Monogenic Parkinson’s Disease: Genotype, Phenotype, Pathophysiology, and Genetic Testing
Abstract
1. Introduction
2. Autosomal Dominant Forms
2.1. SNCA
2.1.1. Genotype-Phenotype
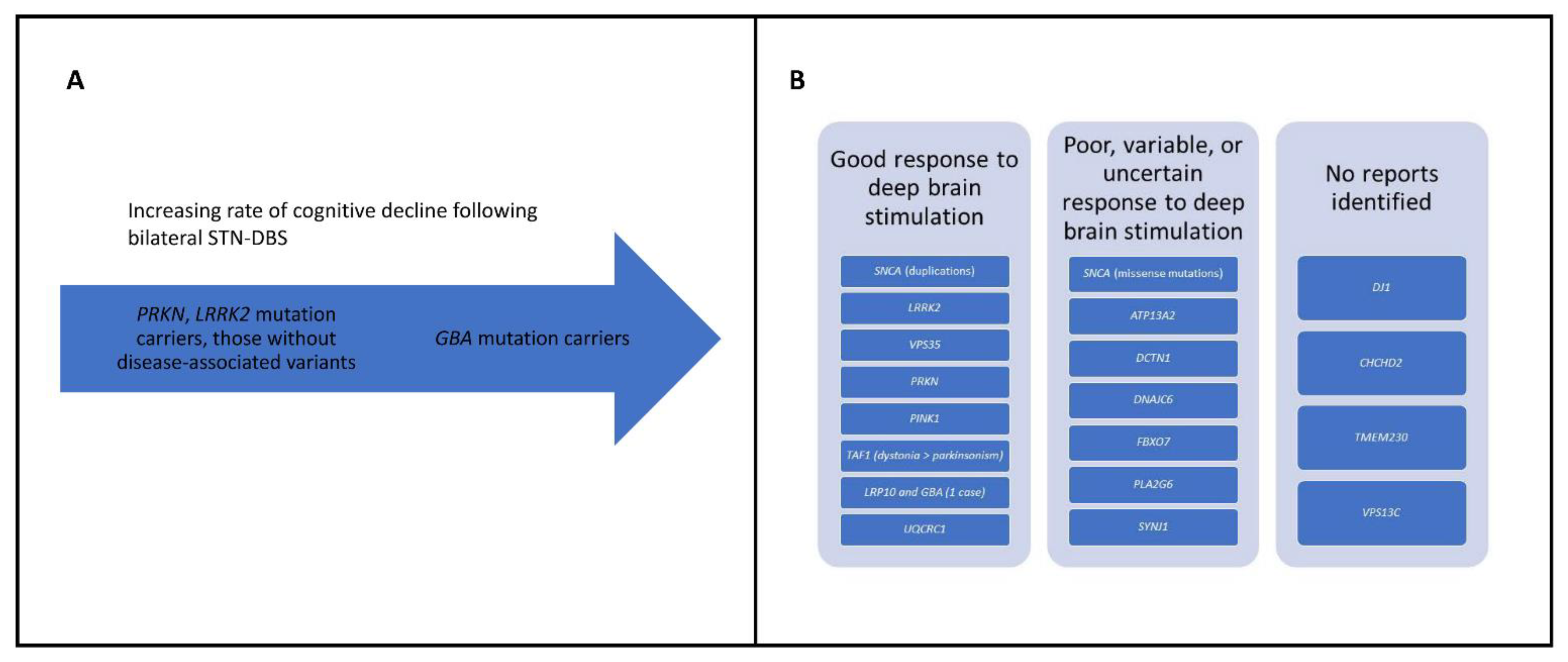
| Gene | Mode of Inheritance | Frequency | Ethnic Population Distribution | Types of Mutations | Clinical Phenotype | Response to PD Medication | Response to DBS | Pathological Findings |
|---|---|---|---|---|---|---|---|---|
| SNCA | AD | Rare, with a frequency from 0.045% to 1.1% in recent studies [9] | Majority European, then Asians and Hispanics [5] | Missense, duplications, and triplications | Range of age at onset, prominent motor fluctuations, range of complications including cognitive impairment and psychiatric manifestations | Initial good response | Few examples, appears to have a good response for duplications, poor response for missense mutations | α-synuclein-positive and LB pathology [10] |
| LRRK2 | AD | 1% of PD but higher in North African Berber Arab and Ashkenazi Jewish populations | The p.G2019S mutation found in Europeans with high prevalence in North African Berbers and Ashkenazi Jews | 7 missense variants described | Resembles idiopathic PD | Vast majority show a good response to levodopa [11] | DBS is effective [12] | Most patients with the p.G2019S mutation show LB pathology, whereas this finding is rare for other mutations |
| VPS35 | AD | Rare (overall prevalence of 0.115%) | European, Asian, Ashkenazi Jewish [5] | 1 missense mutation described, p.D620N | Resembles idiopathic PD | Good response [11] | Small numbers reported, at least 2 had a good outcome [11] | Not available |
| PRKN | AR | Most common cause of EOPD, 12.5% of recessive PD [13] | Majority Asian, followed by Caucasians and Hispanics [14] | Missense mutations, frameshift mutations, structural variants | EOPD, lower limb dystonia, absence of cognitive impairment | Good response to levodopa therapy, frequent motor fluctuations, and dyskinesias | Good outcome in all patients [11] | Substantia nigra pars compacta loss with the notable absence of LB pathology |
| PINK1 | AR | Second most common cause of EOPD, 1.9% of recessive PD [13] | European, Asian, may be frequent in Arab Berber and Polynesian populations [9,14,15] | Missense mutations, nonsense mutations, structural variants | EOPD, typical PD, dyskinesias, dystonia, and motor fluctuations can occur | Vast majority show a good outcome [11] | Good or moderate [11] | LB pathology may or may not be present in the handful of autopsy cases reported |
| PARK7 (DJ1) | AR | 0.16% of recessive PD [13] | Most patients are from Italy, Iran, and Turkey [14] | Missense, splice site, frameshift, and structural variants | EOPD | 50% show a good response, others moderate or minimal [11] | No reports identified [11] | LB pathology [16] |
| TAF1 | X-linked | 0.34 per 100,000 in the Philippines, Island of Panay 5.24 per 100,000 | Philippines, high prevalence on the Island of Panay | Insertion of a SINE-VNTR-Alu type retrotransposon in intron 32 of the TAF1 gene | Parkinsonism, dystonia | May be responsive to levodopa, particularly for those with pure parkinsonism [17] | DBS results in an improvement in dystonia and to a lesser extent parkinsonism [18] | Accumulation of lipofuscin in the neurons and glia, but absence of LB pathology |
| ATP13A2 | AR | Rare | Spread across the globe [19] | Frameshift, missense, and splice site mutations [19] | KRS, clinical triad of spasticity, dementia, and supranuclear gaze palsy [20], facial-faucial-finger mini-myoclonus [21], other phenotypes include HSP | Variable response to levodopa [19] | May respond well, variable [22] | Accumulation of lipofuscin, absence of LB pathology [23] |
| DCTN1 | AD | Rare | Spread across the globe | 10 different heterozygous missense mutations [19] | Perry syndrome—rapidly progressive parkinsonism, depression and mood changes, weight loss, and progressive respiratory changes | May be levodopa-responsive | No reports identified | Selective loss of putative respiratory neurons in the ventrolateral medulla and in the raphe nucleus, no or few LBs, TDP43-positive inclusions [24,25] |
| DNAJC6 | AR | Rare | Mainly found in Middle Eastern populations, although families of European origin have also been found to harbor DNAJC6 mutations | 5 different homozygous mutations, largest family carries a nonsense mutation [19] | Juvenile PD with complicating features, EOPD | Poor | Good outcome [26] | No reports identified |
| FBXO7 | AR | Rare | Reported in the Iranian, Turkish, Italian, Dutch, Pakistani, and Chinese populations | Biallelic missense, splice site, and nonsense mutations | Juvenile PD, EOPD, parknsonian-pyramidal syndrome, can overlap with NBIA [27] | Variable | No reports identified | No reports identified |
| PLA2G6 | AR | Rare | Various ethnic groups, including Indian, Pakistani, European, Japanese, Chinese, and Korean populations | 54 mutations associated with parkinsonism [28] | Adult-onset dystonia-parkinsonism with cognitive and psychiatric symptoms [28], other phenotypes include NBIA | Variable | May benefit from DBS [28] | Mixed Lewy and Tau pathology [28] |
| SYNJ1 | AR | Rare | Reported in Iranian, Italian, German, Algerian, Senegalese, and Chinese populations | Missense, frameshift | Parkinsonism in the third decade of life, complicating features such as dystonia, seizures, or cognitive impairment | Poor | No reports identified | No reports identified |
| CHCHD2 | AD | Rare | Japanese and Chinese patients | Missense, splice site | Typical PD | Good | No reports identified | A brain autopsy revealed widespread α-synuclein pathology with Lewy bodies present in the brainstem, neocortex, and limbic regions [29] |
| LRP10 | AD | Rare | Italy, Taiwan | Loss of function and missense variants | Late onset PD, PD dementia, dementia with Lewy Bodies [30,31,32] | Good [32] | Excellent response for a patient with a LRP10 and GBA variant in trans [33] | Severe LB pathology |
| TMEM230 | AD | Rare | Identified in a Canadian Mennonite family | Missense variant | Typical PD | Responds to levodopa in most cases | No report identified | Typical LB pathology [34] |
