Detection of Target Genes for Drug Repurposing to Treat Skeletal Muscle Atrophy in Mice Flown in Spaceflight
Abstract
:1. Introduction
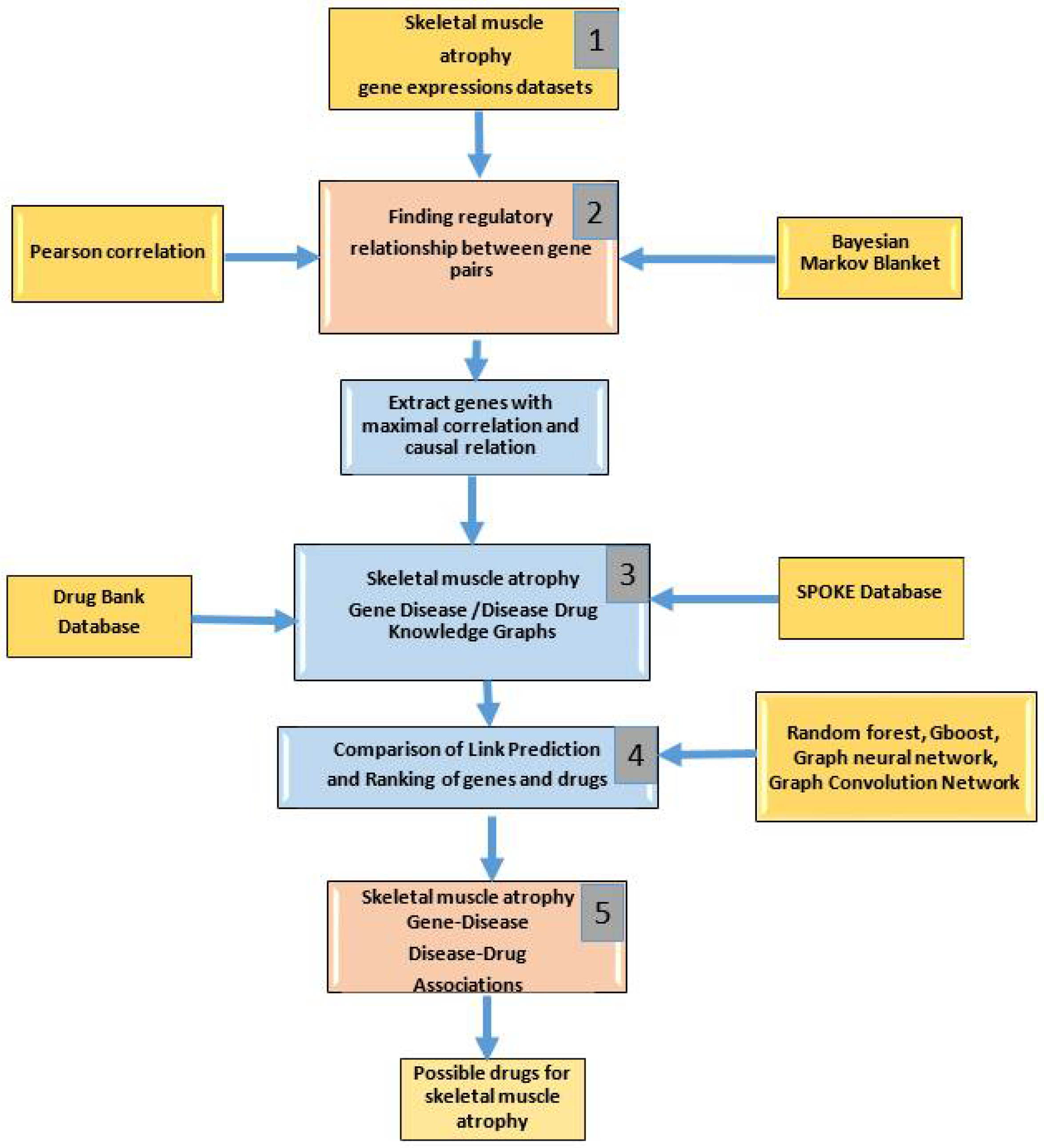
2. Materials and Methods
2.1. GeneLab Datasets
2.2. Finding Regulatory Relationships between Gene Pairs (Stage 1)
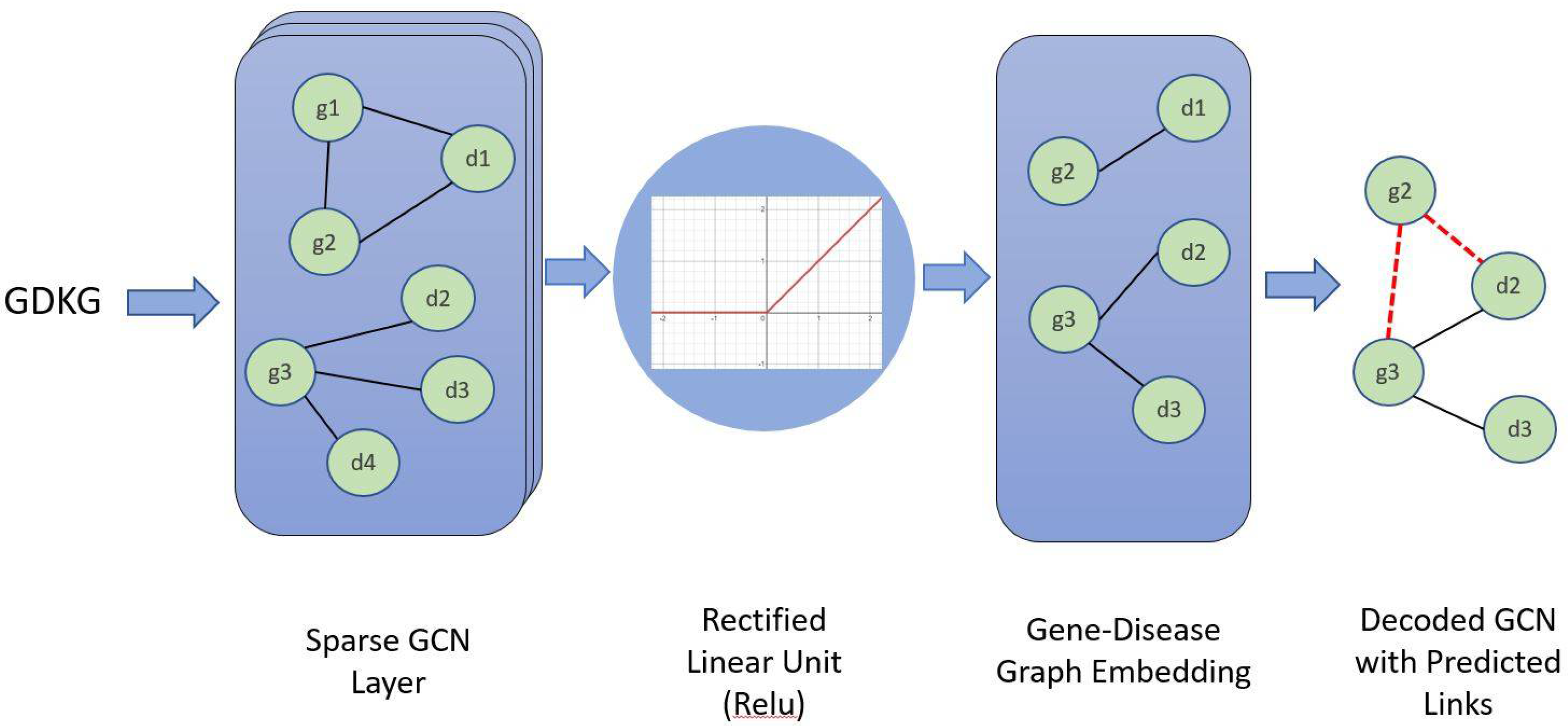
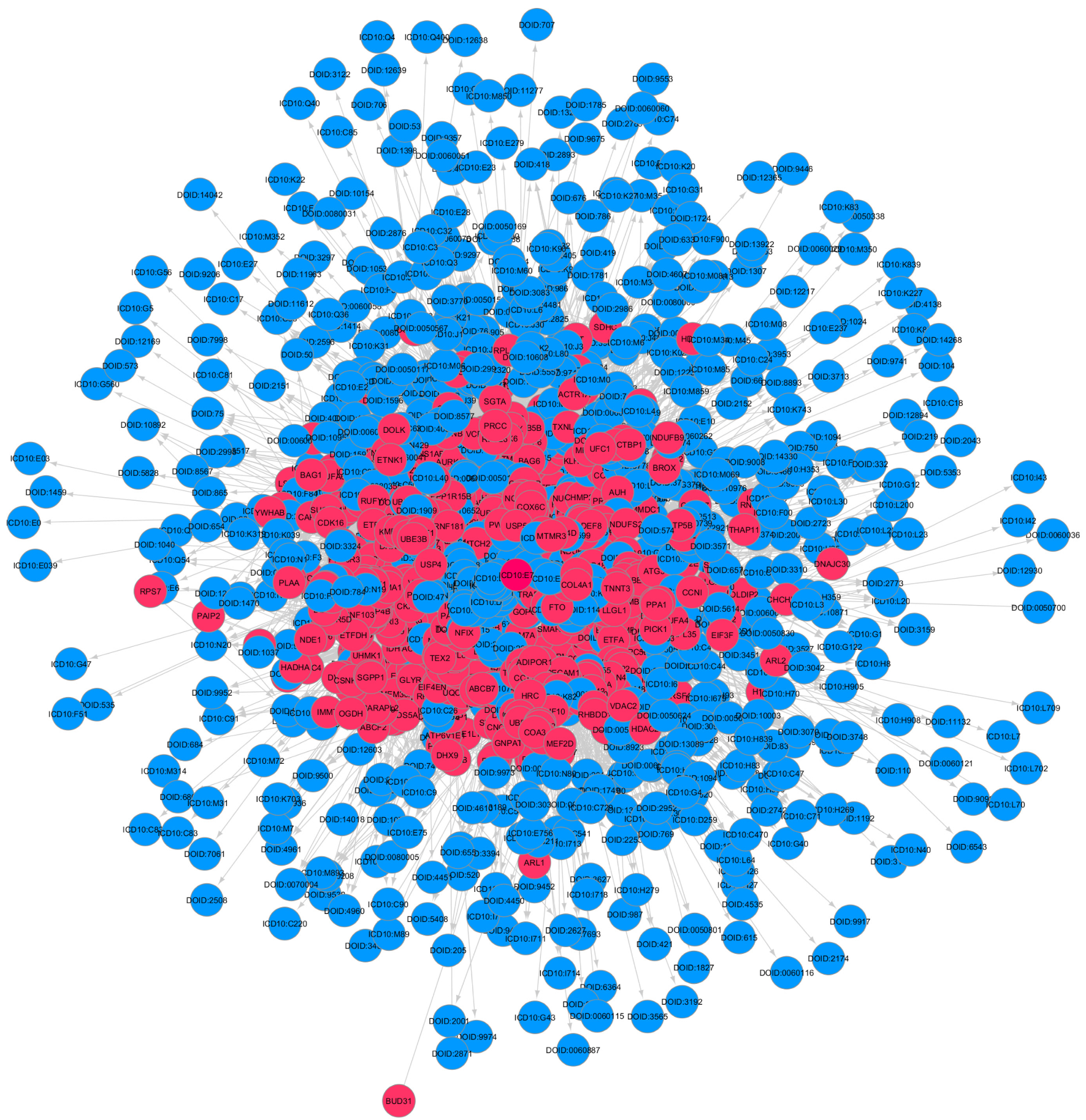
2.3. Construction of Knowledge Graphs (Stage 2)
Graph Concepts and Properties for Analysis of GDKG and DDKG
2.4. ML Methods for Link Prediction (Stage 3)
2.4.1. Random Forest (RF) Method
2.4.2. Gradient Boosting (GB) Method
2.4.3. Graph Neural Network (GNN) Method
2.4.4. Graph Convolution Neural Network (GCN)
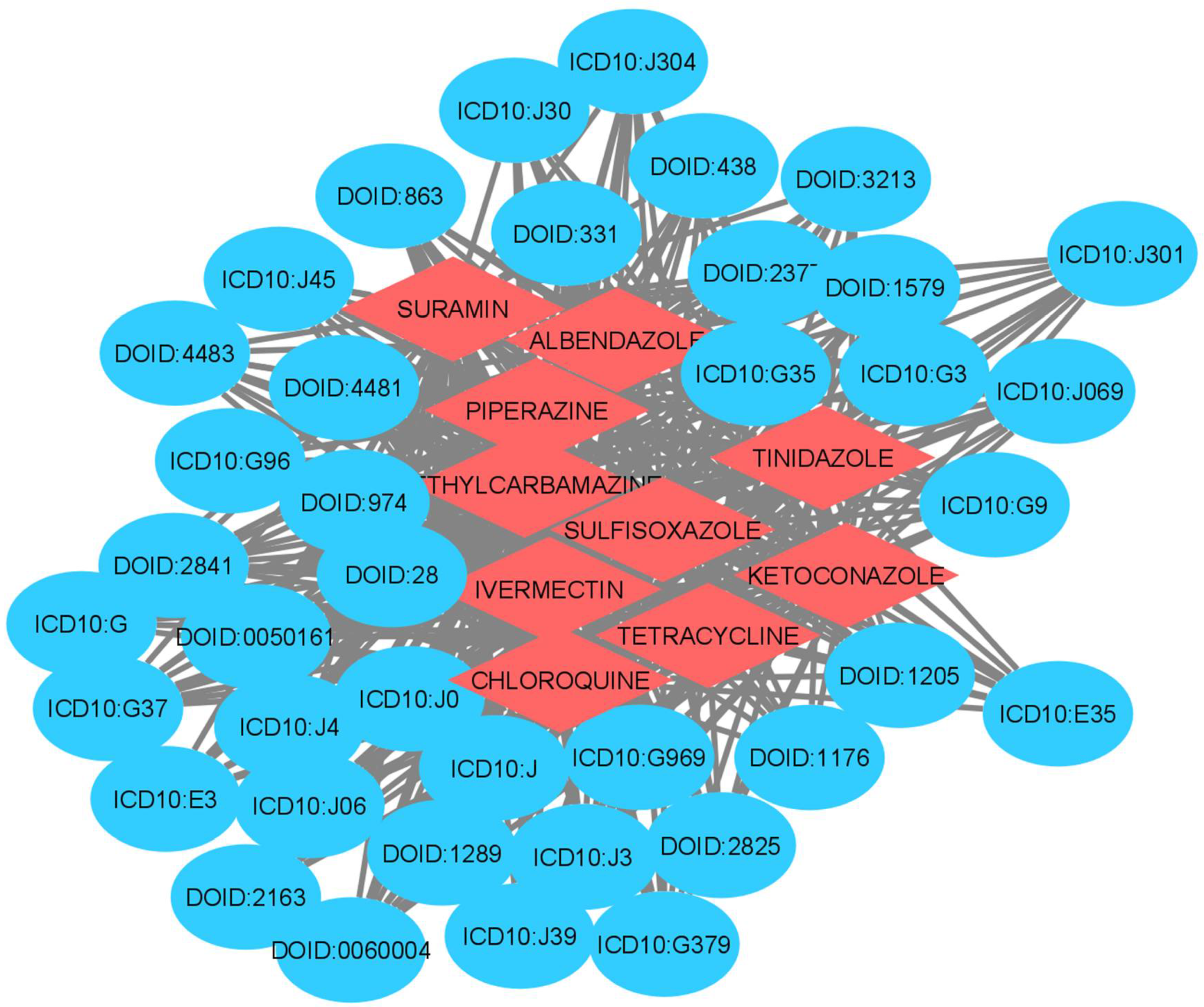
2.5. Gene-Disease and Disease-Drug Associations (Stage 4)
3. Results
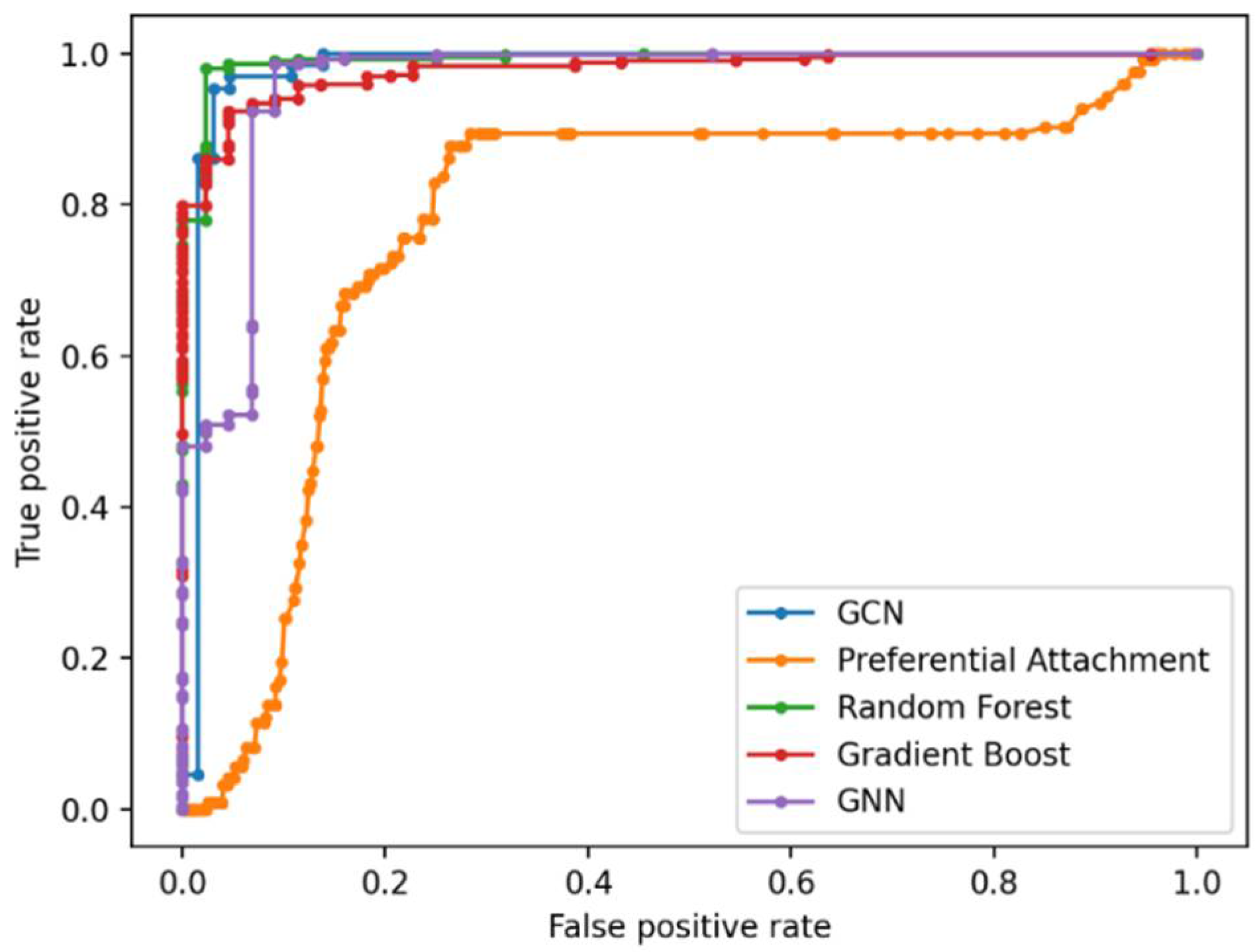
Link Prediction Using GCN and Other ML Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cahill, T.; Cope, H.; Bass, J.J.; Overbey, E.G.; Gilbert, R.; da Silveira, W.A.; Paul, A.M.; Mishra, T.; Herranz, R.; Reinsch, S.S.; et al. Mammalian and invertebrate models as complementary tools for gaining mechanistic insight on muscle responses to spaceflight. Int. J. Mol. Sci. 2021, 22, 9470. [Google Scholar] [CrossRef] [PubMed]
