Abstract
Metopaulias depressus is a non-marine crab endemic to Jamaica that dwells in rainforest bromeliads and exhibits elaborate active parental care behavior. Current genomic resources on M. depressus are rare, limiting the understanding of its adaptation to terrestrial life in species that evolved from marine ancestors. This study reports the complete mitochondrial genome of M. depressus assembled using Sanger sequencing. The AT-rich mitochondrial genome of M. depressus is 15,765 bp in length and comprises 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, and 22 transfer RNA genes. A single 691 bp-long intergenic space is assumed to be the control region (CR) or D-loop. A set of selective pressure analyses indicate that the entirety of the PCGs experience purifying selection. Cox1, cox2, nad5, cox3, and atp6 experience strong purifying selection, and atp8 experiences weak purifying selection compared to the rest of the PCGs. The secondary structures of most tRNA genes exhibit a standard ‘cloverleaf’ structure, with the exception of trnS1, which lacks the dihydroxyuridine (DHU) arm but not the loop, the trnH gene, which lacks the thymine pseudouracil cytosine (T) loop but not the arm, and trnM, which exhibits an overly developed T loop. A maximum likelihood phylogenetic analysis based on all PCGs indicated that M. depressus is more closely related to the genera Clistocoeloma, Nanosesarma, and Parasesarma than to Chiromantes, Geosesarma, and Orisarma. This study contributes to deciphering the phylogenetic relationships within the family Sesarmidae and represents a new genomic resource for this iconic crab species.
1. Introduction
Within the Decapoda, crabs belonging to the Infraorder Brachyura are recognized for their astonishing anatomical, ecological, physiological, and behavioral diversity [1,2]. Among them, the family Sesarmidae is a speciose clade that has successfully colonized marine intertidal and supratidal zones [3,4,5]. Some lineages have even radiated into freshwater and terrestrial habitats, and these non-marine sesarmids often exhibit abbreviated larval development and complex parental–offspring interactions [6]. Metopaulias depressus sets itself apart even within this remarkable family due to its unique forest-dwelling lifestyle and active parental care behavior [3,7,8].
Metopaulias depressus is endemic to Jamaica and inhabits epiphytic and bottom-dwelling large bromeliads in central and western rainforests of the island [7]. As in most decapod crabs, embryos of M. depressus hatch as planktonic larvae, but larval development is abbreviated (9–10 days). Breeding females release their larvae in small water reservoirs located in bromeliad leaf axils, tending their offspring for about eight weeks. Females actively clean the leaf axil of all litter and organic debris except for land snail shells, which are retained. This manipulation in the “nursery axil” improves the dissolved oxygen and the carbon dioxide balance of the axil water and changes the pH from acid to neutral [9,10]. Additionally, female protective behavior reduces the mortality of offspring in the nursery by predatory damselfly nymphs and spiders [11]. Mothers feed their young with prey (i.e., snails, millipedes) caught nearby and carried into the nursery axil [7]. Parental care in this species results in the establishment of a family or a helper group–i.e., a mother and its offspring. This example shows the behavioral plasticity of primarily marine crustaceans when inhabiting unusually harsh, i.e., non-marine, environments.
Active parental care of post-hatching offspring has been observed in other sesarmid crabs that have adapted to adverse terrestrial or semi-terrestrial environments (e.g., Geosesarma notophorum [12], Sesarma jarvisi [13]). In addition, post-hatching parental care is widely recognized among freshwater Astacidea (e.g., Procambarus clarkii [14,15,16]; Orconectes inermis inermis and O. pellucidus [17]; see review in [18]). These examples show the behavioral plasticity and the potential for advanced social behavior in crustaceans evolving from marine ancestors when colonizing unusually harsh environments.
Despite the remarkable lifestyle and behavior of M. depressus and other semi-terrestrial sesarmid crabs, only a limited number of genomic resources exist for these crabs [19,20]), which, in turn, limits the understanding of adaptations to terrestrial life and the genomic mechanisms driving abbreviated development and active parental care. This study forms part of a broader effort aimed at developing genomic resources for comparing marine, semi-terrestrial, and terrestrial crabs, especially those belonging to the subsection Thoracotremata, as it includes most crabs with terrestrial adaptations. This subsection, however, is in need of taxonomic stability, because the most commonly used superfamily classification [21] does not correspond to current knowledge of phylogenetic relationships [22,23,24,25]. Herein, we sequenced and characterized in detail the complete mitochondrial genome of an additional representative of the family Sesarmidae, M. depressus, known as one of the most successful and ecologically specialized crabs that became independent of the sea in a relatively short time frame [8]. The comparison with other known thoracotreme mitogenomes will also help to outline and support the establishment of phylogeny-based groupings.
2. Materials and Methods
2.1. Specimen Collection and Mitochondrial Genome Sequencing
The used specimen was collected during a field trip and visit to the Windor Great House near Sherwood (Trelawny) in Cockpit Country, Jamaica. Collecting permits were obtained beforehand. DNA extraction was conducted using the DNeasy Tissue Kit (Qiagen, Hilden, Germany), following the manufacturer’s protocol. Next, the mitochondrial genome of M. depressus was assembled using a primer-walking strategy with the set of primer pairs developed by [26]. More specifically, the whole mitochondrial genome of M. depressus was first amplified in three long overlapping PCR products. Next, these products were used as templates for amplifying shorter fragments (PCR products > 800 bp) using the Sanger sequencing method, employing a primer-walking strategy. For more details such as primer sequences and PCR conditions, see [26].
2.2. Mitochondrial Genome Annotation and Characterization
The in silico annotation of the mitochondrial genome of M. depressus was conducted with the web servers MITOS (http://mitos.bioinf.uni-leipzig.de/index.py, accessed on 15 May 2021 [27]) and MITOS 2 (http://mitos2.bioinf.uni-leipzig.de/index.py, accessed on 15 May 2021 [28]) using the invertebrate genetic code. Manual curation of the in silico annotation, including start and stop codon corrections, was conducted using the web server Expasy Translate Tool (https://web.expasy.org/translate/, accessed on 15 May 2021 [29]) and the software MEGA 7 [30]. Mitochondrial genome circular visualization was performed with the web server GenomeVx (http://wolfe.ucd.ie/GenomeVx/, accessed on 15 May 2021 [31]).
The nucleotide composition of the entire mitochondrial chromosome and each protein coding gene (PCG) was estimated with the software MEGA 7 [30].
Codon usage of each PCG was estimated using the invertebrate genetic code in the Sequence Manipulation Suite: Codon usage web server (https://www.bioinformatics.org/sms2/codon_usage.html, accessed on 15 May 2021 [32]), and visualization of the Relative Synonymous Codon Usage (RSCU) was performed using the EZcodon tool in the EZmito web server (http://ezmito.unisi.it/ezcodon, accessed on 15 May 2021 [33]).