| UQCRC1 | AD | Rare | Taiwan, may not be in European populations | Missense variants | Parkinsonism with polyneuropathy | Good | No report identified | No report identified |
| VPS13C | AR | Rare | Turkish, French | Truncating mutations | EOPD, rapid progression, complicating features including dysphagia, cognitive impairment, hyperreflexia [19,35] | Initial good response | Poor | Resembles diffuse LB disease [35] |
2.1.2. Pathophysiology
| Good Response to Levodopa | Poor, Variable, or Uncertain Response to Levodopa |
|---|---|
| SNCA | TAF1 |
| LRRK2 | ATP13A2 |
| VPS35 | DCTN1 |
| PRKN | DNAJC6 |
| PINK1 | FBXO7 |
| DJ1 | PLA2G6 |
| CHCHD2 | SYNJ1 |
| LRP10 | |
| TMEM230 | |
| UQCRC1 | |
| VPS13C |
2.2. LRRK2
2.2.1. Genotype-Phenotype
2.2.2. Pathophysiology
2.3. VPS35
2.3.1. Genotype-Phenotype
2.3.2. Pathophysiology
3. Autosomal Recessive Forms
3.1. PRKN
3.1.1. Genotype-Phenotype
3.1.2. Pathophysiology
3.2. PINK1
3.2.1. Genotype-Phenotype
3.2.2. Pathophysiology
3.3. PARK7
3.3.1. Genotype-Phenotype
3.3.2. Pathophysiology
4. X-Linked Dystonia-Parkinsonism
4.1. Genotype-Phenotype
4.2. Pathophysiology
5. Complex or Atypical Forms
5.1. ATP13A2
5.1.1. Genotype-Phenotype
5.1.2. Pathophysiology
5.2. DCTN1
5.2.1. Genotype-Phenotype
5.2.2. Pathophysiology
5.3. DNAJC6
5.3.1. Genotype-Phenotype
5.3.2. Pathophysiology
5.4. FBXO7
5.4.1. Genotype-Phenotype
5.4.2. Pathophysiology
5.5. PLA2G6
5.5.1. Genotype-Phenotype
5.5.2. Pathophysiology
5.6. SYNJ1
5.6.1. Genotype-Phenotype
5.6.2. Pathophysiology
6. Recently Described Parkinson’s Disease Genes
6.1. CHCHD2
6.1.1. Genotype-Phenotype
6.1.2. Pathophysiology
6.2. LRP10
6.2.1. Genotype-Phenotype
6.2.2. Pathophysiology
6.3. TMEM230
6.3.1. Genotype-Phenotype
6.3.2. Pathophysiology
6.4. UQCRC1
6.4.1. Genotype-Phenotype
6.4.2. Pathophysiology
6.5. VPS13C
6.5.1. Genotype-Phenotype
6.5.2. Pathophysiology
7. Rare, Atypical, and Unconfirmed Forms
8. Risk Variants versus Monogenic Forms
9. GBA Variants
10. Genetic Testing in Parkinson’s Disease
10.1. Who Should Be Offered Genetic Testing in Parkinson’s Disease?
10.2. The Implications of a Genetic Diagnosis in Parkinson’s Disease
10.3. Challenges in Genetic Testing
11. Role for Heterozygosity in Autosomal Recessive Parkinson Genes
11.1. PRKN Heterozygotes
11.2. PINK1 Heterozygotes
11.3. Conclusion on Heterozygous Carriers
12. Dual LRRK2 and GBA Mutation Carriers
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, K.R.; Lohmann, K.; Klein, C. Genetics of Parkinson disease and other movement disorders. Curr. Opin. Neurol. 2012, 25, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Panicker, N.; Ge, P.; Dawson, V.L.; Dawson, T.M. The cell biology of Parkinson’s disease. J. Cell Biol. 2021, 220, e202012095. [Google Scholar] [CrossRef] [PubMed]
- Senkevich, K.; Rudakou, U.; Gan-Or, Z. New therapeutic approaches to Parkinson’s disease targeting GBA, LRRK2 and Parkin. Neuropharmacology 2022, 202, 108822. [Google Scholar] [CrossRef] [PubMed]
- Prasuhn, J.; Davis, R.L.; Kumar, K.R. Targeting Mitochondrial Impairment in Parkinson’s Disease: Challenges and Opportunities. Front. Cell Dev. Biol. 2020, 8, 615461. [Google Scholar] [CrossRef]
- Trinh, J.; Zeldenrust, F.M.J.; Huang, J.; Kasten, M.; Schaake, S.; Petkovic, S.; Madoev, H.; Grunewald, A.; Almuammar, S.; Konig, I.R.; et al. Genotype-phenotype relations for the Parkinson’s disease genes SNCA, LRRK2, VPS35: MDS Gene systematic review. Mov. Disord. 2018, 33, 1857–1870. [Google Scholar] [CrossRef]
- Blauwendraat, C.; Kia, D.A.; Pihlstrom, L.; Gan-Or, Z.; Lesage, S.; Gibbs, J.R.; Ding, J.; Alcalay, R.N.; Hassin-Baer, S.; Pittman, A.M.; et al. Insufficient evidence for pathogenicity of SNCA His50Gln (H50Q) in Parkinson’s disease. Neurobiol. Aging 2018, 64, 159.e5–159.e8. [Google Scholar] [CrossRef]
- Liu, H.; Koros, C.; Strohaker, T.; Schulte, C.; Bozi, M.; Varvaresos, S.; Ibanez de Opakua, A.; Simitsi, A.M.; Bougea, A.; Voumvourakis, K.; et al. A Novel SNCA A30G Mutation Causes Familial Parkinson’s Disease. Mov. Disord. 2021, 36, 1624–1633. [Google Scholar] [CrossRef]
- Book, A.; Guella, I.; Candido, T.; Brice, A.; Hattori, N.; Jeon, B.; Farrer, M.J.; SNCA Multiplication Investigators of the GEoPD Consortium. A Meta-Analysis of alpha-Synuclein Multiplication in Familial Parkinsonism. Front. Neurol. 2018, 9, 1021. [Google Scholar] [CrossRef]
- Lesage, S.; Houot, M.; Mangone, G.; Tesson, C.; Bertrand, H.; Forlani, S.; Anheim, M.; Brefel-Courbon, C.; Broussolle, E.; Thobois, S.; et al. Genetic and Phenotypic Basis of Autosomal Dominant Parkinson’s Disease in a Large Multi-Center Cohort. Front. Neurol. 2020, 11, 682. [Google Scholar] [CrossRef]
- Schneider, S.A.; Alcalay, R.N. Neuropathology of genetic synucleinopathies with parkinsonism: Review of the literature. Mov. Disord. 2017, 32, 1504–1523. [Google Scholar] [CrossRef]
- Over, L.; Bruggemann, N.; Lohmann, K. Therapies for Genetic Forms of Parkinson’s Disease: Systematic Literature Review. J. Neuromuscul. Dis. 2021, 8, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Leaver, K.; Viser, A.; Kopell, B.H.; Ortega, R.A.; Miravite, J.; Okun, M.S.; Elango, S.; Raymond, D.; Bressman, S.B.; Saunders-Pullman, R.; et al. Clinical profiles and outcomes of deep brain stimulation in G2019S LRRK2 Parkinson disease. J. Neurosurg. 2021, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lesage, S.; Lunati, A.; Houot, M.; Romdhan, S.B.; Clot, F.; Tesson, C.; Mangone, G.; Toullec, B.L.; Courtin, T.; Larcher, K.; et al. Characterization of recessive Parkinson’s disease in a large multicenter study. Ann. Neurol. 2020, 88, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Kasten, M.; Hartmann, C.; Hampf, J.; Schaake, S.; Westenberger, A.; Vollstedt, E.J.; Balck, A.; Domingo, A.; Vulinovic, F.; Dulovic, M.; et al. Genotype-Phenotype Relations for the Parkinson’s Disease Genes Parkin, PINK1, DJ1: MDSGene Systematic Review. Mov. Disord. 2018, 33, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.G.; Buchanan, C.M.; Mulroy, E.; Simpson, M.; Reid, H.A.; Drake, K.M.; Merriman, M.E.; Phipps-Green, A.; Cadzow, M.; Merriman, T.R.; et al. Potential PINK1 Founder Effect in Polynesia Causing Early-Onset Parkinson’s Disease. Mov. Disord. 2021, 36, 2199–2200. [Google Scholar] [CrossRef]
