Alpha Satellite RNA Levels Are Upregulated in the Blood of Patients with Metastatic Castration-Resistant Prostate Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Isolation and Reverse Transcription
2.3. Quantitative Real-Time PCR (qPCR) Analysis
2.4. Determination of PSA Values in Blood
2.5. Statistical Analyses
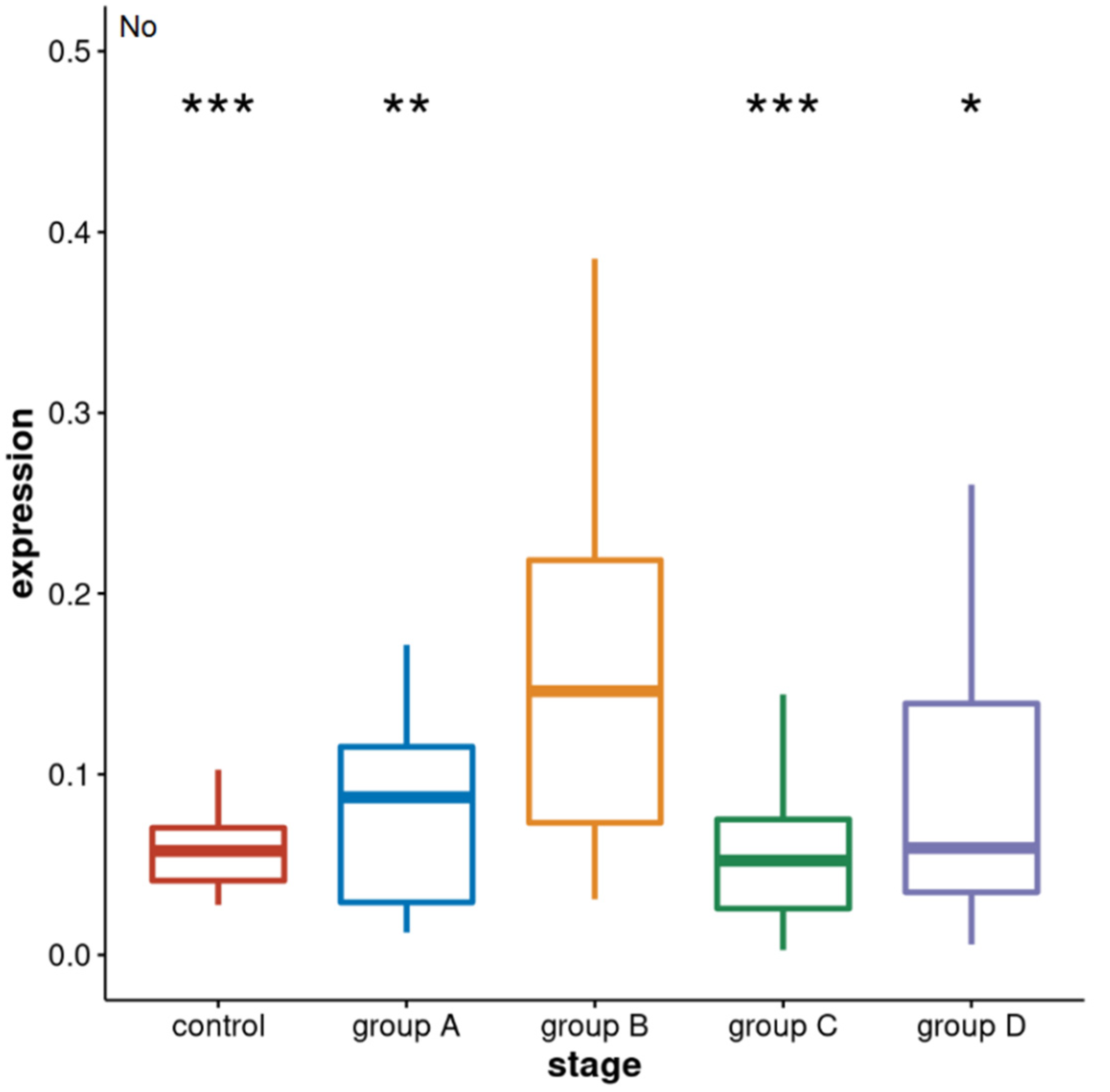
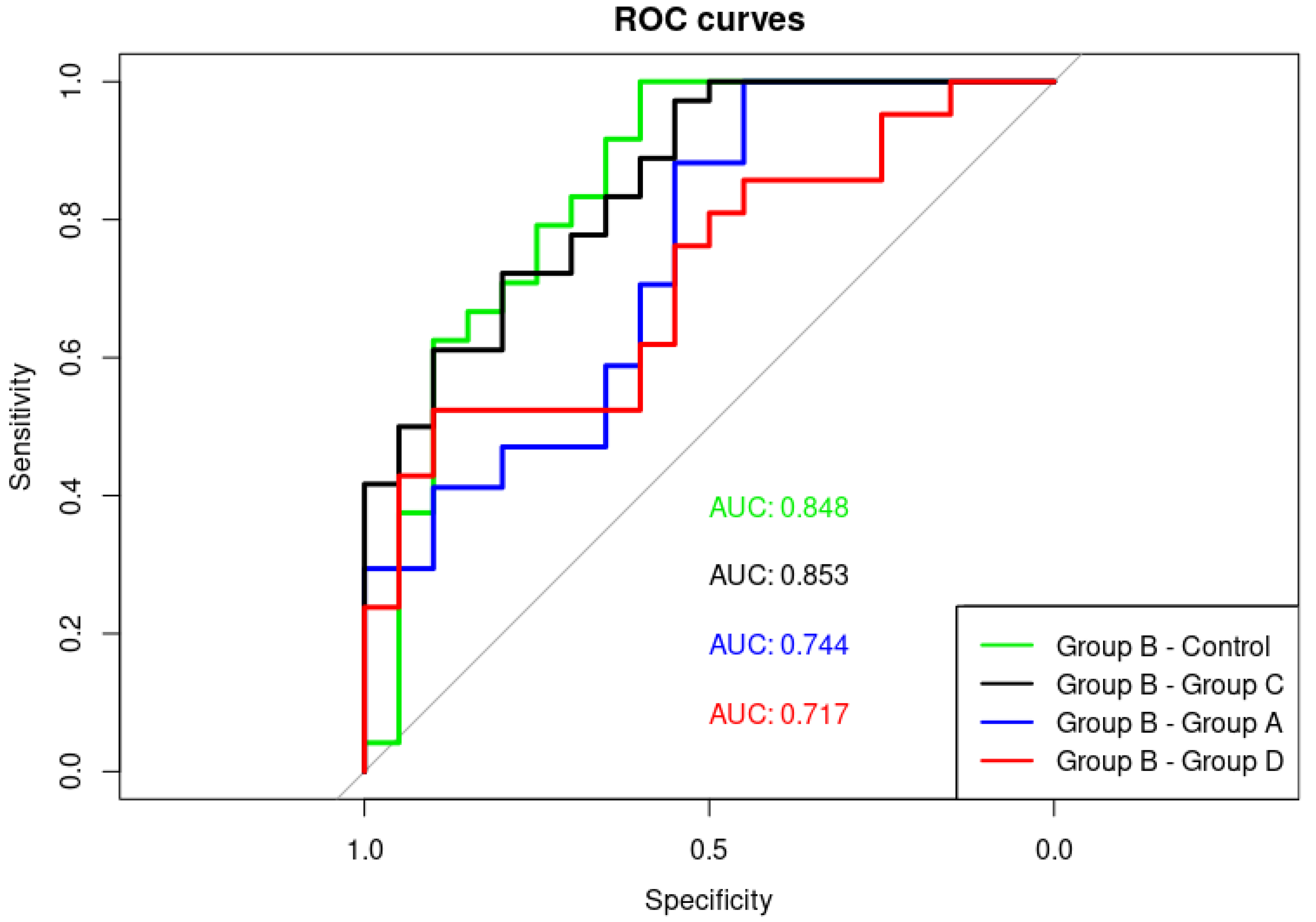
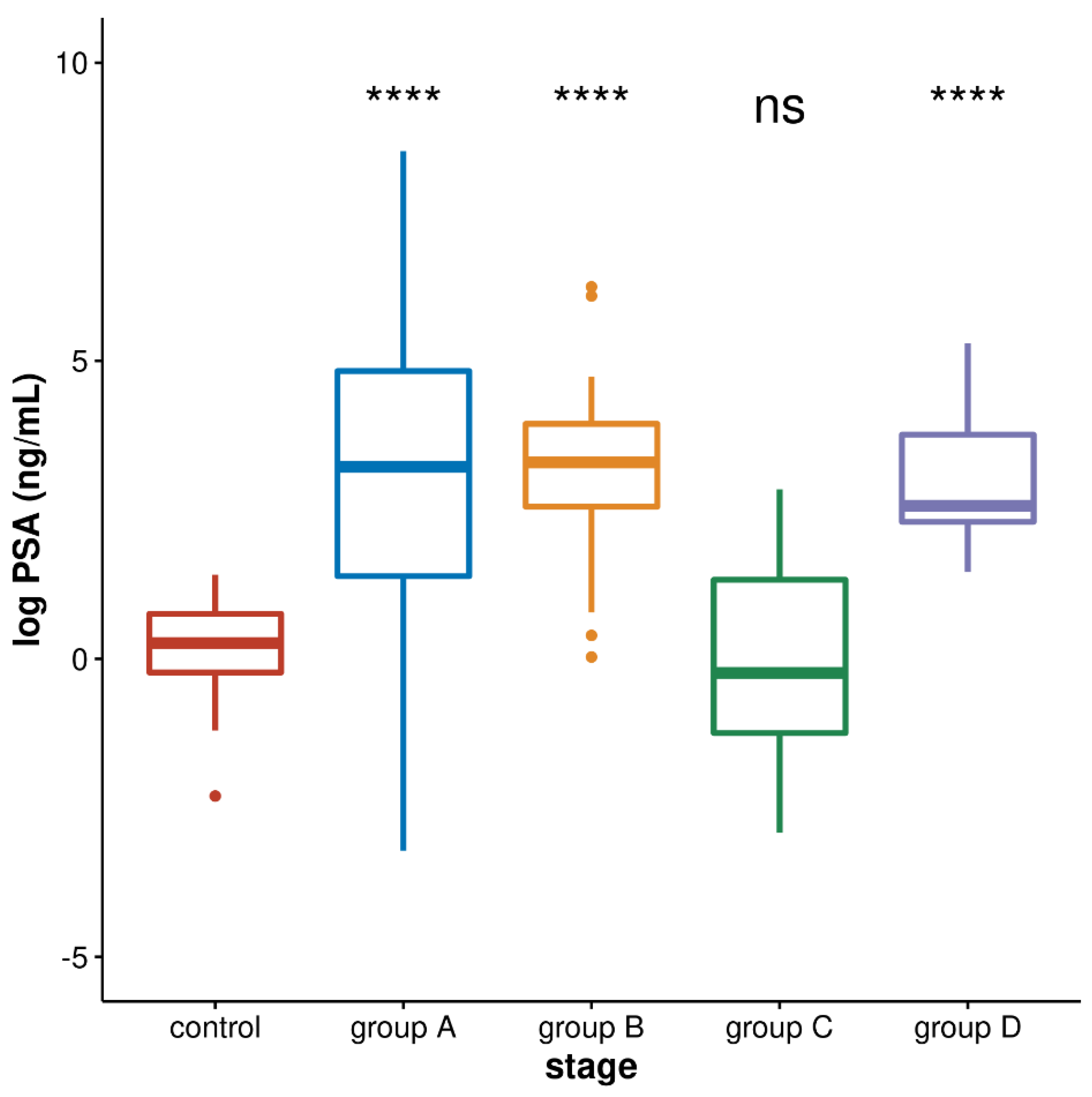
3. Results
3.1. Alpha Satellite RNAs Level in the Blood of Prostate Cancer Patients—qPCR Analysis
3.2. PSA Values in Four Groups of Prostate Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, C.; Wevrick, R.; Fisher, R.B.; Ferguson-Smith, M.A.; Lin, C.C. Human centromeric DNAs. Hum. Genet. 1997, 100, 291–304. [Google Scholar] [CrossRef] [PubMed]
- McNulty, S.M.; Sullivan, B.A. Alpha satellite DNA biology: Finding function in the recesses of the genome. Chromosome Res. 2018, 26, 115–138. [Google Scholar] [CrossRef] [PubMed]
- Saksouk, N.; Simboeck, E.; Déjardin, J. Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin 2015, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, W.L.; Yewdell, W.T.; Bell, J.C.; McNulty, S.M.; Duda, Z.; O’Neill, R.J.; Sullivan, B.A.; Straight, A.F. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. Elife 2017, 6, e25299. [Google Scholar] [CrossRef]