- Kwok, A.T.; Mohamed, N.S.; Plate, J.F.; Yammani, R.R.; Rosas, S.; Bateman, T.A.; Livingston, E.; Moore, J.E.; Kerr, B.A.; Lee, J.; et al. Spaceflight and hind limb unloading induces an arthritic phenotype in knee articular cartilage and menisci of rodents. Sci. Rep. 2021, 11, 10469. [Google Scholar] [CrossRef] [PubMed]
- Sung, M.; Li, J.; Spieker, A.J.; Spatz, J.; Ellman, R.; Ferguson, V.L.; Bateman, T.A.; Rosen, G.D.; Bouxsein, M.; Rutkove, S.B. Spaceflight and hind limb unloading induce similar changes in electrical impedance characteristics of mouse gastrocnemius muscle. J. Musculoskelet. Neuronal Interact. 2013, 13, 405–411. [Google Scholar] [PubMed]
- Romanick, M.; Thompson, L.D.V.; Brown-Borg, H.M. Murine models of atrophy, cachexia, and sarcopenia in skeletal muscle. Biochim. Biophys. Acta-Mol. Basis Dis. 2013, 1832, 1410–1420. [Google Scholar] [CrossRef] [Green Version]
- Allen, D.L.; Bandstra, E.R.; Harrison, B.C.; Thorng, S.; Stodieck, L.S.; Kostenuik, P.J.; Morony, S.; Lacey, D.L.; Hammond, T.G.; Leinwand, L.L.; et al. Effects of spaceflight on murine skeletal muscle gene expression. J. Appl. Physiol. 2009, 106, 582–592. [Google Scholar] [CrossRef] [Green Version]
- Heras, G.; Namuduri, A.V.; Traini, L.; Shevchenko, G.; Falk, A.; Bergström Lind, S.; Jia, M.; Tian, G.; Gastaldello, S. Muscle RING-finger protein-1 (MuRF1) functions and cellular localization are regulated by SUMO1 post-translational modification. J. Mol. Cell Biol. 2019, 11, 356–370. [Google Scholar] [CrossRef] [Green Version]