- Taipa, R.; Pereira, C.; Reis, I.; Alonso, I.; Bastos-Lima, A.; Melo-Pires, M.; Magalhaes, M. DJ-1 linked parkinsonism (PARK7) is associated with Lewy body pathology. Brain 2016, 139, 1680–1687. [Google Scholar] [CrossRef]
- Evidente, V.G.H. X-Linked Dystonia-Parkinsonism. In GeneReviews(R); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Bruggemann, N.; Domingo, A.; Rasche, D.; Moll, C.K.E.; Rosales, R.L.; Jamora, R.D.G.; Hanssen, H.; Munchau, A.; Graf, J.; Weissbach, A.; et al. Association of Pallidal Neurostimulation and Outcome Predictors With X-linked Dystonia Parkinsonism. JAMA Neurol. 2019, 76, 211–216. [Google Scholar] [CrossRef]
- Wittke, C.; Petkovic, S.; Dobricic, V.; Schaake, S.; Group, M.D.-e.P.S.; Respondek, G.; Weissbach, A.; Madoev, H.; Trinh, J.; Vollstedt, E.J.; et al. Genotype-Phenotype Relations for the Atypical Parkinsonism Genes: MDS Gene Systematic Review. Mov. Disord. 2021, 36, 1499–1510. [Google Scholar] [CrossRef]
- Park, J.S.; Blair, N.F.; Sue, C.M. The role of ATP13A2 in Parkinson’s disease: Clinical phenotypes and molecular mechanisms. Mov. Disord. 2015, 30, 770–779. [Google Scholar] [CrossRef]
- Williams, D.R.; Hadeed, A.; al-Din, A.S.; Wreikat, A.L.; Lees, A.J. Kufor Rakeb disease: Autosomal recessive, levodopa-responsive parkinsonism with pyramidal degeneration, supranuclear gaze palsy, and dementia. Mov. Disord. 2005, 20, 1264–1271. [Google Scholar] [CrossRef]
- Wang, D.; Gao, H.; Li, Y.; Jiang, S.; Yang, X. ATP13A2 Gene Variants in Patients with Parkinson’s Disease in Xinjiang. BioMed Res. Int. 2020, 2020, 6954820. [Google Scholar] [CrossRef] [PubMed]
- Chien, H.F.; Rodriguez, R.D.; Bonifati, V.; Nitrini, R.; Pasqualucci, C.A.; Gelpi, E.; Barbosa, E.R. Neuropathologic Findings in a Patient with Juvenile-Onset Levodopa-Responsive Parkinsonism Due to ATP13A2 Mutation. Neurology 2021, 97, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, Y.; Dickson, D.W.; Nabeshima, K.; Schmeichel, A.M.; Wszolek, Z.K.; Yamada, T.; Benarroch, E.E. Neurodegeneration involving putative respiratory neurons in Perry syndrome. Acta Neuropathol. 2008, 115, 263–268. [Google Scholar] [CrossRef]
- Tsuboi, Y.; Mishima, T.; Fujioka, S. Perry Disease: Concept of a New Disease and Clinical Diagnostic Criteria. J. Mov. Disord. 2021, 14, 1–9. [Google Scholar] [CrossRef]
- Kurian, M.A.; Abela, L. DNAJC6 Parkinson Disease. In GeneReviews(R); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Correa-Vela, M.; Lupo, V.; Montpeyo, M.; Sancho, P.; Marce-Grau, A.; Hernandez-Vara, J.; Darling, A.; Jenkins, A.; Fernandez-Rodriguez, S.; Tello, C.; et al. Impaired proteasome activity and neurodegeneration with brain iron accumulation in FBXO7 defect. Ann. Clin. Transl. Neurol. 2020, 7, 1436–1442. [Google Scholar] [CrossRef]
- Magrinelli, F.; Mehta, S.; Di Lazzaro, G.; Latorre, A.; Edwards, M.J.; Balint, B.; Basu, P.; Kobylecki, C.; Groppa, S.; Hegde, A.; et al. Dissecting the Phenotype and Genotype of PLA2G6-Related Parkinsonism. Mov. Disord. 2022, 37, 148–161. [Google Scholar] [CrossRef]
- Ikeda, A.; Nishioka, K.; Meng, H.; Takanashi, M.; Hasegawa, I.; Inoshita, T.; Shiba-Fukushima, K.; Li, Y.; Yoshino, H.; Mori, A.; et al. Mutations in CHCHD2 cause alpha-synuclein aggregation. Hum. Mol. Genet. 2019, 28, 3895–3911. [Google Scholar] [CrossRef]
- Liao, T.W.; Wang, C.C.; Chung, W.H.; Su, S.C.; Chin, S.H.; Fung, H.C.; Wu, Y.R. Role of LRP10 in Parkinson’s disease in a Taiwanese cohort. Parkinsonism Relat. Disord. 2021, 89, 79–83. [Google Scholar] [CrossRef]
- Quadri, M.; Mandemakers, W.; Grochowska, M.M.; Masius, R.; Geut, H.; Fabrizio, E.; Breedveld, G.J.; Kuipers, D.; Minneboo, M.; Vergouw, L.J.M.; et al. LRP10 genetic variants in familial Parkinson’s disease and dementia with Lewy bodies: A genome-wide linkage and sequencing study. Lancet Neurol. 2018, 17, 597–608. [Google Scholar] [CrossRef]
- Manini, A.; Straniero, L.; Monfrini, E.; Percetti, M.; Vizziello, M.; Franco, G.; Rimoldi, V.; Zecchinelli, A.; Pezzoli, G.; Corti, S.; et al. Screening of LRP10 mutations in Parkinson’s disease patients from Italy. Parkinsonism Relat. Disord. 2021, 89, 17–21. [Google Scholar] [CrossRef]
- Neri, M.; Braccia, A.; Panteghini, C.; Garavaglia, B.; Gualandi, F.; Cavallo, M.A.; Scerrati, A.; Ferlini, A.; Sensi, M. Parkinson’s disease-dementia in trans LRP10 and GBA variants: Response to deep brain stimulation. Parkinsonism Relat. Disord. 2021, 92, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.X.; Shi, Y.; Yang, Y.; Ahmeti, K.B.; Miller, N.; Huang, C.; Cheng, L.; Zhai, H.; Deng, S.; Nuytemans, K.; et al. Identification of TMEM230 mutations in familial Parkinson’s disease. Nat. Genet. 2016, 48, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Lesage, S.; Drouet, V.; Majounie, E.; Deramecourt, V.; Jacoupy, M.; Nicolas, A.; Cormier-Dequaire, F.; Hassoun, S.M.; Pujol, C.; Ciura, S.; et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am. J. Hum. Genet. 2016, 98, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Serratos, I.N.; Hernandez-Perez, E.; Campos, C.; Aschner, M.; Santamaria, A. An Update on the Critical Role of alpha-Synuclein in Parkinson’s Disease and Other Synucleinopathies: From Tissue to Cellular and Molecular Levels. Mol. Neurobiol. 2022, 59, 620–642. [Google Scholar] [CrossRef] [PubMed]
- Barbuti, P.A.; Ohnmacht, J.; Santos, B.F.R.; Antony, P.M.; Massart, F.; Cruciani, G.; Dording, C.M.; Pavelka, L.; Casadei, N.; Kwon, Y.J.; et al. Gene-corrected p.A30P SNCA patient-derived isogenic neurons rescue neuronal branching and function. Sci. Rep. 2021, 11, 21946. [Google Scholar] [CrossRef] [PubMed]
- Saunders-Pullman, R.; Raymond, D.; Elango, S. LRRK2 Parkinson Disease. In GeneReviews(R); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Kestenbaum, M.; Alcalay, R.N. Clinical Features of LRRK2 Carriers with Parkinson’s Disease. Adv. Neurobiol. 2017, 14, 31–48. [Google Scholar] [CrossRef]
- Waro, B.J.; Aasly, J.O. Exploring cancer in LRRK2 mutation carriers and idiopathic Parkinson’s disease. Brain Behav. 2018, 8, e00858. [Google Scholar] [CrossRef]
