Huizing, M. Novel pathogenic variants in the HPS1 gene among Puerto Rican patients. Clin. Genet. 2011, 79, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Wei, A.; Yuan, Y.; Bai, D.; Ma, J.; Hao, Z.; Zhang, Y.; Yu, J.; Zhou, Z.; Yang, L.; Yang, X.; et al. NGS-based 100-gene panel of hypopigmentation identifies pathogenic variants in Chinese Hermansky–Pudlak syndrome patients. Pigment. Cell Melanoma Res. 2016, 29, 702–706. [Google Scholar] [CrossRef] [PubMed]
- Wei, A.; Yuan, Y.; Qi, Z.; Liu, T.; Bai, D.; Zhang, Y.; Yu, J.; Yang, L.; Yang, X.; Li, W. Instability of BLOC-2 and BLOC-3 in Chinese patients with Hermansky-Pudlak syndrome. Pigment. Cell Melanoma Res. 2019, 32, 373–380. [Google Scholar] [CrossRef]
- Lasseaux, E.; Plaisant, C.; Michaud, V.; Pennamen, P.; Trimouille, A.; Gaston, L.; Monfermé, S.; Lacombe, D.; Rooryck, C.; Morice-Picard, F.; et al. Molecular characterization of a series of 990 index patients with albinism. Pigment. Cell Melanoma Res. 2018, 31, 466–474. [Google Scholar] [CrossRef]
- Novak, E.K.; Hui, S.W.; Swank, R.T. Platelet storage pool deficiency in mouse pigment pathogenic variants associated with seven distinct genetic loci. Blood 1984, 63, 536–544. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).