To explore selective pressures on each mitochondrial PCG, a pairwise comparison was performed between M. depressus and Clistocoeloma sinense (GenBank: NC_033866). The number of nonsynonymous substitutions per nonsynonymous site (Ka), synonymous substitutions per synonymous site (Ks), and the ratio Ka/Ks (ω) were estimated using the software KaKs_calculator 2.0 [6]. If a PCG experiences neutral selection, then ω = 1. Negative or purifying selection is indicated by values ω < 1, whereas positive or diversifying selection is denoted by values ω > 1. The γ-MYN model was used to account for variable mutation rates along each sequence during calculations [34].
tRNA and their secondary structures were predicted using the program MiTFi [35], as implemented in MITOS and MITOS2. The visualization of the secondary structure for each tRNA was conducted using the FORNA web server (http://rna.tbi.univie.ac.at/forna/, accessed on 15 May 2021 [35,36]).
The control region was examined in detail. First, microsatellites were detected using the web server Microsatellite Repeats Finder (http://insilico.ehu.es/mini_tools/microsatellites/, accessed on 15 May 2021 [37]). Next, the presence of tandem repeats in this region was explored using the web server Tandem Repeats Finder (https://tandem.bu.edu/trf/trf.html, accessed on 15 May 2021 [38]). Lastly, the secondary structure, including the presence of hairpin structures, in the control region was explored using the RNAstructure Secondary Structure web server (https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html, accessed on 15 May 2021 [39]).
2.3. Phylogenetic Position of Metopaulias Depressus
The phylogenetic position of M. depressus among other representatives of the family Sesarmidae was examined based on PCGs. Our analysis was conducted with amino acids instead of nucleotides due to the fact that the phylogenetic signal from nucleotide characters alone has the potential to be saturated. The newly sequenced and annotated mitogenome of M. depressus, together with those of 11 other species (6 genera) belonging to the family Sesarmidae available in GenBank (consulted: 19 December 2021) were used for the phylogenetic analysis conducted using the software MitoPhAST V2.0 [40].
Outgroups included species from each of the families Grapsidae, Gecarcinidae, Ocypodidae, Xenograpsidae, and Varunidae. MitoPhAST first extracted all 13 PCG nucleotide sequences from the species available in GenBank and any others provided by the user (i.e., M. depressus). Next, each PCG nucleotide sequence was translated to amino acids and each PCG amino acid sequence was then aligned using Clustal Omega [41,42]. Poorly aligned regions were removed with trimAl v1.2.0 [43] before the dataset was partitioned and the best fitting models of sequence evolution were selected with ProtTest3 v3.4 [44]. Lastly, the concatenated and partitioned PCG amino acid alignments were used to perform a maximum likelihood phylogenetic tree search in the software IQ-TREE [45]. The robustness of the ML tree topology was ascertained by 1000 bootstrap pseudoreplicates of the tree search.
3. Results and Discussion
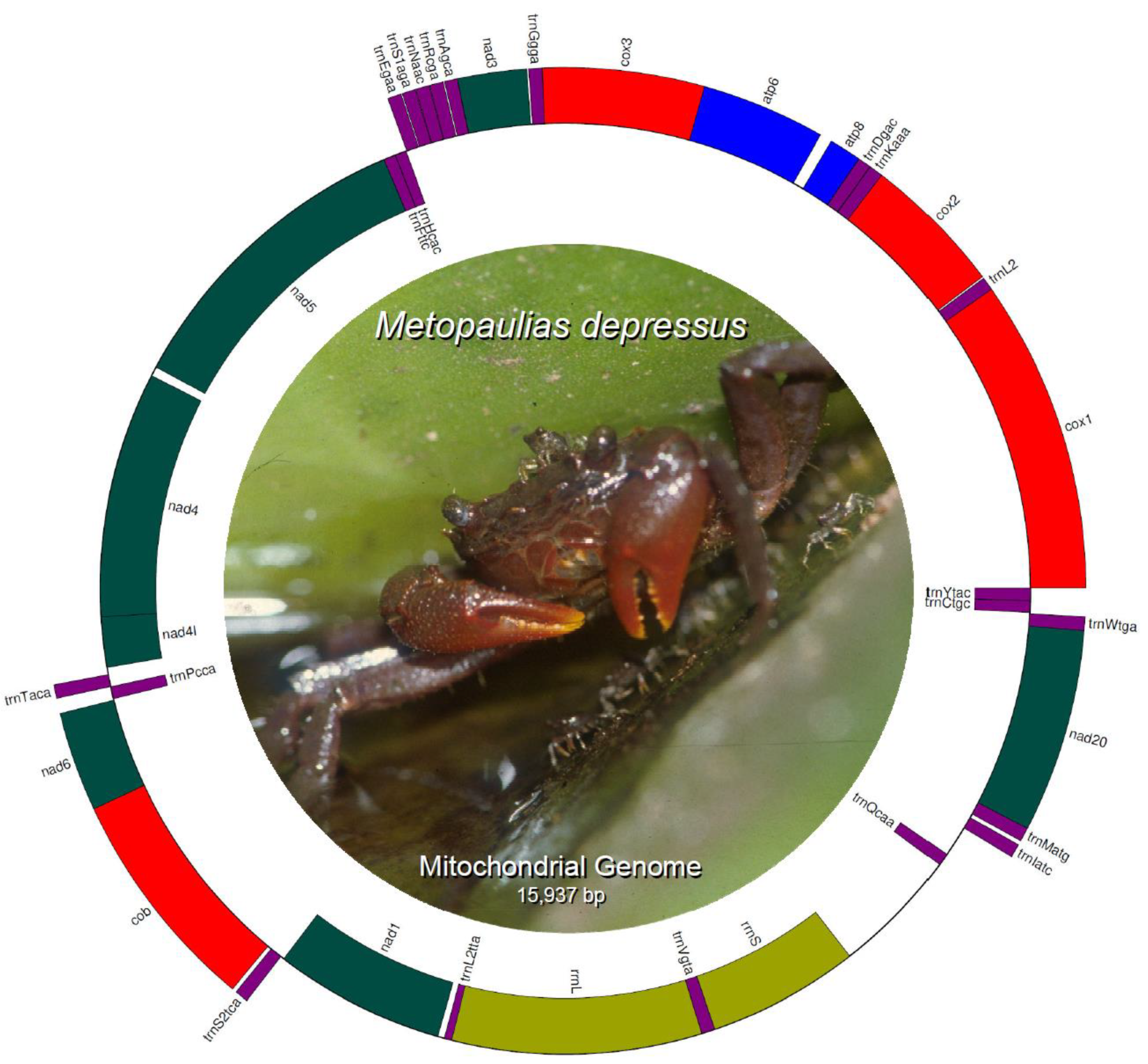
The mitochondrial genome of Metopaulias depressus (KX118277) is 15,765 bp in length and encodes 13 protein coding genes (PCGs), 22 transfer RNA genes, 2 ribosomal RNA genes (rrnL [16S] and rrnS [12S]), and a single, relatively long (691 bp) non-coding putative control region. Most of the PCGs and tRNA genes are encoded on the L-strand, whereas only four PCGs (nad5, nad4, nad4l, and nad1), the two ribosomal RNA genes, and eight tRNA genes (trnH, trnF, trnP, trnL2, trnQ, trnV, trnC, and trnY) are encoded in the H-strand (Table 1) (Figure 1). Gene order and strand arrangement in M. depressus is identical to that reported before in all co-familiar species (except G. penangense [46]) with mitochondrial genomes deposited in GenBank (i.e., O. neglectum, O. sinense, P. bidens, and P. tripectinis, among others [47,48,49,50]). In contrast to the gene arrangement observed in all sesarmid crabs, with a trnQ-trnI-trnM, G. penangense exhibits a trnI-trnQ-trnM gene arrangement [46]. Compared to other decapod infraorders, brachyuran crabs (infraorder Brachyura) contain a translocation of the trnH gene between the trnE and trnF genes, rather than between the nad5 and nad4 genes [5]. This translocation is present in M. depressus and all co-familiar species [5,47,48,49,51].