- Agalliu, I.; San Luciano, M.; Mirelman, A.; Giladi, N.; Waro, B.; Aasly, J.; Inzelberg, R.; Hassin-Baer, S.; Friedman, E.; Ruiz-Martinez, J.; et al. Higher frequency of certain cancers in LRRK2 G2019S mutation carriers with Parkinson disease: A pooled analysis. JAMA Neurol. 2015, 72, 58–65. [Google Scholar] [CrossRef]
- Agalliu, I.; Ortega, R.A.; Luciano, M.S.; Mirelman, A.; Pont-Sunyer, C.; Brockmann, K.; Vilas, D.; Tolosa, E.; Berg, D.; Waro, B.; et al. Cancer outcomes among Parkinson’s disease patients with leucine rich repeat kinase 2 mutations, idiopathic Parkinson’s disease patients, and nonaffected controls. Mov. Disord. 2019, 34, 1392–1398. [Google Scholar] [CrossRef]
- Macias-Garcia, D.; Perinan, M.T.; Munoz-Delgado, L.; Jesus, S.; Jimenez-Jaraba, M.V.; Buiza-Rueda, D.; Bonilla-Toribio, M.; Adarmes-Gomez, A.; Carrillo, F.; Gomez-Garre, P.; et al. Increased Stroke Risk in Patients with Parkinson’s Disease with LRRK2 Mutations. Mov. Disord. 2022, 37, 225–227. [Google Scholar] [CrossRef]
- San Luciano, M.; Tanner, C.M.; Meng, C.; Marras, C.; Goldman, S.M.; Lang, A.E.; Tolosa, E.; Schule, B.; Langston, J.W.; Brice, A.; et al. Nonsteroidal Anti-inflammatory Use and LRRK2 Parkinson’s Disease Penetrance. Mov. Disord. 2020, 35, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Iwaki, H.; Blauwendraat, C.; Makarious, M.B.; Bandres-Ciga, S.; Leonard, H.L.; Gibbs, J.R.; Hernandez, D.G.; Scholz, S.W.; Faghri, F.; International Parkinson’s Disease Genomics Consortium; et al. Penetrance of Parkinson’s Disease in LRRK2 p.G2019S Carriers Is Modified by a Polygenic Risk Score. Mov. Disord. 2020, 35, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Ozelius, L.J.; Senthil, G.; Saunders-Pullman, R.; Ohmann, E.; Deligtisch, A.; Tagliati, M.; Hunt, A.L.; Klein, C.; Henick, B.; Hailpern, S.M.; et al. LRRK2 G2019S as a cause of Parkinson’s disease in Ashkenazi Jews. N. Engl. J. Med. 2006, 354, 424–425. [Google Scholar] [CrossRef] [PubMed]
- Lesage, S.; Durr, A.; Tazir, M.; Lohmann, E.; Leutenegger, A.L.; Janin, S.; Pollak, P.; Brice, A.; French Parkinson’s Disease Genetics Study Group. LRRK2 G2019S as a cause of Parkinson’s disease in North African Arabs. N. Engl. J. Med. 2006, 354, 422–423. [Google Scholar] [CrossRef] [PubMed]
- Healy, D.G.; Falchi, M.; O’Sullivan, S.S.; Bonifati, V.; Durr, A.; Bressman, S.; Brice, A.; Aasly, J.; Zabetian, C.P.; Goldwurm, S.; et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: A case-control study. Lancet Neurol. 2008, 7, 583–590. [Google Scholar] [CrossRef]
- Lim, S.Y.; Tan, A.H.; Ahmad-Annuar, A.; Klein, C.; Tan, L.C.S.; Rosales, R.L.; Bhidayasiri, R.; Wu, Y.R.; Shang, H.F.; Evans, A.H.; et al. Parkinson’s disease in the Western Pacific Region. Lancet Neurol. 2019, 18, 865–879. [Google Scholar] [CrossRef]
- Xie, C.L.; Pan, J.L.; Wang, W.W.; Zhang, Y.; Zhang, S.F.; Gan, J.; Liu, Z.G. The association between the LRRK2 G2385R variant and the risk of Parkinson’s disease: A meta-analysis based on 23 case-control studies. Neurol. Sci. 2014, 35, 1495–1504. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, Q.; Yi, M.; Zhou, X.; Guo, J.; Xu, Q.; Tang, B.; Yan, X. Genetic Analysis of LRRK2 R1628P in Parkinson’s Disease in Asian Populations. Parkinsons Dis. 2017, 2017, 8093124. [Google Scholar] [CrossRef]
- Blauwendraat, C.; Reed, X.; Kia, D.A.; Gan-Or, Z.; Lesage, S.; Pihlstrom, L.; Guerreiro, R.; Gibbs, J.R.; Sabir, M.; Ahmed, S.; et al. Frequency of Loss of Function Variants in LRRK2 in Parkinson Disease. JAMA Neurol. 2018, 75, 1416–1422. [Google Scholar] [CrossRef]
- Bryant, N.; Malpeli, N.; Ziaee, J.; Blauwendraat, C.; Liu, Z.; Consortium, A.P.; West, A.B. Identification of LRRK2 missense variants in the accelerating medicines partnership Parkinson’s disease cohort. Hum. Mol. Genet. 2021, 30, 454–466. [Google Scholar] [CrossRef]
- Berwick, D.C.; Heaton, G.R.; Azeggagh, S.; Harvey, K. LRRK2 Biology from structure to dysfunction: Research progresses, but the themes remain the same. Mol. Neurodegener. 2019, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Zimprich, A.; Benet-Pages, A.; Struhal, W.; Graf, E.; Eck, S.H.; Offman, M.N.; Haubenberger, D.; Spielberger, S.; Schulte, E.C.; Lichtner, P.; et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 2011, 89, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Vilarino-Guell, C.; Wider, C.; Ross, O.A.; Dachsel, J.C.; Kachergus, J.M.; Lincoln, S.J.; Soto-Ortolaza, A.I.; Cobb, S.A.; Wilhoite, G.J.; Bacon, J.A.; et al. VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 2011, 89, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Ando, M.; Funayama, M.; Li, Y.; Kashihara, K.; Murakami, Y.; Ishizu, N.; Toyoda, C.; Noguchi, K.; Hashimoto, T.; Nakano, N.; et al. VPS35 mutation in Japanese patients with typical Parkinson’s disease. Mov. Disord. 2012, 27, 1413–1417. [Google Scholar] [CrossRef]
- Kumar, K.R.; Weissbach, A.; Heldmann, M.; Kasten, M.; Tunc, S.; Sue, C.M.; Svetel, M.; Kostic, V.S.; Segura-Aguilar, J.; Ramirez, A.; et al. Frequency of the D620N mutation in VPS35 in Parkinson disease. Arch. Neurol. 2012, 69, 1360–1364. [Google Scholar] [CrossRef]
- Rahman, A.A.; Morrison, B.E. Contributions of VPS35 Mutations to Parkinson’s Disease. Neuroscience 2019, 401, 1–10. [Google Scholar] [CrossRef]
- Ishiguro, M.; Li, Y.; Yoshino, H.; Daida, K.; Ishiguro, Y.; Oyama, G.; Saiki, S.; Funayama, M.; Hattori, N.; Nishioka, K. Clinical manifestations of Parkinson’s disease harboring VPS35 retromer complex component p.D620N with long-term follow-up. Parkinsonism Relat. Disord. 2021, 84, 139–143. [Google Scholar] [CrossRef]
- Cutillo, G.; Simon, D.K.; Eleuteri, S. VPS35 and the mitochondria: Connecting the dots in Parkinson’s disease pathophysiology. Neurobiol. Dis. 2020, 145, 105056. [Google Scholar] [CrossRef]
- Sassone, J.; Reale, C.; Dati, G.; Regoni, M.; Pellecchia, M.T.; Garavaglia, B. The Role of VPS35 in the Pathobiology of Parkinson’s Disease. Cell. Mol. Neurobiol. 2021, 41, 199–227. [Google Scholar] [CrossRef]