- Cadena, S.M.; Zhang, Y.; Fang, J.; Brachat, S.; Kuss, P.; Giorgetti, E.; Stodieck, L.S.; Kneissel, M.; Glass, D.J. Skeletal muscle in MuRF1 null mice is not spared in low-gravity conditions, indicating atrophy proceeds by unique mechanisms in space. Sci. Rep. 2019, 9, 9397. [Google Scholar] [CrossRef]
- Beehler, B.C.; Sleph, P.G.; Benmassaoud, L.; Grover, G.J. Reduction of Skeletal Muscle Atrophy by a Proteasome Inhibitor in a Rat Model of Denervation. Exp. Biol. Med. 2006, 231, 335–341. [Google Scholar] [CrossRef]
- EI Ilyina-kakueva, P.V. Combined effect of space flight and radiation on skeletal muscles of rats. Aviat. Space Environ. Med. 1977, 48, 115–119. [Google Scholar]
- Harding, C.; Takemoto, J.; Vargis, E. In Vitro Modeling of Microgravity-Induced Muscle Atrophy and Spaceflight Radiation. NASA Sp. Grant Consort. 2016, 1–11. Available online: https://digitalcommons.usu.edu/spacegrant/2016/Session4/3/ (accessed on 17 January 2022).
- Comfort, P.; McMahon, J.J.; Jones, P.A.; Cuthbert, M.; Kendall, K.; Lake, J.P.; Haff, G.G. Effects of Spaceflight on Musculoskeletal Health: A Systematic Review and Meta-analysis, Considerations for Interplanetary Travel. Sports Med. 2021, 51, 2097–2114. [Google Scholar] [CrossRef] [PubMed]