- Feliciello, I.; Sermek, A.; Pezer, Ž.; Matulić, M.; Ugarković, Đ. Heat stress affects H3K9me3 level at human alpha satellite DNA repeats. Genes 2020, 11, 663. [Google Scholar] [CrossRef]
- Frescas, D.; Guardavaccaro, D.; Kuchay, S.M.; Kato, H.; Poleshko, A.; Basrur, V.; Elenitoba-Johnson, K.S.; Katz, R.A.; Pagano, M. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle 2008, 7, 3539–3547. [Google Scholar] [CrossRef] [Green Version]
- Ting, D.T.; Lipson, D.; Paul, S.; Brannigan, B.W.; Akhavanfard, S.; Coffman, E.J.; Contino, G.; Deshpande, V.; Iafrate, A.J.; Letovsky, S.; et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 2011, 331, 593–596. [Google Scholar] [CrossRef] [Green Version]
- Wylie, A.; Jones, A.E.; D’Brot, A.; Lu, W.J.; Kurtz, P.; Moran, J.V.; Rakheja, D.; Chen, K.S.; Hammer, R.E.; Comerford, S.A.; et al. p53 genes function to restrain mobile elements. Genes Dev. 2016, 30, 64–77. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.; Pao, G.M.; Huynh, A.M.; Suh, H.; Tonnu, N.; Nederlof, P.M.; Gage, F.H.; Verma, I.M. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 2011, 477, 179–184. [Google Scholar] [CrossRef]