Table 1.
Mitochondrial genome of Metopaulias depressus. Arrangement and annotation.
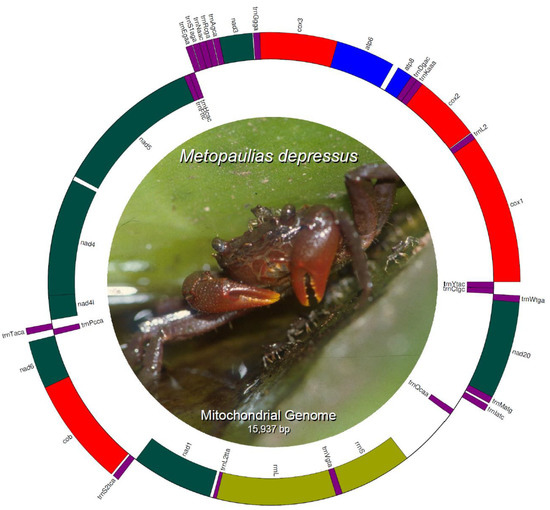
Figure 1.
Circular genome map of Metopaulias depressus mitochondrial DNA. Photo credit: Rudolph Diesel.
The overall nucleotide composition of the mitochondrial genome’s light DNA strand was as follows: A = 37.9%, G = 8.7%, C = 14%, and T = 39.4%, with a GC-content equal to 22.7% and an AT-content equal to 77.3%. This nucleotide usage is within the range reported for other sesarmid crab species (Supplementary Table S1). The highest AT-content value has been reported for Geosesarma penangense (78.44%) [46], whereas the lowest reported AT-content value belongs to Parasesarma tripectinis (74.22%) [50]. AT-skewed mitochondrial genomes are often reported across metazoan clades, including crustaceans and brachyuran crabs [48,52,53].
In the mitochondrial genome of M. depressus, PCGs comprise a total of 3673 codons. Seven (cox1, cox2, atp8, cox3, nad4, cob, and nad2) and five (nad3, nad5, nad6, nad1, and nad4l) of the 13 PCGs use ATG and ATA, respectively, as start codon, whereas atp6 uses ATT as start codon. Nine PCGs use TAA (cox2, atp8, atp6, cox3, nad5, nad4, nad4l, nad6, and nad1) as stop codon and two PCGs use TAG (cox1 and nad2). Lastly, two genes (nad3 and cob) exhibit incomplete (T) stop codons (Table 1). An incomplete stop codon in the cob gene is also observed in the co-familiar species Parasesarma affine, P. pictum, O. neglectum, and E. lafondii [5,6,47,51]. It is assumed that truncated stop codons are completed via post-transcriptional polyadenylation ([54] and references therein).
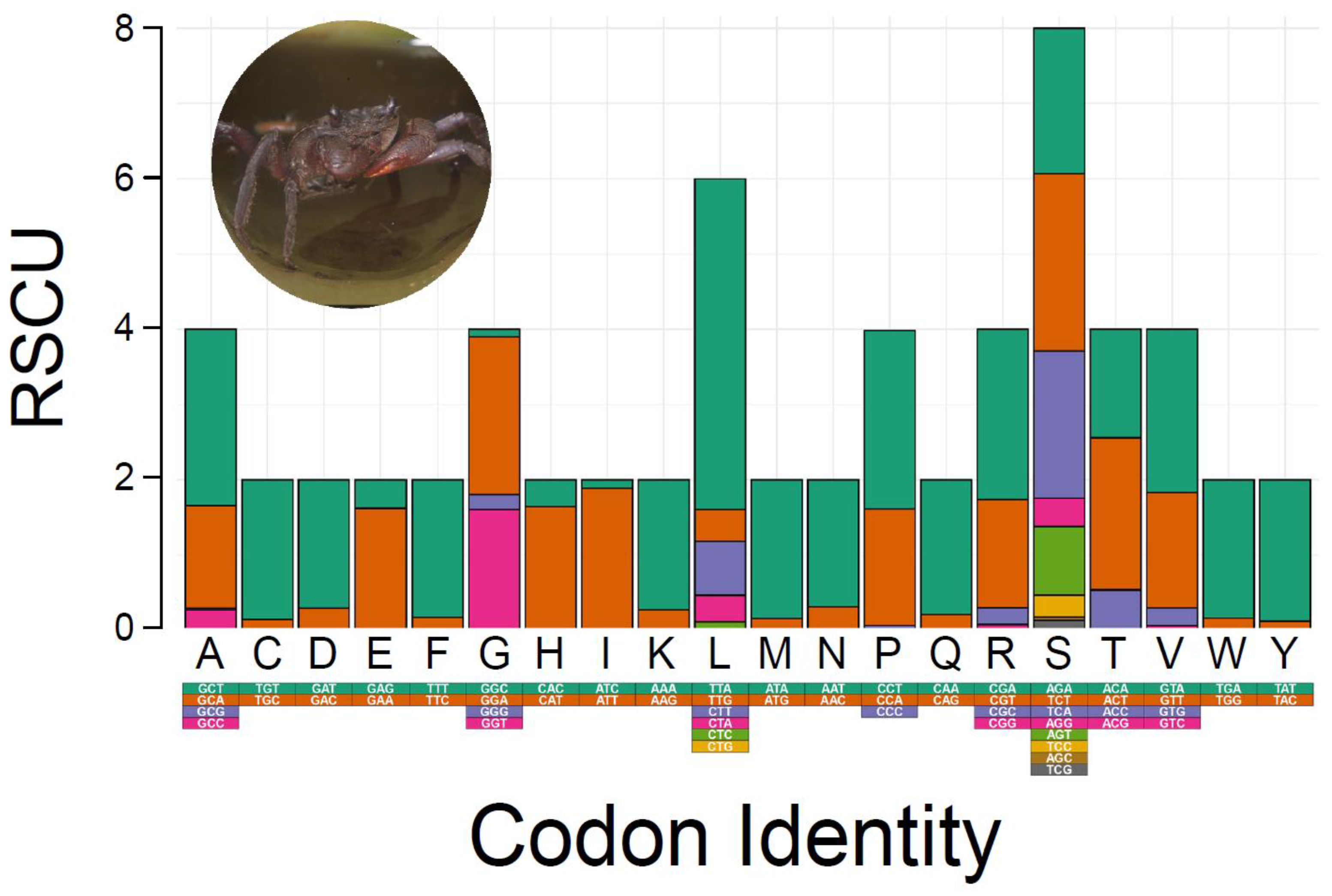
Relative synonymous codon usage (RSCU) and amino acid composition in the PCGs of M. depressus are summarized in Figure 2. The most frequently used codons (amino acids) were: TTA (Leu) used 434 times (73%), ATT (Ile) used 336 times (94%), TTT (Phe) used 317 times (91%), and ATA (Met) used 225 times (92%). Codons (amino acids) that were the least commonly used to encode their respective amino acids (excluding stop codons) included CGC (Ala), used one time (0.01%), CTG (Leu), used one time (undefined %), CGG (Arg), used one time (0.02%), AGC (Ser), used two