- Juhl, O.J.; Buettmann, E.G.; Friedman, M.A.; DeNapoli, R.C.; Hoppock, G.A.; Donahue, H.J. Update on the effects of microgravity on the musculoskeletal system. npj Microgravity 2021, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Van Loon, J.J.W.A.; Bloomfield, S.; Vico, L.; Chopard, A.; Rittweger, J.; Kyparos, A.; Blottner, D.; Vuori, I.; Gerzer, R.; et al. Towards human exploration of space: The THESEUS review series on muscle and bone research priorities. npj Microgravity 2017, 3, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manian, V.; Orozco-Sandoval, J.; Diaz-Martinez, V. An Integrative Network Science and Artificial Intelligence Drug Repurposing Approach for Muscle Atrophy in Spaceflight Microgravity. Front. Cell Dev. Biol. 2021, 9, 2434. [Google Scholar] [CrossRef]
- NASA Gene Lab Data Repository. Available online: Genelab.nasa.gov (accessed on 7 September 2021).
- Radiation Data for GLDS Studies on the International Space Station. Available online: https://genelab.nasa.gov/environmental/radiation/iss (accessed on 27 October 2021).
- Manian, V.; Gangapuram, H.; Orozco, J.; Janwa, H.; Agrinsoni, C. Network analysis of local gene regulators in Arabidopsis thaliana under spaceflight stress. Computers 2021, 10, 18. [Google Scholar] [CrossRef]
- Ram, R.; Chetty, M. A Markov-blanket-based model for gene regulatory network inference. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011, 8, 353–367. [Google Scholar] [CrossRef]
- Scalable Precision Medicine Knowledge Engine. Available online: https://spoke.ucsf.edu/ (accessed on 23 September 2021).