- Lesage, S.; Magali, P.; Lohmann, E.; Lacomblez, L.; Teive, H.; Janin, S.; Cousin, P.Y.; Durr, A.; Brice, A.; French Parkinson Disease Genetics Study Group. Deletion of the parkin and PACRG gene promoter in early-onset parkinsonism. Hum. Mutat. 2007, 28, 27–32. [Google Scholar] [CrossRef]
- Bruggemann, N.; Klein, C. Parkin Type of Early-Onset Parkinson Disease. In GeneReviews(R); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Grunewald, A.; Kumar, K.R.; Sue, C.M. New insights into the complex role of mitochondria in Parkinson’s disease. Prog. Neurobiol. 2019, 177, 73–93. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, A.V.; Campbell, P.; Schapira, A.H.V.; Morris, H.R.; Taanman, J.W. The PINK1-Parkin mitophagy signalling pathway is not functional in peripheral blood mononuclear cells. PLoS ONE 2021, 16, e0259903. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.A.; Klein, C. PINK1 Type of Young-Onset Parkinson Disease. In GeneReviews(R); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Guler, S.; Gul, T.; Guler, S.; Haerle, M.C.; Basak, A.N. Early-Onset Parkinson’s Disease: A Novel Deletion Comprising the DJ-1 and TNFRSF9 Genes. Mov. Disord. 2021, 36, 2973–2976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, J.; Wang, J.; Yang, B.; He, Q.; Weng, Q. Role of DJ-1 in Immune and Inflammatory Diseases. Front. Immunol. 2020, 11, 994. [Google Scholar] [CrossRef]
- Kim, R.H.; Smith, P.D.; Aleyasin, H.; Hayley, S.; Mount, M.P.; Pownall, S.; Wakeham, A.; You-Ten, A.J.; Kalia, S.K.; Horne, P.; et al. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc. Natl. Acad. Sci. USA 2005, 102, 5215–5220. [Google Scholar] [CrossRef]
- Repici, M.; Giorgini, F. DJ-1 in Parkinson’s Disease: Clinical Insights and Therapeutic Perspectives. J. Clin. Med. 2019, 8, 1377. [Google Scholar] [CrossRef]
- Pauly, M.G.; Ruiz Lopez, M.; Westenberger, A.; Saranza, G.; Bruggemann, N.; Weissbach, A.; Rosales, R.L.; Diesta, C.C.; Jamora, R.D.G.; Reyes, C.J.; et al. Expanding Data Collection for the MDSGene Database: X-linked Dystonia-Parkinsonism as Use Case Example. Mov. Disord. 2020, 35, 1933–1938. [Google Scholar] [CrossRef]
- Aneichyk, T.; Hendriks, W.T.; Yadav, R.; Shin, D.; Gao, D.; Vaine, C.A.; Collins, R.L.; Domingo, A.; Currall, B.; Stortchevoi, A.; et al. Dissecting the Causal Mechanism of X-Linked Dystonia-Parkinsonism by Integrating Genome and Transcriptome Assembly. Cell 2018, 172, 897–909.e21. [Google Scholar] [CrossRef]
- Lee, L.V.; Maranon, E.; Demaisip, C.; Peralta, O.; Borres-Icasiano, R.; Arancillo, J.; Rivera, C.; Munoz, E.; Tan, K.; Reyes, M.T. The natural history of sex-linked recessive dystonia parkinsonism of Panay, Philippines (XDP). Parkinsonism Relat. Disord. 2002, 9, 29–38. [Google Scholar] [CrossRef]
- Santiano, R.A.S.; Rosales, R.L. A Cross-Cultural Validation of the Filipino and Hiligaynon Versions of the Parts IIIB (Non-Motor Features) and IV (Activities of Daily Living) of the X-Linked Dystonia-Parkinsonism- MDSP Rating Scale. Clin. Parkinsonism Relat. Disord. 2021, 5, 100100. [Google Scholar] [CrossRef]
- Westenberger, A.; Reyes, C.J.; Saranza, G.; Dobricic, V.; Hanssen, H.; Domingo, A.; Laabs, B.H.; Schaake, S.; Pozojevic, J.; Rakovic, A.; et al. A hexanucleotide repeat modifies expressivity of X-linked dystonia parkinsonism. Ann. Neurol. 2019, 85, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Nybo, C.J.; Gustavsson, E.K.; Farrer, M.J.; Aasly, J.O. Neuropathological findings in PINK1-associated Parkinson’s disease. Parkinsonism Relat. Disord. 2020, 78, 105–108. [Google Scholar] [CrossRef] [PubMed]
- van Veen, S.; Martin, S.; Van den Haute, C.; Benoy, V.; Lyons, J.; Vanhoutte, R.; Kahler, J.P.; Decuypere, J.P.; Gelders, G.; Lambie, E.; et al. ATP13A2 deficiency disrupts lysosomal polyamine export. Nature 2020, 578, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.; McEntagart, M.M.; Isaacs, J.D. DCTN1-related Parkinson-plus disorder (Perry syndrome). Pract. Neurol. 2020, 20, 317–319. [Google Scholar] [CrossRef] [PubMed]
- Wider, C.; Dachsel, J.C.; Farrer, M.J.; Dickson, D.W.; Tsuboi, Y.; Wszolek, Z.K. Elucidating the genetics and pathology of Perry syndrome. J. Neurol. Sci. 2010, 289, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Farrer, M.J.; Hulihan, M.M.; Kachergus, J.M.; Dachsel, J.C.; Stoessl, A.J.; Grantier, L.L.; Calne, S.; Calne, D.B.; Lechevalier, B.; Chapon, F.; et al. DCTN1 mutations in Perry syndrome. Nat. Genet. 2009, 41, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Edvardson, S.; Cinnamon, Y.; Ta-Shma, A.; Shaag, A.; Yim, Y.I.; Zenvirt, S.; Jalas, C.; Lesage, S.; Brice, A.; Taraboulos, A.; et al. A deleterious mutation in DNAJC6 encoding the neuronal-specific clathrin-uncoating co-chaperone auxilin, is associated with juvenile parkinsonism. PLoS ONE 2012, 7, e36458. [Google Scholar] [CrossRef]
- Koroglu, C.; Baysal, L.; Cetinkaya, M.; Karasoy, H.; Tolun, A. DNAJC6 is responsible for juvenile parkinsonism with phenotypic variability. Parkinsonism Relat. Disord. 2013, 19, 320–324. [Google Scholar] [CrossRef]
- Olgiati, S.; Quadri, M.; Fang, M.; Rood, J.P.; Saute, J.A.; Chien, H.F.; Bouwkamp, C.G.; Graafland, J.; Minneboo, M.; Breedveld, G.J.; et al. DNAJC6 Mutations Associated with Early-Onset Parkinson’s Disease. Ann. Neurol. 2016, 79, 244–256. [Google Scholar] [CrossRef]
- Yim, Y.I.; Sun, T.; Wu, L.G.; Raimondi, A.; De Camilli, P.; Eisenberg, E.; Greene, L.E. Endocytosis and clathrin-uncoating defects at synapses of auxilin knockout mice. Proc. Natl. Acad. Sci. USA 2010, 107, 4412–4417. [Google Scholar] [CrossRef]
- Di Fonzo, A.; Dekker, M.C.; Montagna, P.; Baruzzi, A.; Yonova, E.H.; Correia Guedes, L.; Szczerbinska, A.; Zhao, T.; Dubbel-Hulsman, L.O.; Wouters, C.H.; et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology 2009, 72, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Ding, L.; Li, H.; Lin, Y.; Dai, Y.; Xu, X.; Dong, Q.; Lin, Y.; Long, L. Juvenile-onset parkinsonism with pyramidal signs due to compound heterozygous mutations in the F-Box only protein 7 gene. Parkinsonism Relat. Disord. 2018, 47, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Yalcin-Cakmakli, G.; Olgiati, S.; Quadri, M.; Breedveld, G.J.; Cortelli, P.; Bonifati, V.; Elibol, B. A new Turkish family with homozygous FBXO7 truncating mutation and juvenile atypical parkinsonism. Parkinsonism Relat. Disord. 2014, 20, 1248–1252. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, E.; Coquel, A.S.; Honore, A.; Gurvit, H.; Hanagasi, H.; Emre, M.; Leutenegger, A.L.; Drouet, V.; Sahbatou, M.; Guven, G.; et al. A new F-box protein 7 gene mutation causing typical Parkinson’s disease. Mov. Disord. 2015, 30, 1130–1133. [Google Scholar] [CrossRef] [PubMed]