times (0.01%), and CCC (Pro,) used two times (0.02%) (Supplementary Table S2). RSCU and amino acid composition of PCGs in M. depressus is similar to that reported before in other representatives of the family Sesarmidae. For instance, the most frequently used codons in P. affine, O. sinense, and P. bidens were Leu, Ile, and Phe, in agreement with that observed in this study for M. depressus [5,48,49]. In addition to M. depressus, codons for Met are frequently used in P. pictum [6]. All the codons coding for the aforementioned amino acids are AT-rich, in line with the observed overrepresentation of A and T nucleotides in the mitogenome of M. depressus and other co-familiar crabs [5,46].
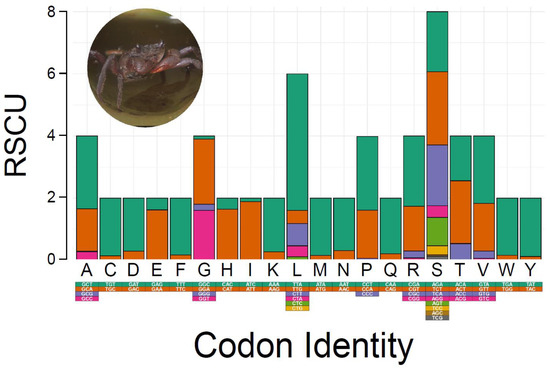
Figure 2.
Relative synonymous codon usage (RSCU) in Metopaulias depressus. Photo credit: Rudolph Diesel.
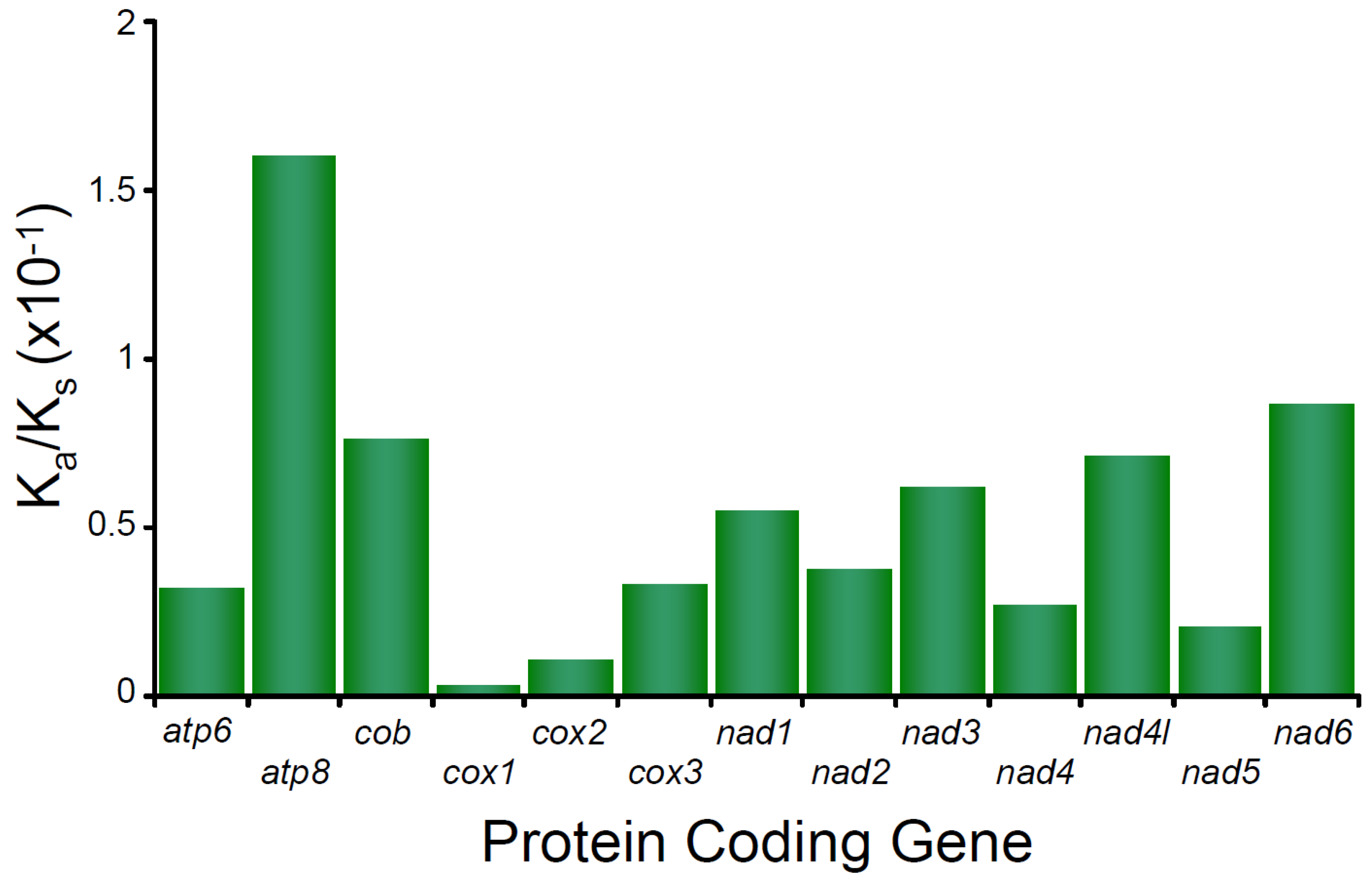
In the mitochondrial genome of M. depressus, the Ka/Ks ratio estimated for all PCGs show values < 1 PCGs (all p values < 0.05), indicating that purifying selection is acting upon all these PCGs. The Ka/Ks ratio estimated for atp8 is the highest observed value (0.16017) compared to the rest of the PCGs and indicates that the purifying selection was relatively weak in this gene. In turn, Ka/Ks ratios calculated for cox1, cox2, nad5, cox3, and atp6 are the lowest observed values (0.029, 0.01069, 0.02027, 0.03302, and 0.03196, respectively) and indicate strong selective pressure affecting the latter PCGs (Figure 3). Selective pressure in PCGs has not been studied before in any other crab belonging to the family Sesarmidae. However, a strong pattern of purifying selection has been reported for many other brachyuran crabs, crustaceans, and arthropods in general ([34] and references therein). A recent study of caridean shrimps (genus Synalpheus) found a relationship between PCG length and the strength of purifying selection, with short genes (e.g., atp8) being subject to weaker purifying selection than longer PCGs [55]. Our observations are in agreement with the aforementioned pattern. Whether or not an association between gene length and the strength of purifying selection exists in sesarmid and other brachyuran crabs remains to be addressed.