- Nelson, C.A.; Acuna, A.U.; Paul, A.M.; Scott, R.T.; Butte, A.J.; Cekanaviciute, E.; Baranzini, S.E.; Costes, S.V. Knowledge network embedding of transcriptomic data from spaceflown mice uncovers signs and symptoms associated with terrestrial diseases. Life 2021, 11, 42. [Google Scholar] [CrossRef]
- Janwa, H.; Massey, S.E.; Velev, J.; Mishra, B. On the origin of biomolecular networks. Front. Genet. 2019, 10, 240–257. [Google Scholar] [CrossRef] [Green Version]
- Biggs, N. Algebraic Graph Theory, 2nd ed.; Cambridge Mathematical Library; Cambridge University Press: Cambridge, UK, 1993. [Google Scholar]
- Serre, J.-P. Trees; Springer: Berlin/Heidelberg, Germany, 1980. [Google Scholar]
- Loscalzo, J.; Barabási, A.-L. Network Science, 3rd ed.; Cambridge University Press: Cambridge, UK, 2016. [Google Scholar]
- Thulasiraman, K.; Arumugam, S.; Nishizeki, T.; Brandstädt, A. Handbook of Graph Theory, Combinatorial Optimization, and Algorithms; Taylor & Francis: Abingdon, UK, 2016. [Google Scholar]
- Barabási, A.-L. Network Science, 1st ed.; Royal Society: London, UK, 2014. [Google Scholar]
- Barabási, A.L.; Loscalzo, J.; Silverman, E.K. (Eds.) Network Medicine: Complex Systems in Human Disease and Therapeutic; Harvard University Press: Harvard, MA, USA, 2017. [Google Scholar]
- Abbas, K.; Abbasi, A.; Dong, S.; Niu, L.; Yu, L.; Chen, B.; Cai, S.-M.; Hasan, Q. Application of network link prediction in drug discovery. BMC Bioinform. 2021, 22, 187. [Google Scholar] [CrossRef]
- Grover, A.; Leskovec, J. Node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 855–864. [Google Scholar]
- Liben-Nowell, D.; Kleinberg, J. The link-prediction problem for social networks. J. Am. Soc. Inf. Sci. Technol. 2007, 58, 1019–1031. [Google Scholar] [CrossRef] [Green Version]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. In Proceedings of the 5th International Conference on Learning Representations, Toulon, France, 22 February 2017. [Google Scholar]
- Manian, V.; Orozco-sandoval, J.; Diaz-Martinez, V. Detection of Genes in Arabidopsis thaliana L. Responding to DNA Damage from Radiation and Other Stressors in Spaceflight. Genes 2021, 12, 938. [Google Scholar] [CrossRef] [PubMed]
- DrugBank Online. Available online: https://go.drugbank.com/ (accessed on 23 September 2021).
- GeneCards®: The Human Gene Database. Available online: https://doi.org/10.1093/database/baq020 (accessed on 23 September 2021).
- Kenny, H.C.; Tascher, G.; Ziemianin, A.; Rudwill, F.; Zahariev, A.; Chery, I.; Gauquelin-Koch, G.; Barielle, M.; Heer, M.; Blanc, S.; et al. Effectiveness of Resistive Vibration Exercise and Whey Protein Supplementation Plus Alkaline Salt on the Skeletal Muscle Proteome Following 21 Days of Bed Rest in Healthy Males. J. Proteome Res. Am. Chem. Soc. 2020, 19, 3438–3451. [Google Scholar] [CrossRef] [PubMed]
- Zatyka, M.; Sarkar, S.; Barrett, T. Autophagy in Rare (NonLysosomal) Neurodegenerative Diseases. J. Mol. Biol. 2020, 432, 2735–2753. [Google Scholar] [CrossRef] [PubMed]
- Calura, E.; Cagnin, S.; Raffaello, A.; Laveder, P.; Lanfranchi, G.; Romualdi, C. Meta-analysis of expression signatures of muscle atrophy: Gene interaction networks in early and late stages. BMC Genom. 2008, 9, 630. [Google Scholar] [CrossRef] [Green Version]
- Gomes-Duarte, A.; Lacerda, R.; Menezes, J.; Romão, L. eIF3: A factor for human health and disease. RNA Biol. 2018, 15, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Sandri, M. Protein breakdown in muscle wasting: Role of autophagy-lysosome and ubiquitin-proteasome. Int. J. Biochem. Cell Biol. 2013, 45, 2121–2129. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.V.; Peralta, S.; Arguello, T.; Bacman, S.R.; Diaz, F.; Moraes, C.T. Myopathy reversion in mice after restauration of mitochondrial complex I. EMBO Mol. Med. 2020, 12, e10674. [Google Scholar] [CrossRef]
