Alternative Promoters of GRIK2 (GluR6) Gene in Human Carcinoma Cell Lines Are Regulated by Differential Methylation of CpG Dinucleotides
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA Extraction, Reverse Transcription and Polymerase Chain Reaction (RT-PCR)
2.3. DNA Extraction
2.4. Southern Hybridization
2.5. Treatment with 5-Azacytidine and Trichostatin (TSA)
2.6. Bisulfite Sequencing
2.7. In Vitro Methylation of Promoter Constructs in Reporter Plasmid
3. Results
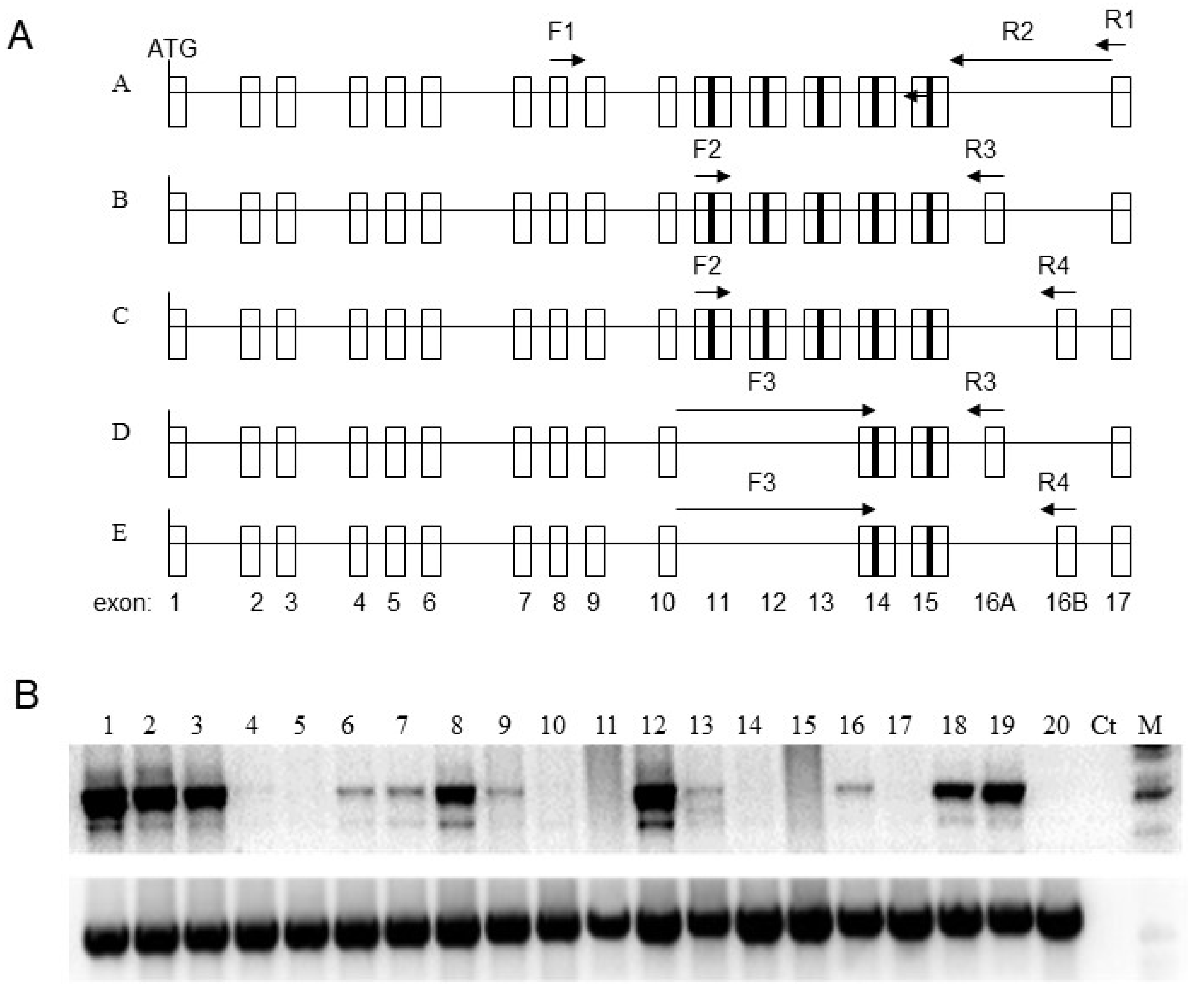
3.1. Expression Pattern of GluR6 in Tumor Cell Lines
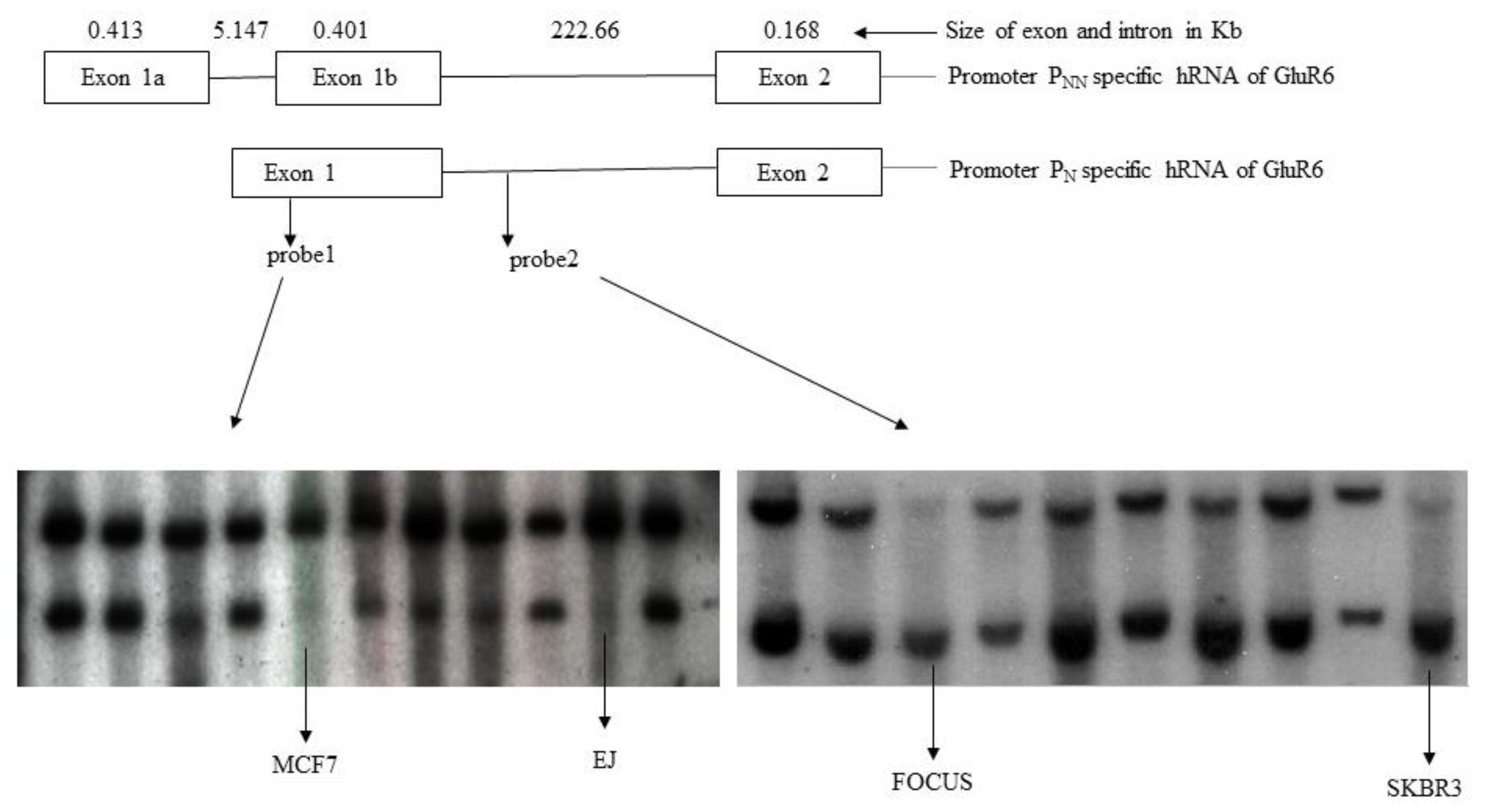
3.2. Rearrangements of GluR6 Gene Locus in Tumor Cells
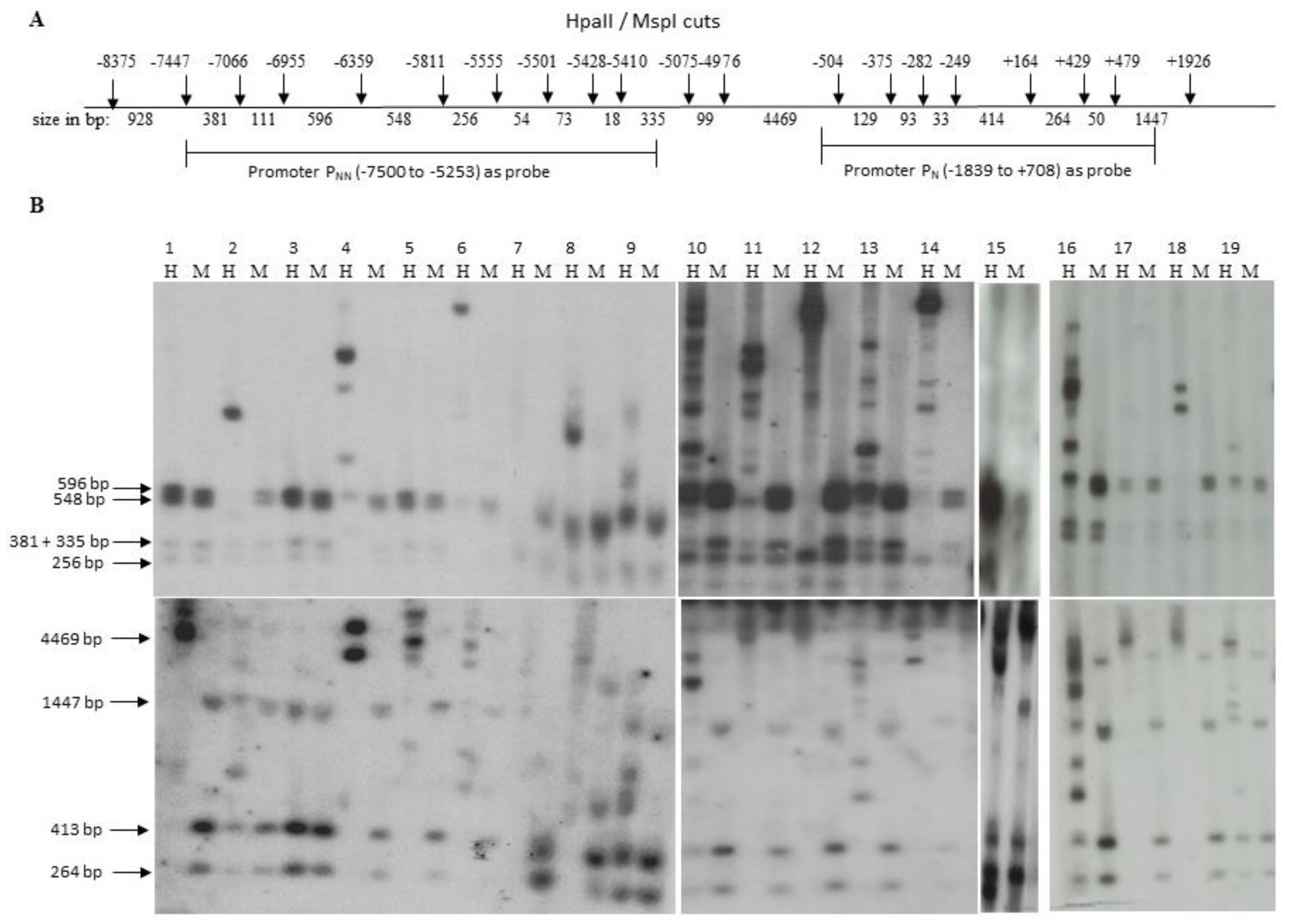
3.3. Identification of CpG Rich Regions in Promoters of GluR6
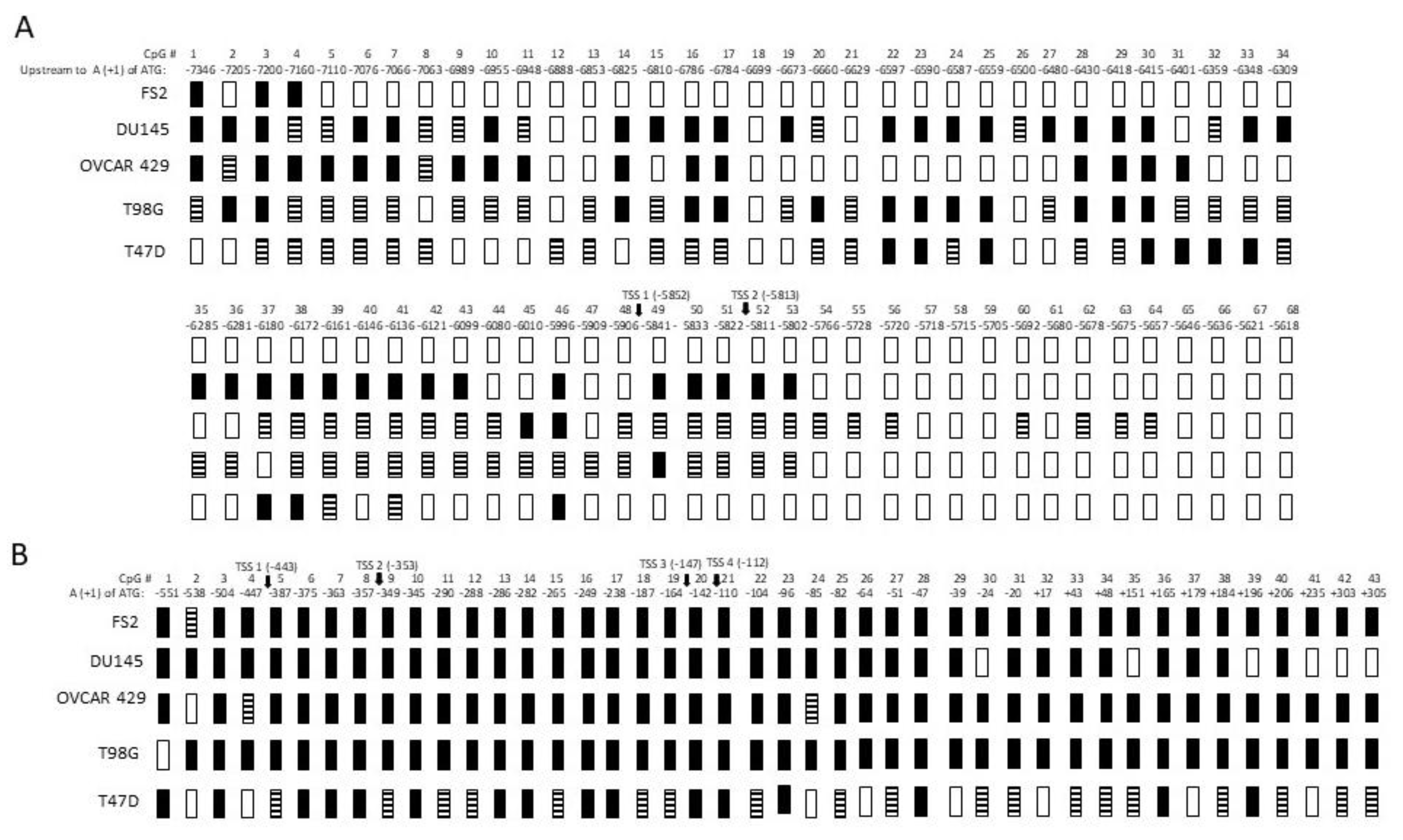
3.4. Methylation Status of CpG Dinucleotides in Non-Neuronal Promoter (PNN) and Neuronal Promoter (PN)
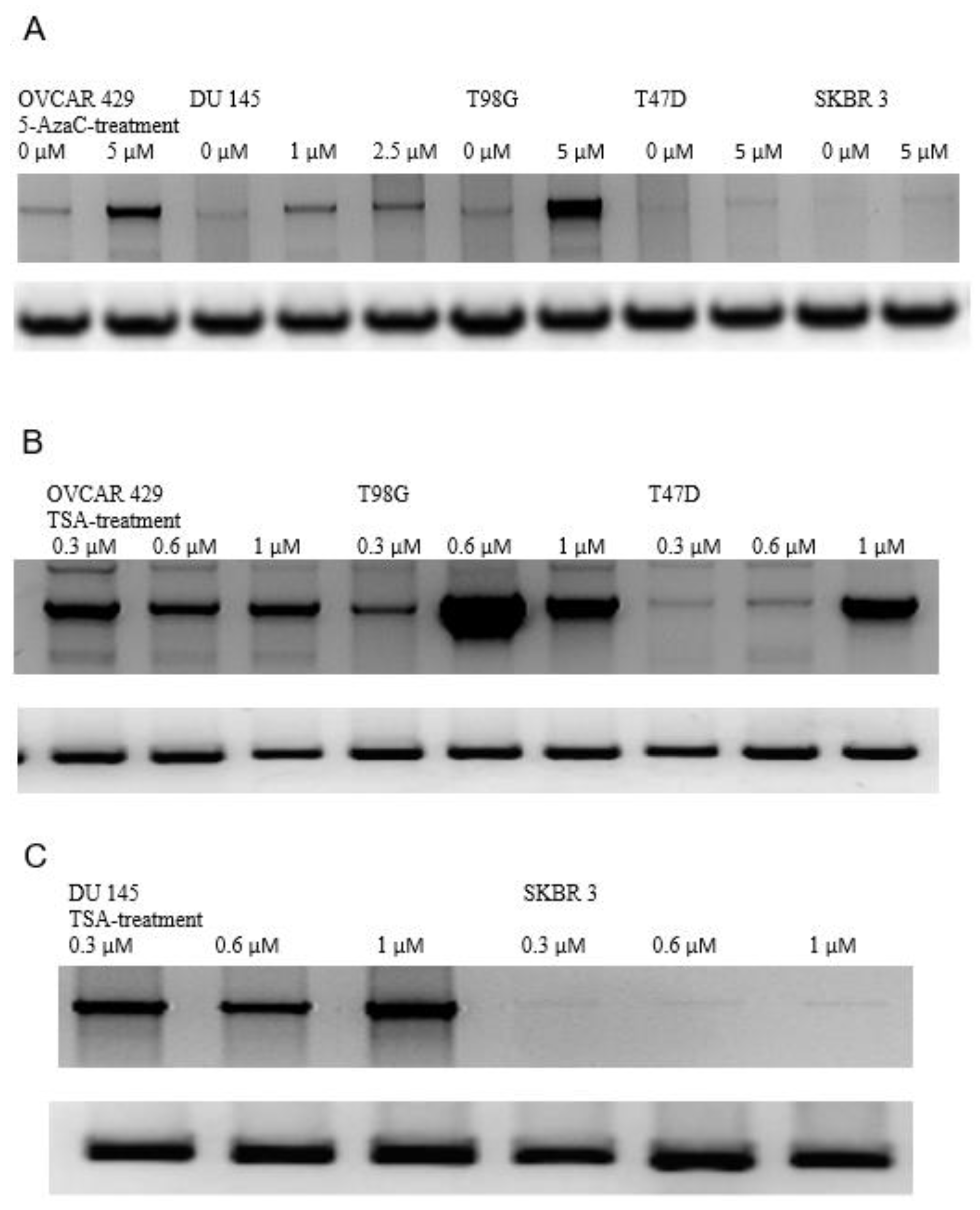
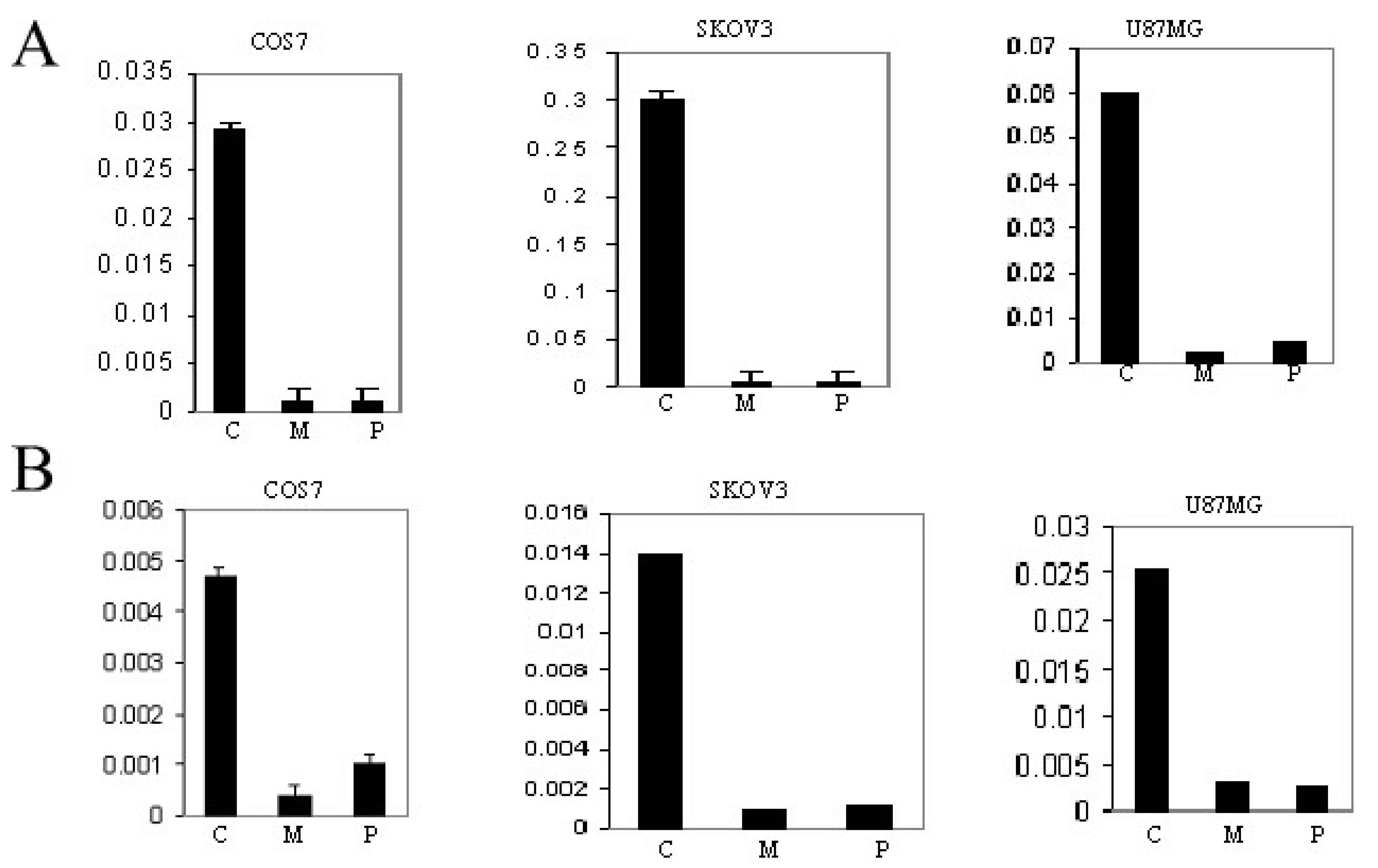
3.5. Induction of GluR6 Expression by 5-Azacytidine (5-Aza) and Trichostatin (TSA)
3.6. Inactivation of Promoter PNN (−7500pGL3) and Promoter PN (−710pGL3) by CpG Methylation