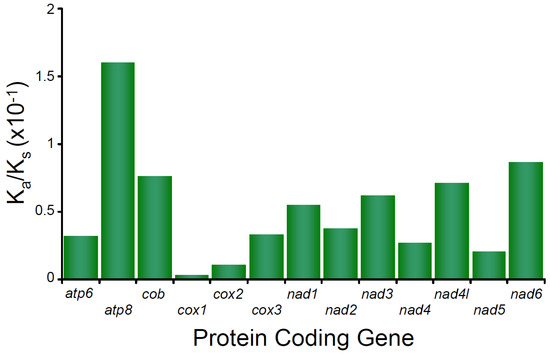
Figure 3.
Selective pressure analysis in the protein coding genes of Metopaulias depressus.
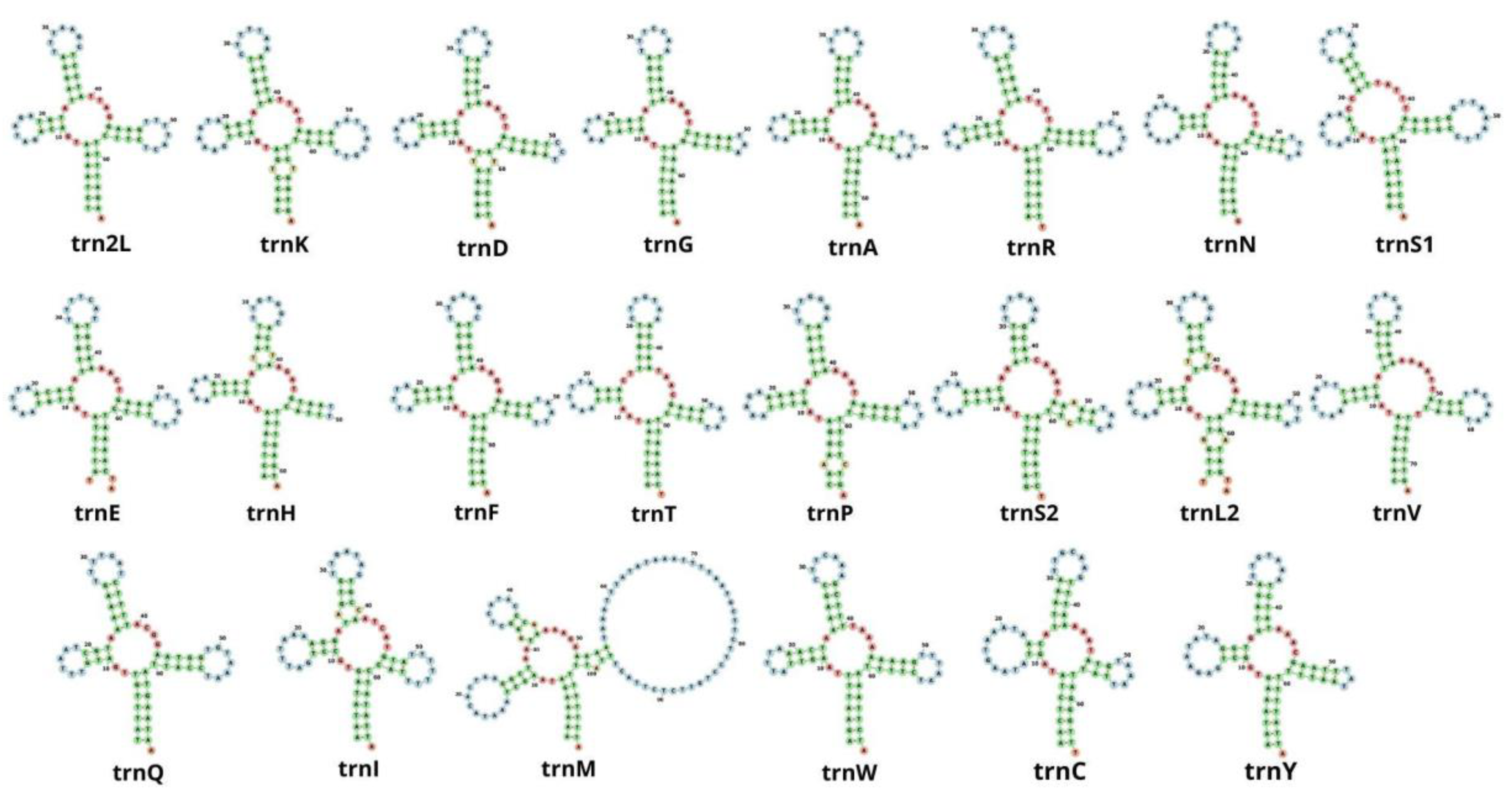
In the mitochondrial genome of M. depressus, 19 out of the 22 tRNA genes exhibited a cloverleaf secondary structure (Figure 4). The trnS1 gene exhibited a deletion of the dihydroxyuridine (DHU) arm, having only its loop. Other co-familiar crabs, including O. sinense, P. pictum, P. affine, P. bidens, G. faustum, G. penangense, C. sinense, and C. haematocheir, presented the same deletion of the DHU arm in the trnS1 gene [5,6,46,48,49,51,52,53,54,55,56,57,58], with the exception of O. neglectum [47], in which all tRNAs exhibited the typical cloverleaf secondary structure. A truncated trnS1 gene represents a conserved mitochondrial feature in eumetazoans, including crabs and other decapod crustaceans [6,49,56].