- Burchell, V.S.; Nelson, D.E.; Sanchez-Martinez, A.; Delgado-Camprubi, M.; Ivatt, R.M.; Pogson, J.H.; Randle, S.J.; Wray, S.; Lewis, P.A.; Houlden, H.; et al. The Parkinson’s disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat. Neurosci. 2013, 16, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Paisan-Ruiz, C.; Bhatia, K.P.; Li, A.; Hernandez, D.; Davis, M.; Wood, N.W.; Hardy, J.; Houlden, H.; Singleton, A.; Schneider, S.A. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann. Neurol. 2009, 65, 19–23. [Google Scholar] [CrossRef]
- Fasano, D.; Parisi, S.; Pierantoni, G.M.; De Rosa, A.; Picillo, M.; Amodio, G.; Pellecchia, M.T.; Barone, P.; Moltedo, O.; Bonifati, V.; et al. Alteration of endosomal trafficking is associated with early-onset parkinsonism caused by SYNJ1 mutations. Cell Death Dis. 2018, 9, 385. [Google Scholar] [CrossRef]
- Funayama, M.; Ohe, K.; Amo, T.; Furuya, N.; Yamaguchi, J.; Saiki, S.; Li, Y.; Ogaki, K.; Ando, M.; Yoshino, H.; et al. CHCHD2 mutations in autosomal dominant late-onset Parkinson’s disease: A genome-wide linkage and sequencing study. Lancet Neurol. 2015, 14, 274–282. [Google Scholar] [CrossRef]
- Karkheiran, S.; Shahidi, G.A.; Walker, R.H.; Paisan-Ruiz, C. PLA2G6-associated Dystonia-Parkinsonism: Case Report and Literature Review. Tremor Other Hyperkinet. Mov. 2015, 5, 317. [Google Scholar] [CrossRef]
- Lesage, S.; Mangone, G.; Tesson, C.; Bertrand, H.; Benmahdjoub, M.; Kesraoui, S.; Arezki, M.; Singleton, A.; Corvol, J.C.; Brice, A. Clinical Variability of SYNJ1-Associated Early-Onset Parkinsonism. Front. Neurol. 2021, 12, 648457. [Google Scholar] [CrossRef]
- Cao, M.; Wu, Y.; Ashrafi, G.; McCartney, A.J.; Wheeler, H.; Bushong, E.A.; Boassa, D.; Ellisman, M.H.; Ryan, T.A.; De Camilli, P. Parkinson Sac Domain Mutation in Synaptojanin 1 Impairs Clathrin Uncoating at Synapses and Triggers Dystrophic Changes in Dopaminergic Axons. Neuron 2017, 93, 882–896.e5. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.H.; Mao, C.Y.; Zhang, S.Y.; Yang, J.; Song, B.; Wu, P.; Zuo, C.T.; Liu, Y.T.; Ji, Y.; Yang, Z.H.; et al. CHCHD2 gene mutations in familial and sporadic Parkinson’s disease. Neurobiol. Aging 2016, 38, 217.e9–217.e13. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhao, Q.; An, R.; Zheng, J.; Tian, S.; Chen, Y.; Xu, Y. Mutational scanning of the CHCHD2 gene in Han Chinese patients with Parkinson’s disease and meta-analysis of the literature. Parkinsonism Relat. Disord. 2016, 29, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Jansen, I.E.; Bras, J.M.; Lesage, S.; Schulte, C.; Gibbs, J.R.; Nalls, M.A.; Brice, A.; Wood, N.W.; Morris, H.; Hardy, J.A.; et al. CHCHD2 and Parkinson’s disease. Lancet Neurol. 2015, 14, 678–679. [Google Scholar] [CrossRef]
- Lee, R.G.; Sedghi, M.; Salari, M.; Shearwood, A.J.; Stentenbach, M.; Kariminejad, A.; Goullee, H.; Rackham, O.; Laing, N.G.; Tajsharghi, H.; et al. Early-onset Parkinson disease caused by a mutation in CHCHD2 and mitochondrial dysfunction. Neurol. Genet. 2018, 4, e276. [Google Scholar] [CrossRef] [PubMed]
- Bannwarth, S.; Ait-El-Mkadem, S.; Chaussenot, A.; Genin, E.C.; Lacas-Gervais, S.; Fragaki, K.; Berg-Alonso, L.; Kageyama, Y.; Serre, V.; Moore, D.G.; et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 2014, 137, 2329–2345. [Google Scholar] [CrossRef]
- Meng, H.; Yamashita, C.; Shiba-Fukushima, K.; Inoshita, T.; Funayama, M.; Sato, S.; Hatta, T.; Natsume, T.; Umitsu, M.; Takagi, J.; et al. Loss of Parkinson’s disease-associated protein CHCHD2 affects mitochondrial crista structure and destabilizes cytochrome c. Nat. Commun. 2017, 8, 15500. [Google Scholar] [CrossRef]
- Tesson, C.; Brefel-Courbon, C.; Corvol, J.C.; Lesage, S.; Brice, A.; French Parkinson’s Disease Genetics Study Group. LRP10 in alpha-synucleinopathies. Lancet Neurol. 2018, 17, 1034. [Google Scholar] [CrossRef]
- Chen, Y.; Cen, Z.; Zheng, X.; Pan, Q.; Chen, X.; Zhu, L.; Chen, S.; Wu, H.; Xie, F.; Wang, H.; et al. LRP10 in autosomal-dominant Parkinson’s disease. Mov. Disord. 2019, 34, 912–916. [Google Scholar] [CrossRef]
- Daida, K.; Nishioka, K.; Li, Y.; Yoshino, H.; Kikuchi, A.; Hasegawa, T.; Funayama, M.; Hattori, N. Mutation analysis of LRP10 in Japanese patients with familial Parkinson’s disease, progressive supranuclear palsy, and frontotemporal dementia. Neurobiol. Aging 2019, 84, 235.e11–235.e16. [Google Scholar] [CrossRef]
- Li, C.; Chen, Y.; Ou, R.; Gu, X.; Wei, Q.; Cao, B.; Zhang, L.; Hou, Y.; Liu, K.; Chen, X.; et al. Mutation analysis of LRP10 in a large Chinese familial Parkinson disease cohort. Neurobiol. Aging 2021, 99, 99.e1–99.e6. [Google Scholar] [CrossRef] [PubMed]
- Grochowska, M.M.; Carreras Mascaro, A.; Boumeester, V.; Natale, D.; Breedveld, G.J.; Geut, H.; van Cappellen, W.A.; Boon, A.J.W.; Kievit, A.J.A.; Sammler, E.; et al. LRP10 interacts with SORL1 in the intracellular vesicle trafficking pathway in non-neuronal brain cells and localises to Lewy bodies in Parkinson’s disease and dementia with Lewy bodies. Acta Neuropathol. 2021, 142, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Vilarino-Guell, C.; Rajput, A.; Milnerwood, A.J.; Shah, B.; Szu-Tu, C.; Trinh, J.; Yu, I.; Encarnacion, M.; Munsie, L.N.; Tapia, L.; et al. DNAJC13 mutations in Parkinson disease. Hum. Mol. Genet. 2014, 23, 1794–1801. [Google Scholar] [CrossRef] [PubMed]
- Farrer, M.J. Doubts about TMEM230 as a gene for parkinsonism. Nat. Genet. 2019, 51, 367–368. [Google Scholar] [CrossRef]
- Deng, H.X.; Pericak-Vance, M.A.; Siddique, T. Reply to ‘TMEM230 variants in Parkinson’s disease’ and ‘Doubts about TMEM230 as a gene for parkinsonism’. Nat. Genet. 2019, 51, 369–371. [Google Scholar] [CrossRef]