3.7. Bisulfite Sequencing of GluR6 Promoters in Tumor Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hollmann, M.; Heinemann, S. Cloned glutamate receptors. Annu. Rev. Neurosci. 1994, 17, 31–108. [Google Scholar] [CrossRef] [PubMed]
- Lerma, J. Ionotropic glutamate receptors in the CNS. In Handbook of Experimental Pharmacology; Jonas, P., Monyer, H., Eds.; Springer: Berlin, Germany, 1999; Volume 141, pp. 275–307. [Google Scholar]
- Fogarty, D.J.; Perez-Cerda, F.; Matute, C. KA1-like kainate receptor subunit immunoreactivity in neurons and glia using a novel anti-peptide antibody. Mol. Brain Res. 2000, 81, 164–176. [Google Scholar] [CrossRef]
- Savinainen, A.; Garcia, E.P.; Dorow, D.; Marshall, J.; Lin, Y.F. Kainate receptor activation induces mixed lineage kinase-mediated cellular signaling cascades via Post-synaptic Density Protein 95. J. Biol. Chem. 2001, 276, 11382–11386. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.D.; Kuan, C.Y.; Whitmarsh, A.J.; Rinócn, M.; Zheng, T.S.; Davis, R.J.; Rakic, P.; Flavell, R.A. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the jnk3 gene. Nature 1997, 389, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Paschen, W.; Blackstone, C.D.; Huganir, R.L.; Ross, C.A. Human GluR6 kainate receptor (GRIK2): Molecular cloning, expression, polymorphism, and chromosomal assignment. Genomics 1994, 20, 435–440. [Google Scholar] [CrossRef]
- Zhawar, V.K.; Kaur, G.; deRiel, J.K.; Kaur, G.P.; Kandpal, R.P.; Athwal, R.S. Novel spliced variants of ionotropic glutamate receptor glur6 in normal human fibroblast and brain cells are transcribed by tissue specific promoters. Gene 2010, 459, 1–10. [Google Scholar] [CrossRef]
- Noviello, C.; Courjal, F.; Theillet, C. Loss of heterozygosity on the long arm of chromosome 6 in breast cancer: Possibly four regions of deletion. Clin. Cancer. Res. 1996, 2, 1601–1606. [Google Scholar]
- Konishi, N.; Nakamura, M.; Kishi, M.; Ishida, E.; Shimada, K.; Matsuyoshi, S.; Nagai, H.; Emi, M. Genetic mapping of allelic loss on chromosome 6q within heterogeneous prostate carcinoma. Cancer Sci. 2003, 94, 764–768. [Google Scholar] [CrossRef]
- Abe, T.; Makino, N.; Furukawa, T.; Ouyang, H.; Kimura, M.; Yatsuoka, T.; Yokoyama, T.; Inoue, H.; Fukushige, S.; Hoshi, M.; et al. Identification of three commonly deleted regions on chromosome arm 6q in human pancreatic cancer. Genes Chromosomes Cancer 1999, 25, 60–64. [Google Scholar] [CrossRef]