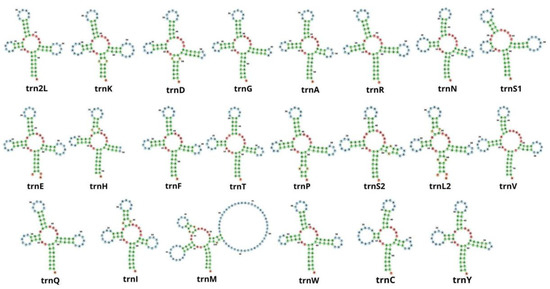
Figure 4.
Secondary structures of 22 transfer RNA genes in Metopaulias depressus.
Unexpectedly, we found two other tRNA genes with a secondary structure that deviates from the expected ‘cloverleaf’ shape: the trnH gene lacks the thymine pseudouracil cytosine (T) loop, and the trnM exhibits an overly developed T loop (Supplementary Figure S1). Some studies have examined the secondary structure of mitochondrial tRNA genes in co-familiar species (Orisarma sinense as C. haematocheir [57], O. sinense [48], P. pictum [6], P. affine [5], P. bidens [49], E. lafondii [51], G. penangense and G. faustum [46], O. neglectum [47], and C. sinense [58]), and truncated arms have also been observed in G. penangense (trnC), G. faustum (trnD, trnH, and trnR) [46], and C. haematocheir (trnS) [57]. Whether or not truncated tRNAs are functional remains to be explored. It has been hypothesized that tRNA editing after the translation of truncated tRNAs might make them functional [59].
In M. depressus, the 691 bp-long control region (CR) is located between the rrnS and trnQ genes, starting at position 13,480 and ending at position 14,170. The length of the CR was similar in range (630 to 751 bp) to that previously reported in other crabs belonging to the family Sesarmidae [5,6,47,48,49,58]. The Microsatellite Repeats Finder analysis found 18 TA-rich microsatellites (SSRs) distributed from position 57 to 680. Most SSRs exhibited TA, AA, and TT di-nucleotide repeats (Supplementary Table S3). The tandem repeat analysis identified one TA-rich tandem repeat, 17 bp in length, repeated four times and located between positions 572 and 637 of the CR. The RNA structure Web Server tool revealed 20 possible secondary structures (Gibbs free energy (ΔG) ranged from −77.8 to −76.7 kcal/mol, Supplementary Figure S2), and in all of them, hairpin structures were observed along most of the entire length of this region. A detailed characterization of the CR is not available for any other sesarmid crab. However, the presence of SSRs, tandem repeats, and numerous hairpin secondary structures are often observed in the CR of other brachyuran crabs as well as other closely or more distantly related decapod crustaceans (e.g., [34] and references therein).
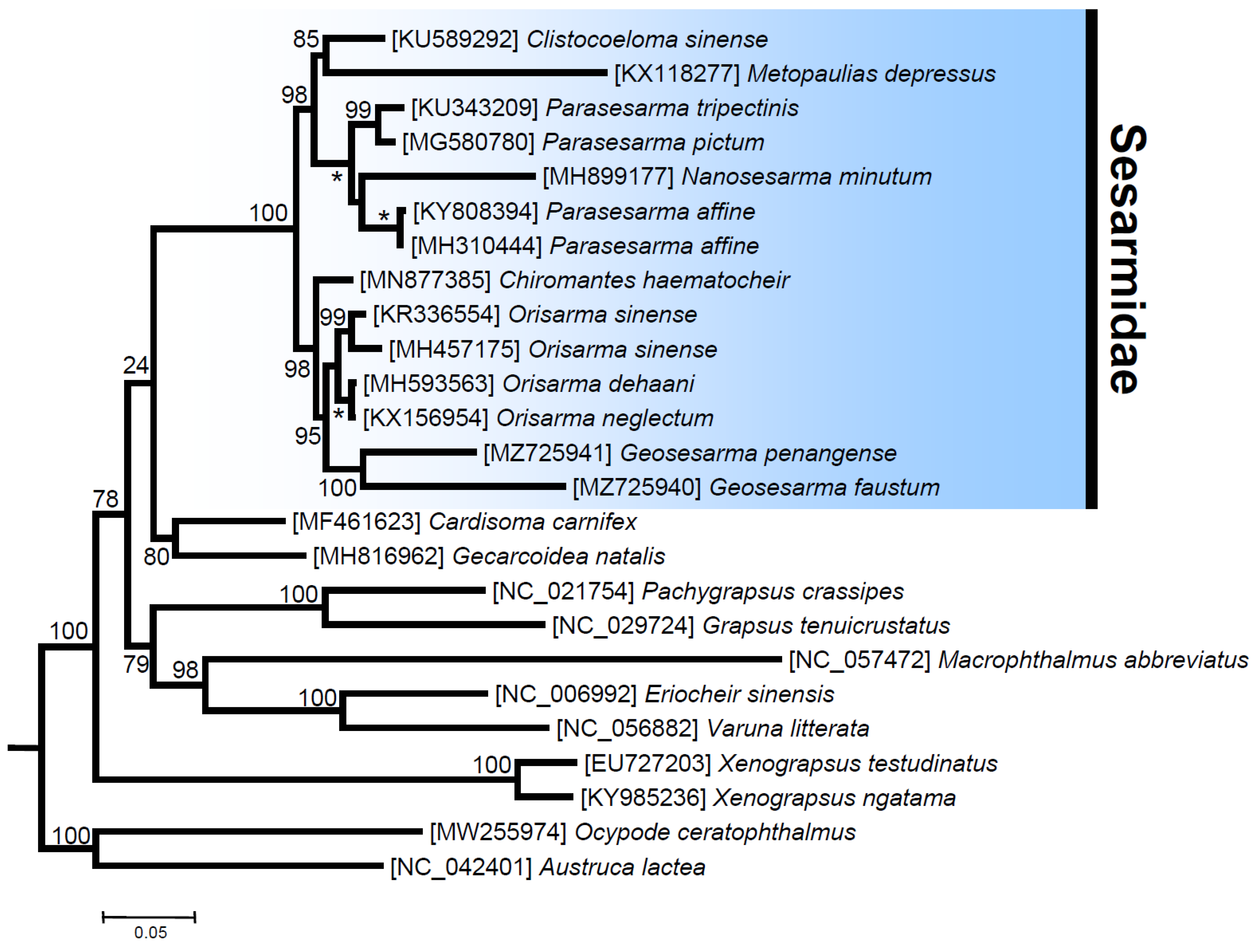
The ML phylogenetic tree with various representatives of the Thoracotremata (25 terminals, 3695 amino acid characters, and 1074 informative sites) fully supports the monophyly of the family Sesarmidae and the other selected crab families (Ocypodidae, Grapsidae, Varunidae, Xenograpsidae, and Gecarcinidae), with bootstrap values (bv) of 100 (except for the Gecarcinidae with bv = 80). Even if inter-familiar relationships are not fully resolved, clear trends become visible. The Ocypodidae, with fiddler and ghost crabs, splits off first, so that all other included families group together as a clear-cut monophylum. This serves as additional evidence that the former superfamily Ocypodoidea has to be redefined with exclusion of the family Macrophthalmidae, for which we provide additional evidence that the latter forms a sister taxon to the Varunidae (bv = 98) (see also [23,24,25]). This will require redefinition of the Grapsoidea at the same time, and one solution is to create a separate superfamily for the Sesarmidae. Within this family, two well-supported clades comprise representatives belonging to the genera Clistocoeloma + Metopaulias + Nanosesarma + Parasesarma (CMNPP clade, bv = 98) and Chiromantes + Geosesarma + Orisarma (CCGO clade, bv = 97). In the first CMNPP clade, Metopaulias and Clistocoeloma form a well-supported clade (bv = 85), sister to