- Zhu, Q.; Hoong, N.; Aslanian, A.; Hara, T.; Benner, C.; Heinz, S.; Miga, K.H.; Ke, E.; Verma, S.; Soroczynski, J.; et al. Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer. Mol. Cell 2018, 70, 842–853. [Google Scholar] [CrossRef] [Green Version]
- Enukashvily, N.I.; Donev, R.; Waisertreiger, I.S.; Podgornaya, O.I. Human chromosome 1 satellite 3 DNA is decondensed, demethylated and transcribed in senescent cells and in A431 epithelial carcinoma cells. Cytogenet. Genome Res. 2007, 118, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Smurova, K.; De Wulf, P. Centromere and Pericentromere Transcription: Roles and Regulation … in Sickness and in Health. Front. Genet. 2018, 9, 674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vourc’h, C.; Biamonti, G. Transcription of Satellite DNAs in Mammals. Prog. Mol. Subcell. Biol. 2011, 51, 95–118. [Google Scholar] [PubMed]
- Pezer, Ž.; Ugarković, Đ. Satellite DNA-associated siRNAs as mediators of heat shock response in insects. RNA Biol. 2012, 9, 587–595. [Google Scholar] [CrossRef] [Green Version]
- Feliciello, I.; Akrap, I.; Ugarković, Đ. Satellite DNA Modulates Gene Expression in the Beetle Tribolium castaneum after Heat Stress. PLoS Genet. 2015, 11, e1005466. [Google Scholar]
- Sermek, A.; Feliciello, I.; Ugarković, Đ. Distinct Regulation of the Expression of Satellite DNAs in the Beetle Tribolium castaneum. Int. J. Mol. Sci. 2021, 22, 296. [Google Scholar] [CrossRef]