- Ezaki, T.; Watanabe, M.; Inoue, N.; Kanai, T.; Ogata, H.; Iwao, Y.; Ishii, H.; Hibi, T. A specific genetic alteration on chromosome 6 in ulcerative colitis-associated colorectal cancers. Cancer Res. 2003, 63, 3747–3749. [Google Scholar]
- Yoon, J.; Ko, Y.H. Deletion mapping of the long arm of chromosome 6 in peripheral T and NK cell lymphomas. Leuk. Lymphoma 2003, 44, 2077–2082. [Google Scholar] [CrossRef]
- Sinclair, P.B.; Sorour, A.; Martineau, M.; Harrison, C.J.; Mitchell, W.A.; O’Neill, E.; Foroni, L. A fluorescence in situ hybridization map of 6q deletions in acute lymphocytic leukemia: Identification and analysis of a candidate tumor suppressor gene. Cancer Res. 2004, 64, 4089–4098. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, A.K.; Hubbard, K.; Kaur, G.P.; Jha, K.K.; Ozer, H.L.; Athwal, R.S. Senescence of immortal human fibroblasts by the introduction of normal human chromosome 6. Proc. Natl. Acad. Sci. USA 1994, 91, 5498–5502. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, A.K.; Kaur, G.P.; Reddy, D.E.; Rane, N.S.; Athwal, R.S. A gene on 6q 14-21 restores senescence to immortal ovarian tumor cells. Oncogene 1996, 12, 247–252. [Google Scholar] [PubMed]