- Hunt, L.C.; Graca, F.A.; Pagala, V.; Wang, Y.D.; Li, Y.; Yuan, Z.F.; Fan, Y.; Labelle, M.; Peng, J.; Demontis, F. Integrated genomic and proteomic analyses identify stimulus-dependent molecular changes associated with distinct modes of skeletal muscle atrophy. Cell Rep. 2021, 37, 109971. [Google Scholar] [CrossRef]
- Nath, S.R.; Lieberman, M.L.; Yu, Z.; Marchioretti, C.; Jones, S.T.; Danby, E.C.E.; Van Pelt, K.M.; Sorarù, G.; Robins, D.M.; Bates, G.P.; et al. MEF2 impairment underlies skeletal muscle atrophy in polyglutamine disease. Acta Neuropathol. 2020, 140, 63–80. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.T.; Yang, Y.J.; Huang, R.H.; Zhang, Z.H.; Lin, X. Myostatin Activates the Ubiquitin-Proteasome and Autophagy-Lysosome Systems Contributing to Muscle Wasting in Chronic Kidney Disease. Oxid. Med. Cell. Longev. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Salanova, M.; Gambara, G.; Moriggi, M.; Vasso, M.; Ungethuem, U.; Belavý, D.L.; Felsenberg, D.; Cerretelli, P.; Gelfi, C.; Blottner, D. Vibration mechanosignals superimposed to resistive exercise result in baseline skeletal muscle transcriptome profiles following chronic disuse in bed rest. Sci. Rep. 2015, 5, 17027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, D.M.; Cannavino, J.; Li, H.; Anderson, K.M.; Nelson, B.R.; McAnally, J.; Bezprozvannaya, S.; Liu, Y.; Lin, W.; Liu, N.; et al. Severe muscle wasting and denervation in mice lacking the RNA-binding protein ZFP106. Proc. Natl. Acad. Sci. USA 2016, 113, E4494–E4503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geister, K.A.; Lopez-Jimenez, A.J.; Houghtaling, S.; Ho, T.H.; Vanacore, R.; Beier, D.R. Loss of function of Colgalt1 disrupts collagen post-translational modification and causes musculoskeletal defects. DMM Dis. Model. Mech. 2019, 12, dmm037176. [Google Scholar] [CrossRef] [Green Version]
- Massart, I.S.; Paulissen, G.; Loumaye, A.; Lause, P.; Pötgens, S.A.; Thibaut, M.M.; Balan, E.; Deldicque, L.; Atfi, A.; Louis, E.; et al. Marked increased production of acute phase reactants by skeletal muscle during cancer cachexia. Cancers 2020, 12, 3221. [Google Scholar] [CrossRef] [PubMed]
- So, H.K.; Kim, S.; Kang, J.S.; Lee, S.J. Role of Protein Arginine Methyltransferases and Inflammation in Muscle Pathophysiology. Front. Physiol. 2021, 12, 712389. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Haider, S. Protein Network Analysis to Prioritize Key Genes in Amyotrophic Lateral Sclerosis. IBRO Neurosci. Rep. 2021, 12, 25–44. [Google Scholar] [CrossRef]
- Kan, J.; Hu, Y.; Ge, Y.; Zhang, W.S.; Lu, S.; Zhao, C.; Zhang, R.; Liu, Y. Declined expressions of vast mitochondria-related genes represented by CYCS and transcription factor ESRRA in skeletal muscle aging. Bioengineered 2021, 12, 3485–3502. [Google Scholar] [CrossRef]
- Montesano, A.; Senesi, P.; Luzi, L.; Benedini, S.; Terruzzi, I. Potential therapeutic role of L-carnitine in skeletal muscle oxidative stress and atrophy conditions. Oxid. Med. Cell. Longev. 2015, 2015, 646171. [Google Scholar] [CrossRef]
- Costantini, A.; Trevi, E.; Pala, M.I.; Fancellu, R. Can long-term thiamine treatment improve the clinical outcomes of myotonic dystrophy type 1? Neural Regen. Res. 2016, 11, 1487–1491. [Google Scholar] [CrossRef]
- Li, Y.; Chen, M.; Zhao, Y.; Li, M.; Qin, Y.; Cheng, S.; Yang, Y.; Yin, P.; Zhang, L.; Tang, P. Advance in Drug Delivery for Ageing Skeletal Muscle. Front. Pharmacol. 2020, 11, 1016. [Google Scholar] [CrossRef]
- Marchetti, N. Anabolic medications for muscle wasting in chronic obstructive pulmonary disease: Is the evidence getting stronger? Am. J. Respir. Crit. Care Med. 2019, 199, 256–258. [Google Scholar] [CrossRef] [PubMed]