- Kim, M.J.; Deng, H.X.; Wong, Y.C.; Siddique, T.; Krainc, D. The Parkinson’s disease-linked protein TMEM230 is required for Rab8a-mediated secretory vesicle trafficking and retromer trafficking. Hum. Mol. Genet. 2017, 26, 729–741. [Google Scholar] [CrossRef]
- Lin, C.H.; Tsai, P.I.; Lin, H.Y.; Hattori, N.; Funayama, M.; Jeon, B.; Sato, K.; Abe, K.; Mukai, Y.; Takahashi, Y.; et al. Mitochondrial UQCRC1 mutations cause autosomal dominant parkinsonism with polyneuropathy. Brain 2020, 143, 3352–3373. [Google Scholar] [CrossRef]
- Senkevich, K.; Bandres-Ciga, S.; Gan-Or, Z.; Krohn, L.; International Parkinson’s Disease Genomics Consortium. Lack of evidence for association of UQCRC1 with Parkinson’s disease in Europeans. Neurobiol. Aging 2021, 101, 297.e1–297.e4. [Google Scholar] [CrossRef]
- Shan, W.; Li, J.; Xu, W.; Li, H.; Zuo, Z. Critical role of UQCRC1 in embryo survival, brain ischemic tolerance and normal cognition in mice. Cell. Mol. Life Sci. 2019, 76, 1381–1396. [Google Scholar] [CrossRef]
- Hung, Y.C.; Huang, K.L.; Chen, P.L.; Li, J.L.; Lu, S.H.; Chang, J.C.; Lin, H.Y.; Lo, W.C.; Huang, S.Y.; Lee, T.T.; et al. UQCRC1 engages cytochrome c for neuronal apoptotic cell death. Cell Rep. 2021, 36, 109729. [Google Scholar] [CrossRef]
- Rudakou, U.; Ruskey, J.A.; Krohn, L.; Laurent, S.B.; Spiegelman, D.; Greenbaum, L.; Yahalom, G.; Desautels, A.; Montplaisir, J.Y.; Fahn, S.; et al. Analysis of common and rare VPS13C variants in late-onset Parkinson disease. Neurol. Genet. 2020, 6, 385. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Mari, M.; Parashar, S.; Liu, D.; Cui, Y.; Reggiori, F.; Novick, P.J.; Ferro-Novick, S. Vps13 is required for the packaging of the ER into autophagosomes during ER-phagy. Proc. Natl. Acad. Sci. USA 2020, 117, 18530–18539. [Google Scholar] [CrossRef] [PubMed]
- Morales-Briceno, H.; Mohammad, S.S.; Post, B.; Fois, A.F.; Dale, R.C.; Tchan, M.; Fung, V.S.C. Clinical and neuroimaging phenotypes of genetic parkinsonism from infancy to adolescence. Brain 2020, 143, 751–770. [Google Scholar] [CrossRef] [PubMed]
- Khodadadi, H.; Azcona, L.J.; Aghamollaii, V.; Omrani, M.D.; Garshasbi, M.; Taghavi, S.; Tafakhori, A.; Shahidi, G.A.; Jamshidi, J.; Darvish, H.; et al. PTRHD1 (C2orf79) mutations lead to autosomal-recessive intellectual disability and parkinsonism. Mov. Disord. 2017, 32, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.R.; Sim, J.C.; McLean, C.; Giannandrea, M.; Galea, C.A.; Riseley, J.R.; Stephenson, S.E.; Fitzpatrick, E.; Haas, S.A.; Pope, K.; et al. Mutations in RAB39B cause X-linked intellectual disability and early-onset Parkinson disease with alpha-synuclein pathology. Am. J. Hum. Genet. 2014, 95, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Rudakou, U.; Yu, E.; Ruskey, J.A.; Asayesh, F.; Laurent, S.B.; Spiegelman, D.; Fahn, S.; Waters, C.; Monchi, O.; et al. Association study of DNAJC13, UCHL1, HTRA2, GIGYF2, and EIF4G1 with Parkinson’s disease. Neurobiol. Aging 2021, 100, 119.e7–119.e13. [Google Scholar] [CrossRef]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef]
- Skrahina, V.; Gaber, H.; Vollstedt, E.J.; Forster, T.M.; Usnich, T.; Curado, F.; Bruggemann, N.; Paul, J.; Bogdanovic, X.; Zulbahar, S.; et al. The Rostock International Parkinson’s Disease (ROPAD) Study: Protocol and Initial Findings. Mov. Disord. 2021, 36, 1005–1010. [Google Scholar] [CrossRef]
- Neumann, J.; Bras, J.; Deas, E.; O’Sullivan, S.S.; Parkkinen, L.; Lachmann, R.H.; Li, A.; Holton, J.; Guerreiro, R.; Paudel, R.; et al. Glucocerebrosidase mutations in clinical and pathologically proven Parkinson’s disease. Brain 2009, 132, 1783–1794. [Google Scholar] [CrossRef]
- Lim, J.L.; Lohmann, K.; Tan, A.H.; Tay, Y.W.; Ibrahim, K.A.; Abdul Aziz, Z.; Mawardi, A.S.; Puvanarajah, S.D.; Lim, T.T.; Looi, I.; et al. Glucocerebrosidase (GBA) gene variants in a multi-ethnic Asian cohort with Parkinson’s disease: Mutational spectrum and clinical features. J. Neural Transm. 2022, 129, 37–48. [Google Scholar] [CrossRef]
- Stoker, T.B.; Camacho, M.; Winder-Rhodes, S.; Liu, G.; Scherzer, C.R.; Foltynie, T.; Evans, J.; Breen, D.P.; Barker, R.A.; Williams-Gray, C.H. Impact of GBA1 variants on long-term clinical progression and mortality in incident Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2020, 91, 695–702. [Google Scholar] [CrossRef] [PubMed]
- McNeill, A.; Duran, R.; Hughes, D.A.; Mehta, A.; Schapira, A.H. A clinical and family history study of Parkinson’s disease in heterozygous glucocerebrosidase mutation carriers. J. Neurol. Neurosurg. Psychiatry 2012, 83, 853–854. [Google Scholar] [CrossRef] [PubMed]
- Anheim, M.; Elbaz, A.; Lesage, S.; Durr, A.; Condroyer, C.; Viallet, F.; Pollak, P.; Bonaiti, B.; Bonaiti-Pellie, C.; Brice, A.; et al. Penetrance of Parkinson disease in glucocerebrosidase gene mutation carriers. Neurology 2012, 78, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Rana, H.Q.; Balwani, M.; Bier, L.; Alcalay, R.N. Age-specific Parkinson disease risk in GBA mutation carriers: Information for genetic counseling. Genet. Med. 2013, 15, 146–149. [Google Scholar] [CrossRef]
- Alcalay, R.N.; Dinur, T.; Quinn, T.; Sakanaka, K.; Levy, O.; Waters, C.; Fahn, S.; Dorovski, T.; Chung, W.K.; Pauciulo, M.; et al. Comparison of Parkinson risk in Ashkenazi Jewish patients with Gaucher disease and GBA heterozygotes. JAMA Neurol. 2014, 71, 752–757. [Google Scholar] [CrossRef]
- Balestrino, R.; Tunesi, S.; Tesei, S.; Lopiano, L.; Zecchinelli, A.L.; Goldwurm, S. Penetrance of Glucocerebrosidase (GBA) Mutations in Parkinson’s Disease: A Kin Cohort Study. Mov. Disord. 2020, 35, 2111–2114. [Google Scholar] [CrossRef]
- Blauwendraat, C.; Reed, X.; Krohn, L.; Heilbron, K.; Bandres-Ciga, S.; Tan, M.; Gibbs, J.R.; Hernandez, D.G.; Kumaran, R.; Langston, R.; et al. Genetic modifiers of risk and age at onset in GBA associated Parkinson’s disease and Lewy body dementia. Brain 2020, 143, 234–248. [Google Scholar] [CrossRef]
- Dinur, T.; Becker-Cohen, M.; Revel-Vilk, S.; Zimran, A.; Arkadir, D. Parkinson’s Clustering in Families of Non-Neuronopathic N370S GBA Mutation Carriers Indicates the Presence of Genetic Modifiers. J. Parkinsons Dis. 2021, 11, 615–618. [Google Scholar] [CrossRef]