- Reddy, D.E.; Sandhu, A.K.; DeRiel, J.K.; Athwal, R.S.; Kaur, G.P. Identification of a gene at 16q24.3 that restores cellular senescence in immortal mammary tumor cells. Oncogene 1999, 18, 5100–5107. [Google Scholar] [CrossRef]
- Zhawar, V.K.; Kandpal, R.P.; Athwal, R.S. Senescence of normal human fibroblasts relates to the expression of ionotropic glutamate receptor glur6/grik2. Cancer Genom. Proteom. 2020, 17, 707–714. [Google Scholar] [CrossRef]
- Zhawar, V.K.; Kandpal, R.P.; Athwal, R.S. Isoforms of ionotropic glutamate receptor grik2 induce senescence of carcinoma cells. Cancer Genom. Proteom. 2019, 16, 59–64. [Google Scholar] [CrossRef]
- Clark, S.J.; Harrison, J.; Paul, C.L.; Frommer, M. High sensitivity mapping of methylated cytosines. Nucleic Acid Res. 1994, 22, 2990–2997. [Google Scholar] [CrossRef]
- Neumeister, P.; Albanese, C.; Balent, B.; Greally, J.; Pestell, R.G. Senescence and epigenetic dysregulation in cancer. Int. J. Biochem. Cell Biol. 2002, 34, 1475–1490. [Google Scholar] [CrossRef]