- Prensner, J.R.; Rubin, M.A.; Wei, J.T.; Chinnaiyan, A.M. Beyond PSA: The next generation of prostate cancer biomarkers. Sci. Transl. Med. 2012, 4, 127rv3. [Google Scholar] [CrossRef] [Green Version]
- Saini, S. PSA and beyond: Alternative prostate cancer biomarkers. Cell Oncol. 2016, 39, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Berry, S.J.; Coffey, D.S.; Walsh, P.C.; Ewing, L.L. The development of human benign prostatic hyperplasia with age. J. Urol. 1984, 132, 474–479. [Google Scholar] [CrossRef]
- Choo, K.H.; Vissel, B.; Nagy, A.; Earle, E.; Kalitsis, P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 1991, 19, 1179–1182. [Google Scholar] [CrossRef] [Green Version]
- Aerts, J.L.; Gonzales, M.I.; Topalian, S.L. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. BioTechniques 2004, 36, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruijter, J.M.; Pfaffl, M.W.; Zhao, S.; Spiess, A.N.; Boggy, G.; Blom, J.; Rutledge, R.G.; Sisti, D.; Lievens, A.; De Preter, K.; et al. Evaluation of qPCR curve analysis methods for reliable biomarker discovery: Bias, resolution, precision and implications. Methods 2013, 59, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 20 December 2021).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 11 January 2022).