representatives of the genera Parasesarma and Nanosesarma (bv = 100). Within the second clade, the genus Parasesarma appears paraphyletic due to the position of Nanosesarma minutum, but the latter genus is in need of revision, because it currently includes all small-sized representatives of the family (Figure 5). In the first CCGO clade, the real Chiromantes haematocheir (as Cristarma eulimene in GenBank, moleculary re-assigned in [60]) is sister to all other species comprised in this clade. The two species of Geosesarma used in this analysis cluster together as a fully supported monophyletic clade. With the species re-assignment [60], the monophyly of the genus Orisarma becomes well supported, considering that the record of “Chiromantes haematocheir” is shown to be another representative of Orisarma sinense [60] and the two together are sister to a second clade that comprises O. dehaani and O. neglectum (Figure 5). Overall, the phylogenetic relationships among genera and families reported in this study are not in full agreement with inferences drawn by previous phylogenetic studies that used complete mitochondrial genomes. However, these included a smaller number of species belonging to the family Sesarmidae and other and fewer members of the Thoracotremata than were included in the present study ([6] and references therein).
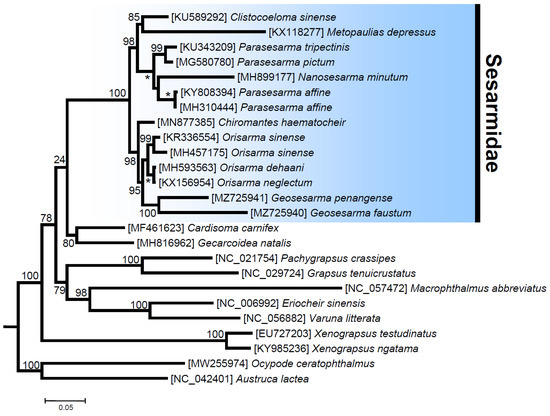
Figure 5.
Total evidence phylogenetic tree obtained from ML analysis based on a concatenated alignment of amino acids of the 13 protein-coding genes present in the mitochondrial genome of Metopaulias depressus and other representatives of the family Sesarmidae. Outgroups included a total of four species belonging to the families Gecarcinidae and Xenograpsidae. The robustness of the ML tree topology was ascertained by 1000 bootstrap pseudoreplicates (numbers above or below the nodes) of the tree search. *: full support, boostrap value = 100.
4. Conclusions
This study sequenced and characterized in detail the mitochondrial genome of the bromeliad crab Metopaulias depressus. Characterization of the complete mitochondrial genome of M. depressus enhances the genomic resources available for the family Sesarmidae and the Thoracotremata and Brachyura in general, particularly its radiation into semi-terrestrial and terrestrial environments. Present and future mitochondrial genomes assembled for other species in these taxa will permit the exploration of the interlink between the colonization of harsh, i.e., non-marine, including terrestrial, environments from marine ancestral species and selective pressures and rates of molecular evolution in mitochondrial genomes.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes13020299/s1, Figure S1: tRNA-M gene secondary structure of Metopaulias depressus exhibiting an unusually developed loop in the T arm; Figure S2: Secondary structure prediction of the control region (CR) in the mitochondrial genome of Metopaulias depressus. Table S1: Nucleotide usage, AT-content, and GC-content in crabs belonging to the family Sesarmidae; Table S2: Codon usage analysis of protein coding genes (PCGs) in the mitochondrial genome; Table S3: Microsatellites present in the control region (CR) of Metopaulias depressus.
Author Contributions
M.A.R.-P., P.L. and J.A.B. analyzed the data. M.A.R.-P., P.L. and J.A.B. drafted the manuscript. J.A.B., L.P. and C.D.S. provided supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This project was partially funded by Creative Inquiry and Clemson Thinks 2, Clemson University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The mitochondrial genome has been deposited in the NCBI with accession number KX118277.