- Zheng, S.; Chen, P.; McMillan, A.; Lafuente, A.; Lafuente, M.J.; Ballesta, A.; Trias, M.; Wiencke, J.K. Correlations of partial and extensive methylation at the p14(ARF) locus with reduced mRNA expression in colorectal cancer cell lines and clinicopathological features in primary tumors. Carcinogenesis 2000, 21, 2057–2064. [Google Scholar] [CrossRef]
- Esteller, M.; Sparks, A.; Toyota, M.; Sanchez-Cespedes, M.; Capella, G.; Peinado, M.A.; Gonzalez, S.; Tarafa, G.; Sidransky, D.; Meltzer, S.J.; et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000, 60, 4366–4371. [Google Scholar] [PubMed]
- Stefansson, O.A.; Hermanowicz, S.; van der Horst, J.; Hilmarsdottir, H.; Staszczak, Z.; Jonasson, J.G.; Tryggvadottir, L.; Gudjonsson, T.; Sigurdsson, S. CpG promoter methylation of the ALKBH3 alkylation repair gene in breast cancer. BMC Cancer 2017, 17, 469. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wade, P.A.; Gegonne, A.; Jones, P.L.; Ballestar, E.; Aubry, F.; Wolffe, A.P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 1999, 23, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Coussen, F.; Normand, E.; Marchal, C.; Costet, P.; Choquet, D.; Lambert, M.; Mege, R.M.; Mulle, C. Recruitment of the kainate receptor subunit glutamate receptor 6 by cadherin/catenin complexes. J. Neurosci. 