- Karantanos, T.; Corn, P.G.; Thompson, T.C. Prostate cancer progression after androgen deprivation therapy: Mechanisms of castrate resistance and novel therapeutic approaches. Oncogene 2013, 32, 5501–5511. [Google Scholar] [CrossRef] [PubMed]
- Feldman, B.J.; Feldman, D. The development of androgen-independent prostate cancer. Nat. Rev. Cancer 2001, 1, 34–45. [Google Scholar] [CrossRef]
- Debes, J.D.; Tindal, J.D. Mechanisms of androgen-refractory prostate cancer. N. Engl. J. Med. 2004, 351, 1488–1490. [Google Scholar] [CrossRef]
- Zhu, M.L.; Kyprianou, N. Androgen receptor and growth factor signaling crosstalk in prostate cancer cells. Endocr. Relat. Cancer 2008, 15, 841–849. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef] [Green Version]
- Mateo, J.; Seed, G.; Bertan, C.; Rescigno, P.; Dolling, D.; Figueiredo, I.; Miranda, S.; Rodrigues, D.N.; Gurel, B.; Clarke, M.; et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J. Clin. Investig. 2020, 130, 1743–1751. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Mateo, J.; Walsh, M.F.; De Sarkar, N.; Abida, W.; Beltran, H.; Garofalo, A.; Gulati, R.; Carreira, S.; Eeles, R.; et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N. Engl. J. Med. 2016, 375, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Bluemn, E.G.; Coleman, I.M.; Lucas, J.M.; Coleman, R.T.; Hernandez-Lopez, S.; Tharakan, R.; Bianchi-Frias, D.; Dumpit, R.F.; Kaipainen, A.; Corella, A.N.; et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 2017, 32, 474–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feliciello, I.; Pezer, Ž.; Sermek, A.; Bruvo Mađarić, B.; Ljubić, S.; Ugarković, Đ. Satellite DNA-mediated gene expression regulation: Physiological and evolutionary implication. Prog. Mol. Subcell. Biol. 2021, 60, 145–168. [Google Scholar] [PubMed]
- Onishi-Seebacher, M.; Erikson, G.; Sawitzki, Z.; Ryan, D.; Greve, G.; Lübbert, M.; Jenuwein, T. Repeat to gene expression ratios in leukemic blast cells can stratify risk prediction in acute myeloid leukemia. BMC Med. Genom. 2021, 14, 166. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Bosch-Presegué, L.; Vaquero, A. Sirtuins in stress response: Guardians of the genome. Oncogene 2014, 33, 3764–3775. [Google Scholar] [CrossRef] [Green Version]
- Korotkov, A.; Seluanov, A.; Gorbunova, V. Sirtuin 6: Linking longevity with genome and epigenome stability. Trends Cell Biol. 2021, 31, 994–1006. [Google Scholar] [CrossRef]
- Santos-Barriopedro, I.; Vaquero, A. Complex role of SIRT6 in NF-κB pathway regulation. Mol. Cell Oncol. 2018, 5, e1445942. [Google Scholar] [CrossRef] [Green Version]
- Tasselli, L.; Xi, Y.; Zheng, W.; Tennen, R.I.; Odrowaz, Z.; Simeoni, F.; Li, W.; Chua, K.F. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016, 23, 434–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, L.; Lin, G.; Sun, L.; Liu, Y.; Huang, X.; Cao, C.; Guo, Y.; Xie, C. Upregulation of SIRT6 predicts poor prognosis and promotes metastasis of non-small cell lung cancer via the ERK1/2/MMP9 pathway. Oncotarget 2016, 7, 40377–40386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galletti, G.; Portella, L.; Tagawa, S.T.; Kirby, B.J.; Giannakakou, P.; Nanus, D.M. Circulating tumor cells in prostate cancer diagnosis and monitoring: An appraisal of clinical potential. Mol. Diagn. Ther. 2014, 18, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Kishikawa, T.; Otsuka, M.; Yoshikawa, T.; Ohno, M.; Yamamoto, K.; Yamamoto, N.; Kotani, A.; Koike, K. Quantitation of circulating satellite RNAs in pancreatic cancer patients. JCI Insight 2016, 1, e86646. [Google Scholar] [CrossRef] [Green Version]
- Özgür, E.; Mayer, Z.; Keskin, M.; Yörüker, E.E.; Holdenrieder, S.; Gezer, U. Satellite 2 repeat DNA in blood plasma as a candidate biomarker for the detection of cancer. Clin. Chim. Acta 2021, 514, 74–79. [Google Scholar] [CrossRef]
- Bersani, F.; Lee, E.; Kharchenko, P.V.; Xu, A.W.; Liu, M.; Xega, K.; MacKenzie, O.C.; Brannigan, B.W.; Wittner, B.S.; Jung, H.; et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 15148–15153. [Google Scholar] [CrossRef] [Green Version]
- de Lima, L.G.; Howe, E.; Singh, V.P.; Potapova, T.; Li, H.; Xu, B.; Castle, J.; Crozier, S.; Harrison, C.J.; Clifford, S.C.; et al. PCR amplicons identify widespread copy number variation in human centromeric arrays and instability in cancer. Cell Genom. 2021, 1, 100064. [Google Scholar] [CrossRef]
| Group | Characteristics |
|---|---|
| Healthy controls | |
| n | 27 |
| average age/y | 39.4 |
| age min–max/y | 19–59 |
| Group A patients | |
| n | 19 |
| average age/y | 74.5 |
| age min–max/y | 62–85 |
| Group B patients | |
| n | 20 |
| average age/y | 67.4 |
| age min–max/y | 51–87 |
| Group C patients | |
| n | 34 |
| average age/y | 69.9 |
| age min–max/y | 57–83 |
| Group D patients | |
| n | 21 |
| average age/y | 71.9 |
| age min–max/y | 47–83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ljubić, S.; Sermek, A.; Prgomet Sečan, A.; Prpić, M.; Jakšić, B.; Murgić, J.; Fröbe, A.; Ugarković, Đ.; Feliciello, I. Alpha Satellite RNA Levels Are Upregulated in the Blood of Patients with Metastatic Castration-Resistant Prostate Cancer. Genes 2022, 13, 383. https://doi.org/10.3390/genes13020383
Ljubić S, Sermek A, Prgomet Sečan A, Prpić M, Jakšić B, Murgić J, Fröbe A, Ugarković Đ, Feliciello I. Alpha Satellite RNA Levels Are Upregulated in the Blood of Patients with Metastatic Castration-Resistant Prostate Cancer. Genes. 2022; 13(2):383. https://doi.org/10.3390/genes13020383
Chicago/Turabian StyleLjubić, Sven, Antonio Sermek, Angela Prgomet Sečan, Marin Prpić, Blanka Jakšić, Jure Murgić, Ana Fröbe, Đurđica Ugarković, and Isidoro Feliciello. 2022. "Alpha Satellite RNA Levels Are Upregulated in the Blood of Patients with Metastatic Castration-Resistant Prostate Cancer" Genes 13, no. 2: 383. https://doi.org/10.3390/genes13020383
APA StyleLjubić, S., Sermek, A., Prgomet Sečan, A., Prpić, M., Jakšić, B., Murgić, J., Fröbe, A., Ugarković, Đ., & Feliciello, I. (2022). Alpha Satellite RNA Levels Are Upregulated in the Blood of Patients with Metastatic Castration-Resistant Prostate Cancer. Genes, 13(2), 383. https://doi.org/10.3390/genes13020383