- Guasconi, V.; Puri, P.L. Epigenetic drugs in the treatment of skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 2008, 11, 233–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Lee, J.H.; Kim, S.W.; Lee, S.H.; Jung, D.W.; Williams, D.R. Investigation of niclosamide as a repurposing agent for skeletal muscle atrophy. PLoS ONE 2021, 16, e0252135. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.C.; Chiu, C.Y.; Yang, R.S.; Chan, D.C.; Liu, S.H.; Chiang, C.K. Preventing muscle wasting by osteoporosis drug alendronate in vitro and in myopathy models via sirtuin-3 down-regulation. J. Cachexia. Sarcopenia Muscle 2018, 9, 585–602. [Google Scholar] [CrossRef] [Green Version]
- Campins, L.; Camps, M.; Riera, A.; Pleguezuelos, E.; Yebenes, J.C.; Serra-Prat, M. Oral drugs related with muscle wasting and sarcopenia. A review. Pharmacology 2017, 99, 1–8. [Google Scholar] [CrossRef]

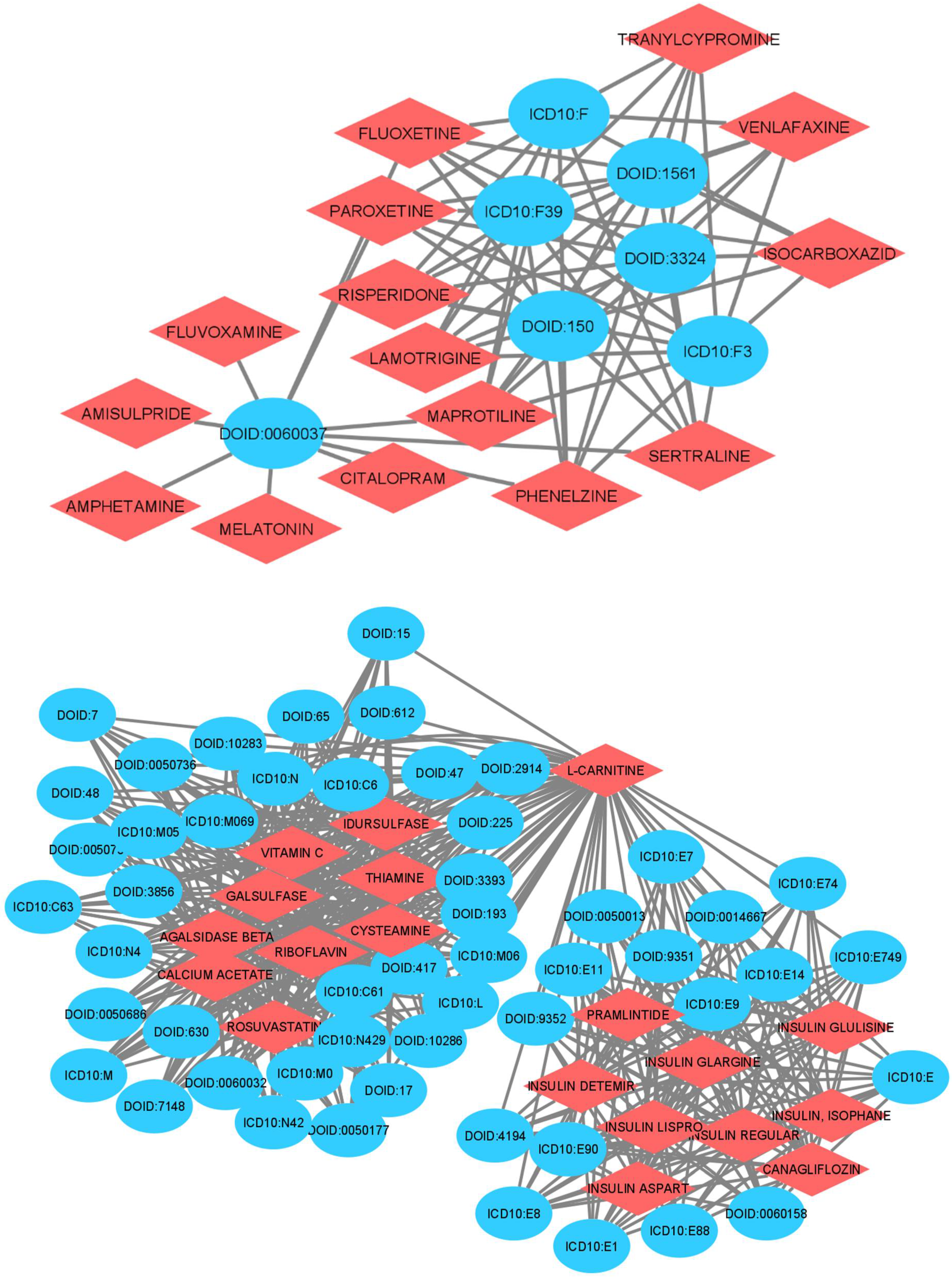
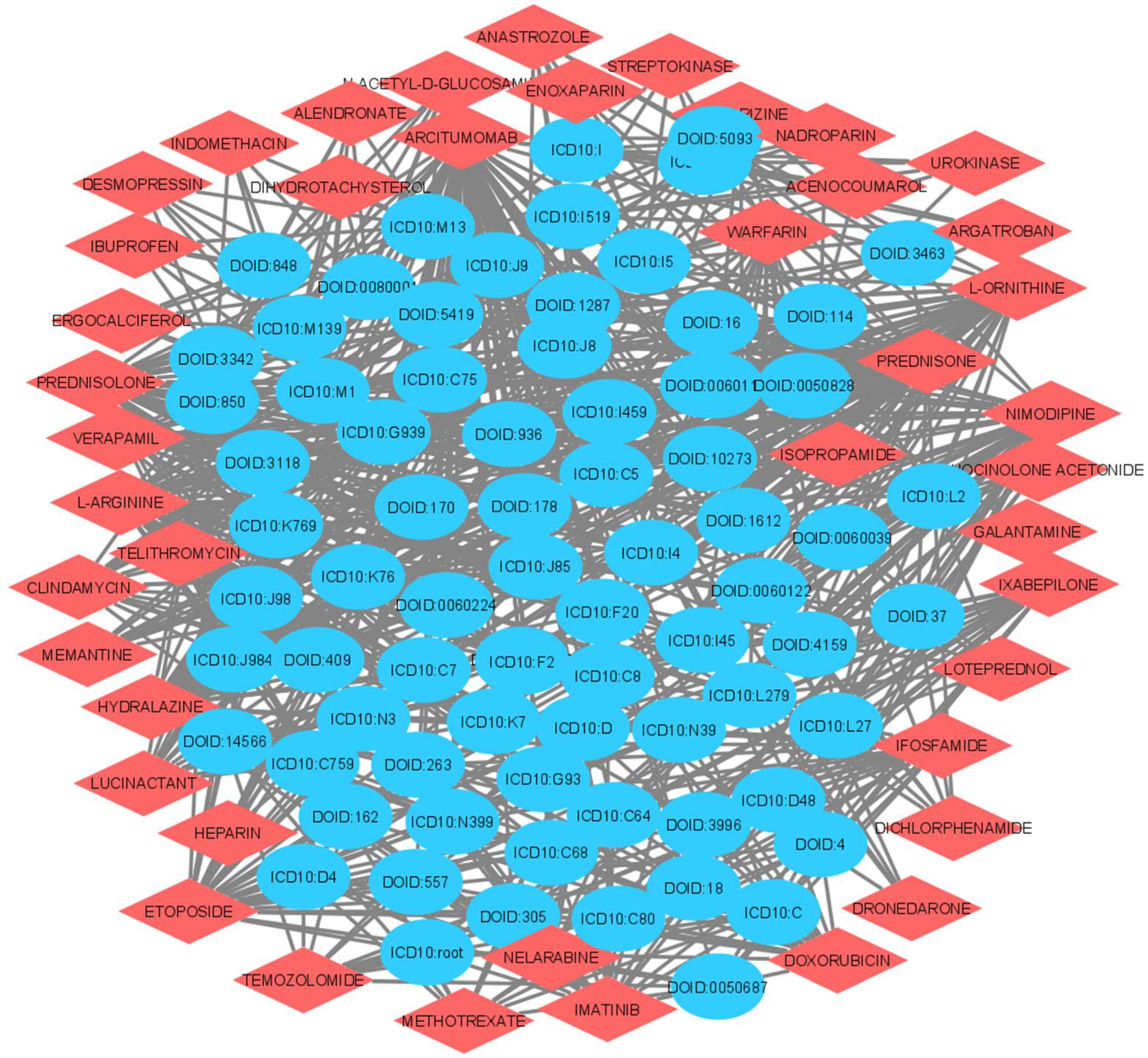
| Gene | Disease Code | Link Prediction Probabilities | Disease Name |
|---|---|---|---|
| EIF3H | ICD10:C22 | 0.92 | Malignant neoplasm of liver and intrahepatic bile ducts |
| SNF8 | DOID:178 | 0.83 | vascular disease |
| RPS25 | ICD10:G969 | 0.77 | Disorder of central nervous system |
| NDUFB2 | DOID:0050589 | 0.79 | inflammatory bowel disease |
| MTCH2 | DOID:10273 | 0.95 | heart conduction disease |
| FTO | DOID:1289 | 0.79 | neurodegenerative disease |
| NDUFS3 | ICD10:I5 | 0.97 | Non-ischemic myocardial injury (non-traumatic) |
| MEF2A | ICD10:C25 | 0.79 | Malignant neoplasm of the pancreas |
| DDA1 | DOID:8857 | 0.84 | lupus erythematosus |
| ATG3 | ICD10:H8 | 0.8 | disorder of vestibular function |
| COG6 | ICD10:N429 | 0.73 | Disorder of prostate |
| ELK4 | DOID:6364 | 0.71 | migraine |
| MRPL4 | DOID:2007 | 0.73 | Pesticide residues in food |
| ZFR | ICD10:N399 | 0.78 | Disorder of urinary system |
| ELK4 | ICD10:G93 | 0.89 | brain disorder |
| COLGALT1 | DOID:0050890 | 0.84 | synucleinopathy |
| RPL7A | ICD10:K0 | 0.81 | Diseases of the oral cavity and salivary glands |
| PRMT5 | ICD10:N399 | 0.86 | Disorder of urinary system |
| MRPS21 | DOID:0050687 | 0.89 | cell type cancer |
| SNW1 | ICD10:C64 | 0.83 | Malignant neoplasm of kidney |
| Methods | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | AUROC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RF | 89.39 | 88.64 | 90.54 | 91.76 | 89.09 | 88.57 | 86.04 | 91.03 | 87.66 | 90.94 | 88.75 |
| GB | 85.28 | 84.62 | 87.03 | 86.82 | 85.62 | 86.52 | 83.22 | 87.33 | 82.89 | 88.06 | 85.69 |
| GNN | 87.70 | 90.13 | 89.78 | 89.96 | 88.79 | 90.59 | 85.60 | 89.44 | 88.35 | 90.26 | 88.63 |
| GCN | 88.95 | 90.87 | 91.21 | 92.79 | 93.00 | 93.20 | 94.29 | 95.05 | 95.72 | 96.00 | 96.11 |
| Drugs | Disease | Link Prediction Probability |
|---|---|---|
| L-CARNITINE | Metabolic disease | 1 |
| THIAMINE | Autoimmune disease of the musculoskeletal system | 1 |
| TELITHROMYCIN | Breast cancer | 0.98 |
| FLUOCINOLONE ACETONIDE | Uterine disease | 0.96 |
| RIBOFLAVIN | Autoimmune disease of the musculoskeletal system | 0.94 |
| AZATHIOPRINE | Cardiovascular system disease | 0.94 |
| IVERMECTIN | Allergic rhinitis | 0.9 |
| INSULIN LISPRO | Urinary system disease | 0.9 |
| NELARABINE | Hypervitaminosis | 0.9 |
| SURAMIN | Allergic rhinitis | 0.89 |
| TETRACYCLINE | Male reproductive organ cancer | 0.86 |
| INSULIN DETEMIR | Urinary system disease | 0.85 |
| PRAMLINTIDE | Type 2 diabetes mellitus | 0.84 |
| ARCITUMOMAB | Breast cancer | 0.83 |
| CLINDAMYCIN | Influenza and pneumonia | 0.83 |
| L-ORNITHINE | Vasomotor and allergic rhinitis | 0.83 |
| BUDESONIDE | Autoimmune thyroiditis | 0.82 |
| GOLIMUMAB | Benign neoplasm | 0.82 |
| ARCITUMOMAB | Skin disease | 0.82 |
| INSULIN, ISOPHANE | Unspecified diabetes mellitus | 0.82 |
| HYDROCORTISONE | Integumentary system cancer | 0.82 |
| CHLOROQUINE | Bone inflammation disease | 0.82 |
| L-CARNITINE | Malignant neoplasm | 0.82 |
| INSULIN GLARGINE | Disease of the genitourinary system | 0.81 |
| KETOCONAZOLE | Allergic rhinitis | 0.8 |
| WARFARIN | Generalized skin eruption | 0.79 |
| ARCITUMOMAB | Nasal cavity disease | 0.79 |
| KETOCONAZOLE | Malignant neoplasm of prostate | 0.79 |