2002, 22, 6426–6436. [Google Scholar] [CrossRef]
- Mehta, S.; Wu, H.; Garners, C.C.; Marshall, J. Molecular mechanism regulating the differential association of kainate receptor subunits with SAP90/PSD95 and SAP97. J. Biol. Chem. 2001, 276, 16092–16099. [Google Scholar] [CrossRef]
- Tanabe, Y.; Masu, M.; Ishii, T.; Shigemoto, R.; Nakanishi, S. A family of metabotropic glutamate receptors. Neuron 1992, 8, 169–179. [Google Scholar] [CrossRef]
- Yu, L.J.; Wall, B.A.; Wangari-Talbot, J.; Chen, S. Metabotropic glutamate receptors in cancer. Neuropharmacology 2017, 115, 193–202. [Google Scholar] [CrossRef]
- Deng, W.; Wang, H.; Rosenberg, P.A.; Volpe, J.J.; Jensen, F. Role of metabotropic glutamate receptors in oligodendrocyte excitotoxicity and oxidative stress. Proc. Natl. Acad. Sci. USA 2004, 101, 7751–7756. [Google Scholar] [CrossRef]
- Pollock, P.M.; Cohen-Solal, K.; Sood, R.; Namkoong, J.; Martino, J.J.; Koganti, A.; Zhu, H.; Robbins, C.; Makalowska, I.; Shin, S.S.; et al. Melanoma mouse model implicates metabotropic glutamate signaling in melanocytic neoplasia. Nat. Genet. 