- Alcalay, R.N.; Kehoe, C.; Shorr, E.; Battista, R.; Hall, A.; Simuni, T.; Marder, K.; Wills, A.M.; Naito, A.; Beck, J.C.; et al. Genetic testing for Parkinson disease: Current practice, knowledge, and attitudes among US and Canadian movement disorders specialists. Genet. Med. 2020, 22, 574–580. [Google Scholar] [CrossRef]
- Cook, L.; Schulze, J.; Kopil, C.; Hastings, T.; Naito, A.; Wojcieszek, J.; Payne, K.; Alcalay, R.N.; Klein, C.; Saunders-Pullman, R.; et al. Genetic Testing for Parkinson Disease: Are We Ready. Neurol. Clin. Pract. 2021, 11, 69–77. [Google Scholar] [CrossRef]
- Mangone, G.; Bekadar, S.; Cormier-Dequaire, F.; Tahiri, K.; Welaratne, A.; Czernecki, V.; Pineau, F.; Karachi, C.; Castrioto, A.; Durif, F.; et al. Early cognitive decline after bilateral subthalamic deep brain stimulation in Parkinson’s disease patients with GBA mutations. Parkinsonism Relat. Disord. 2020, 76, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Pal, G.; Mangone, G.; Hill, E.J.; Ouyang, B.; Liu, Y.; Lythe, V.; Ehrlich, D.; Saunders-Pullman, R.; Shanker, V.; Bressman, S.; et al. Parkinson Disease and Subthalamic Nucleus Deep Brain Stimulation: Cognitive Effects in GBA Mutation Carriers. Ann. Neurol. 2022, 91, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.; Oyama, G.; Hattori, N.; Shimo, Y.; Kuusimaki, T.; Kaasinen, V.; Antonini, A.; Kim, D.; Lee, J.I.; Cho, K.R.; et al. Subthalamic deep brain stimulation in Parkinson’s disease with SNCA mutations: Based on the follow-up to 10 years. Brain Behav. 2022, 12, e2503. [Google Scholar] [CrossRef] [PubMed]
- Cook, L.; Schulze, J.; Verbrugge, J.; Beck, J.C.; Marder, K.S.; Saunders-Pullman, R.; Klein, C.; Naito, A.; Alcalay, R.N.; ClinGen Parkinson’s Disease Gene Curation Expert Panel; et al. The commercial genetic testing landscape for Parkinson’s disease. Parkinsonism Relat. Disord. 2021, 92, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Castelo Rueda, M.P.; Raftopoulou, A.; Gogele, M.; Borsche, M.; Emmert, D.; Fuchsberger, C.; Hantikainen, E.M.; Vukovic, V.; Klein, C.; Pramstaller, P.P.; et al. Frequency of Heterozygous Parkin (PRKN) Variants and Penetrance of Parkinson’s Disease Risk Markers in the Population-Based CHRIS Cohort. Front. Neurol. 2021, 12, 706145. [Google Scholar] [CrossRef]
- Lubbe, S.J.; Bustos, B.I.; Hu, J.; Krainc, D.; Joseph, T.; Hehir, J.; Tan, M.; Zhang, W.; Escott-Price, V.; Williams, N.M.; et al. Assessing the relationship between monoallelic PRKN mutations and Parkinson’s risk. Hum. Mol. Genet. 2021, 30, 78–86. [Google Scholar] [CrossRef]
- Yu, E.; Rudakou, U.; Krohn, L.; Mufti, K.; Ruskey, J.A.; Asayesh, F.; Estiar, M.A.; Spiegelman, D.; Surface, M.; Fahn, S.; et al. Analysis of Heterozygous PRKN Variants and Copy-Number Variations in Parkinson’s Disease. Mov. Disord. 2021, 36, 178–187. [Google Scholar] [CrossRef]
- Eggers, C.; Schmidt, A.; Hagenah, J.; Bruggemann, N.; Klein, J.C.; Tadic, V.; Kertelge, L.; Kasten, M.; Binkofski, F.; Siebner, H.; et al. Progression of subtle motor signs in PINK1 mutation carriers with mild dopaminergic deficit. Neurology 2010, 74, 1798–1805. [Google Scholar] [CrossRef]
- Puschmann, A.; Fiesel, F.C.; Caulfield, T.R.; Hudec, R.; Ando, M.; Truban, D.; Hou, X.; Ogaki, K.; Heckman, M.G.; James, E.D.; et al. Heterozygous PINK1 p.G411S increases risk of Parkinson’s disease via a dominant-negative mechanism. Brain 2017, 140, 98–117. [Google Scholar] [CrossRef]
- Krohn, L.; Grenn, F.P.; Makarious, M.B.; Kim, J.J.; Bandres-Ciga, S.; Roosen, D.A.; Gan-Or, Z.; Nalls, M.A.; Singleton, A.B.; Blauwendraat, C.; et al. Comprehensive assessment of PINK1 variants in Parkinson’s disease. Neurobiol. Aging 2020, 91, 168.e1–168.e5. [Google Scholar] [CrossRef]
- Ysselstein, D.; Nguyen, M.; Young, T.J.; Severino, A.; Schwake, M.; Merchant, K.; Krainc, D. LRRK2 kinase activity regulates lysosomal glucocerebrosidase in neurons derived from Parkinson’s disease patients. Nat. Commun. 2019, 10, 5570. [Google Scholar] [CrossRef]
- Sanyal, A.; DeAndrade, M.P.; Novis, H.S.; Lin, S.; Chang, J.; Lengacher, N.; Tomlinson, J.J.; Tansey, M.G.; LaVoie, M.J. Lysosome and Inflammatory Defects in GBA1-Mutant Astrocytes Are Normalized by LRRK2 Inhibition. Mov. Disord. 2020, 35, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Ortega, R.A.; Wang, C.; Raymond, D.; Bryant, N.; Scherzer, C.R.; Thaler, A.; Alcalay, R.N.; West, A.B.; Mirelman, A.; Kuras, Y.; et al. Association of Dual LRRK2 G2019S and GBA Variations with Parkinson Disease Progression. JAMA Netw. Open 2021, 4, e215845. [Google Scholar] [CrossRef] [PubMed]
- Yahalom, G.; Greenbaum, L.; Israeli-Korn, S.; Fay-Karmon, T.; Livneh, V.; Ruskey, J.A.; Ronciere, L.; Alam, A.; Gan-Or, Z.; Hassin-Baer, S. Carriers of both GBA and LRRK2 mutations, compared to carriers of either, in Parkinson’s disease: Risk estimates and genotype-phenotype correlations. Parkinsonism Relat. Disord. 2019, 62, 179–184. [Google Scholar] [CrossRef] [PubMed]
- International Parkinson Disease Genomics Consortium. Ten Years of the International Parkinson Disease Genomics Consortium: Progress and Next Steps. J. Parkinsons Dis. 2020, 10, 19–30. [Google Scholar] [CrossRef]
- Global Parkinson’s Genetics Program. GP2: The Global Parkinson’s Genetics Program. Mov. Disord. 2021, 36, 842–851. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, F.; Fellner, A.; Kumar, K.R. Monogenic Parkinson’s Disease: Genotype, Phenotype, Pathophysiology, and Genetic Testing. Genes 2022, 13, 471. https://doi.org/10.3390/genes13030471
Jia F, Fellner A, Kumar KR. Monogenic Parkinson’s Disease: Genotype, Phenotype, Pathophysiology, and Genetic Testing. Genes. 2022; 13(3):471. https://doi.org/10.3390/genes13030471
Chicago/Turabian StyleJia, Fangzhi, Avi Fellner, and Kishore Raj Kumar. 2022. "Monogenic Parkinson’s Disease: Genotype, Phenotype, Pathophysiology, and Genetic Testing" Genes 13, no. 3: 471. https://doi.org/10.3390/genes13030471
APA StyleJia, F., Fellner, A., & Kumar, K. R. (2022). Monogenic Parkinson’s Disease: Genotype, Phenotype, Pathophysiology, and Genetic Testing. Genes, 13(3), 471. https://doi.org/10.3390/genes13030471