| VITAMIN C | Lung disease | 0.78 |
| GALSULFASE | Malignant neoplasm of other endocrine glands | 0.77 |
| L-ORNITHINE | Arterial fibrillation | 0.75 |
| LUCINACTANT | Mood disorder | 0.75 |
| VITAMIN C | Mental, behavioral and neurodevelopmental disorders | 0.74 |
| TETRACYCLINE | Allergic rhinitis | 0.74 |
| SURAMIN | Other disorders of central nervous system | 0.73 |
| SULFASALAZINE | Other and unspecified noninfective gastroenteritis and colitis | 0.71 |
| TINIDAZOLE | Bronchial disease | 0.71 |
| Drugs. | Disease Name | Link Prediction Probability |
|---|---|---|
| MEMANTINE | Carcinoma | 0.98 |
| CINNARIZINE | Carcinoma | 0.97 |
| MEMANTINE | Heart Disease | 0.97 |
| IXABEPILONE | Complications Additionally, Ill-Defined Descriptions Of Heart Disease | 0.96 |
| PREDNISOLONE | Malignant Neoplasm of Other Additionally, Unspecified Urinary Organs | 0.95 |
| CLINDAMYCIN | Artery Disease | 0.93 |
| CLINDAMYCIN | Urinary System Disease | 0.93 |
| LUCINACTANT | Malignant Neoplasm of Other Additionally, Unspecified Major Salivary Glands | 0.93 |
| CINNARIZINE | Cancer | 0.93 |
| ETOPOSIDE | Artery Disease | 0.93 |
| L-ORNITHINE | Carcinoma | 0.92 |
| LUCINACTANT | Disorder Of Urinary System | 0.92 |
| IMATINIB | Heart Conduction Disease | 0.91 |
| L-ORNITHINE | Heart Disease | 0.89 |
| NELARABINE | Heart Conduction Disease | 0.88 |
| NIMODIPINE | Abscess Of Lung Additionally, Mediastinum | 0.87 |
| METHOTREXATE | Integumentary System Cancer | 0.86 |
| PREDNISOLONE | In Situ Neoplasms | 0.85 |
| MELATONIN | Cognitive Disorder | 0.85 |
| TEMOZOLOMIDE | Other Disorders of Urinary System | 0.84 |
| ANASTROZOLE | Malignant Neoplasm of Other Endocrine Glands Additionally, Related Structures | 0.82 |
| FLUOCINOLONE ACETONIDE | Other Diseases of Liver | 0.79 |
| AGALSIDASE β | Carbohydrate Metabolism Disease | 0.77 |
| CALCIUM ACETATE | Type 2 Diabetes Mellitus | 0.75 |
| CYSTEAMINE | Other Disorders of Carbohydrate Metabolism | 0.74 |
| VITAMIN C | Type 2 Diabetes Mellitus | 0.74 |
| L-CARNITINE | Autosomal Dominant Disease | 0.70 |
| IBUPROFEN | Cardiovascular System Disease | 0.70 |
| Methods | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | AUROC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RF | 96.69 | 99.44 | 99.60 | 98.05 | 99.88 | 99.65 | 98.34 | 98.86 | 99.68 | 99.52 | 98.09 |
| GB | 92.10 | 97.12 | 99.80 | 91.60 | 99.69 | 96.83 | 97.07 | 94.86 | 97.32 | 98.39 | 96.19 |
| GNN | 95.55 | 99.36 | 95.56 | 95.42 | 98.62 | 99.22 | 97.98 | 95.18 | 99.86 | 100.00 | 97.70 |
| GCN | 99.75 | 100.00 | 99.75 | 99.872 | 99.87 | 100.00 | 99.75 | 100.00 | 100.00 | 99.87 | 99.19 |
| Network Measure | GDKG | DDKG |
|---|---|---|
| Spectral gap | 37.5218 | 99.7221 |
| Density | 0.0221 | 0.0452 |
| Average number of neighbors | 26.423 | 13.345 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manian, V.; Orozco-Sandoval, J.; Diaz-Martinez, V.; Janwa, H.; Agrinsoni, C. Detection of Target Genes for Drug Repurposing to Treat Skeletal Muscle Atrophy in Mice Flown in Spaceflight. Genes 2022, 13, 473. https://doi.org/10.3390/genes13030473
Manian V, Orozco-Sandoval J, Diaz-Martinez V, Janwa H, Agrinsoni C. Detection of Target Genes for Drug Repurposing to Treat Skeletal Muscle Atrophy in Mice Flown in Spaceflight. Genes. 2022; 13(3):473. https://doi.org/10.3390/genes13030473
Chicago/Turabian StyleManian, Vidya, Jairo Orozco-Sandoval, Victor Diaz-Martinez, Heeralal Janwa, and Carlos Agrinsoni. 2022. "Detection of Target Genes for Drug Repurposing to Treat Skeletal Muscle Atrophy in Mice Flown in Spaceflight" Genes 13, no. 3: 473. https://doi.org/10.3390/genes13030473
APA StyleManian, V., Orozco-Sandoval, J., Diaz-Martinez, V., Janwa, H., & Agrinsoni, C. (2022). Detection of Target Genes for Drug Repurposing to Treat Skeletal Muscle Atrophy in Mice Flown in Spaceflight. Genes, 13(3), 473. https://doi.org/10.3390/genes13030473