2003, 34, 108–112. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhawar, V.K.; Kandpal, R.P.; Athwal, R.S. Alternative Promoters of GRIK2 (GluR6) Gene in Human Carcinoma Cell Lines Are Regulated by Differential Methylation of CpG Dinucleotides. Genes 2022, 13, 490. https://doi.org/10.3390/genes13030490
Zhawar VK, Kandpal RP, Athwal RS. Alternative Promoters of GRIK2 (GluR6) Gene in Human Carcinoma Cell Lines Are Regulated by Differential Methylation of CpG Dinucleotides. Genes. 2022; 13(3):490. https://doi.org/10.3390/genes13030490
Chicago/Turabian StyleZhawar, Vikramjit K., Raj P. Kandpal, and Raghbir S. Athwal. 2022. "Alternative Promoters of GRIK2 (GluR6) Gene in Human Carcinoma Cell Lines Are Regulated by Differential Methylation of CpG Dinucleotides" Genes 13, no. 3: 490. https://doi.org/10.3390/genes13030490
APA StyleZhawar, V. K., Kandpal, R. P., & Athwal, R. S. (2022). Alternative Promoters of GRIK2 (GluR6) Gene in Human Carcinoma Cell Lines Are Regulated by Differential Methylation of CpG Dinucleotides. Genes, 13(3), 490. https://doi.org/10.3390/genes13030490