Acknowledgments
J.A.B. thanks Vincent P. Richards for bioinformatic support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Martin, J.W.; Davis, G.E. An Updated Classification of the Recent Crustacea; Natural History Museum of L.A. County: Los Angeles, CA, USA, 2001; pp. 1–124. [Google Scholar]
- Davie, P.J. Crabs; Princeton University Press: Princeton, NJ, USA, 2022. [Google Scholar]
- Schubart, C.D.; Koller, P. Genetic diversity of freshwater crabs (Brachyura: Sesarmidae) from central Jamaica with description of a new species. J. Nat. Hist. 2005, 39, 469–481. [Google Scholar] [CrossRef]
- Diez, Y.L. Notes on the crabs of Cuba II: The family Sesarmidae (Decapoda, Brachyura) in the Oriental region. J. Mar. Res. 2018, 38, 159–167. [Google Scholar]
- Wang, Z.; Wang, Z.; Shi, X.; Wu, Q.; Tao, Y.; Guo, H.; Ji, C.; Bai, Y. Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): Gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura. Int. J. Biol. Macromol. 2018, 118, 31–40. [Google Scholar] [CrossRef]
- Wang, Z.; Shi, X.; Tao, Y.; Wu, Q.; Bai, Y.; Guo, H.; Tang, D. The complete mitochondrial genome of Parasesarma pictum (Brachyura: Grapsoidea: Sesarmidae) and comparison with other Brachyuran crabs. Genomics 2019, 111, 799–807. [Google Scholar] [CrossRef]
- Diesel, R. Parental care in an unusual environment: Metopaulias depressus (Decapoda: Grapsidae), a crab that lives in epiphytic bromeliads. Anim. Behav. 1989, 38, 561–575. [Google Scholar] [CrossRef]
- Schubart, C.D.; Diesel, R.; Hedges, S.B. Rapid evolution to terrestrial life in Jamaican crabs. Nature 1998, 393, 363–365. [Google Scholar] [CrossRef]
- Diesel, R.; Schuh, M. Maternal care in the bromeliad crab, Metopaulias depressus: Maintaining oxygen, pH and calcium levels optimal for the larvae. Behav. Ecol. Sociobiol. 1993, 32, 11–15. [Google Scholar] [CrossRef]
- Diesel, R. Maternal control of calcium concentration in the larval nursery of the bromeliad crab, Metopaulias depressus (Grapsidae). Proc. R. Soc. Ldn. Ser. B Biol. Sci. 1997, 264, 1403–1406. [Google Scholar] [CrossRef]
- Diesel, R. Maternal care in the bromeliad crab, Metopaulias depressus: Protection of larvae from predation by damselfly nymphs. Anim. Behav. 1992, 43, 803–812. [Google Scholar] [CrossRef]
- Ng, P.K.L.; Tan, C.G.S. Geosesarma notophorum sp. nov. (Decapoda, Brachyura, Grapsidae, Sesarminae), a terrestrial crab from Sumatra, with novel brooding behaviour. Crustaceana 1995, 68, 390–395. [Google Scholar]
- Diesel, R.; Horst, D. Breeding in a snail shell: Ecology and biology of the Jamaican montane crab Sesarma jarvisi (Decapoda: Grapsidae). J. Crustac. Biol. 1995, 15, 179–195. [Google Scholar] [CrossRef]
- Little, E.E. Chemical communication of maternal behavior in crayfish. Nature 1975, 255, 400–401. [Google Scholar] [CrossRef]
- Little, E.E. Ontogeny of maternal behavior and brood pheromone in crayfish. J. Comp. Physiol. 1976, 112, 133–142. [Google Scholar] [CrossRef]
- Figler, M.H.; Grants, S.B.; Peeke, H.V.S. Maternal aggression and post-hatch care in red swamp crayfish, Procambarus clarkii (Girard): The influences of presence of offspring, fostering, and maternal molting. Mar. Freshw. Behav. Physiol. 1997, 30, 173–194. [Google Scholar] [CrossRef]
- Bechler, D.L. Copulatory and maternal offspring behavior in the hypogean crayfish Orconectes inermis inermis Cope and Orconectes pellucidus (Tell Kampf) (Decapoda, Astacidea). Crustaceana 1981, 40, 136–143. [Google Scholar] [CrossRef]
- Hazlett, B.A. Parental Behavior in Decapod Crustacea. In Studies in Adaptation: The Behavior of Higher Crustacea; Rebach, S., Dunham, D.W., Eds.; John Wiley & Sons: New York, NY, USA, 1983; pp. 171–193. [Google Scholar]
- Leese, F.; Brand, P.; Rozenberg, A.; Mayer, C.; Agrawal, S.; Dambach, J.; Dietz, L.; Doemel, J.S.; Goodall-Copstake, W.P.; Held, C.; et al. Exploring Pandora’s box: Potential and pitfalls of low coverage genome surveys for evolutionary biology. PLoS ONE 2012, 7, e49202. [Google Scholar] [CrossRef]
- Rozenberg, A.; Brand, P.; Rivera, N.; Leese, F.; Schubart, C.D. Characterization of fossilized relatives of the White Spot Syndrome Virus in genomes of decapod crustaceans. BMC Evol. Biol. 2015, 15, 142. [Google Scholar] [CrossRef]
- Ng, P.K.L.; Guinot, D.; Davie, P. Systema Brachyurorum: Part I. An annotated checklist of extant Brachyuran crabs of the world. Raffles Bull. Zool. 2008, 17, 1–296. [Google Scholar]
- Schubart, C.D.; Cuesta, J.A.; Felder, D.L. Glyptograpsidae, a new brachyuran family from Central America: Larval and adult morphology, and a molecular phylogeny of the Grapsoidea. J. Crustacean Biol. 2002, 22, 28–44. [Google Scholar] [CrossRef]
- Kitaura, J.; Wada, K.; Nishida, M. Molecular phylogeny of grapsoid and ocypodoid crabs with special reference to the genera Metaplax and Macrophthalmus. J. Crustac. Biol. 2002, 22, 682–693. [Google Scholar] [CrossRef]
- Schubart, C.D.; Cannicci, S.; Vannini, M.; Fratini, S. Molecular phylogeny of grapsoid crabs (Decapoda, Brachyura) and allies based on two mitochondrial genes and a proposal for refraining from current superfamily classification. J. Zool. Syst. Evol. Res. 2006, 44, 193–199. [Google Scholar] [CrossRef]
- Tsang, L.M.; Schubart, C.D.; Ahyong, S.T.; Lai, J.C.Y.; Au, E.; Chan, T.Y.; Ng, P.K.L.; Chu, K.H. Evolutionary history of true crabs (Crustacea: Decapoda: Brachyura) and the origins of freshwater crabs. Mol. Biol. Evol. 2014, 31, 1173–1187. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, M.M.; Miya, M.U.; Machida, R.J.; Nishida, M. PCR-based approach for sequencing mitochondrial genomes of decapod crustaceans, with a practical example from kuruma prawn (Marsupenaeus japonicus). Mar. Biotechnol. 2004, 6, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogen. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Donath, A.; Jühling, F.; Al-Ara, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics 2007, 24, 861–862. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Lee, B.D. Python implementation of codon adaptation index. J. Open Source Softw. 2018, 3, 905. [Google Scholar] [CrossRef]
- Conrad, I.; Craft, A.; Thurman, C.L.; Baeza, J.A. The complete mitochondrial genome of the red-jointed brackish-water fiddler crab Minuca minax (LeConte 1855) (Brachyura: Ocypodidae): New family gene order, and purifying selection and phylogenetic informativeness of protein coding genes. Genomics 2021, 11, 565–572. [Google Scholar] [CrossRef]
- Jühling, F.; Püt, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef] [PubMed]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef] [PubMed]
- Bikandi, J.; San Millán, R.; Rementeria, A.; Garaizar, J. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR, and endonuclease restriction. Bioinformatics 2004, 20, 798–799. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Tan, M.H.; Gan, H.; Schultz, M.B.; Austin, C.M. MitoPhAST, a new automated mitogenomic phylogeny tool in the post-genomic era with a case study of 89 decapod mitogenomes including eight new freshwater crayfish mitogenomes. Mol. Phylogenet. Evol. 2015, 85, 180–188. [Google Scholar] [CrossRef]
- Siervers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.S.; Sam, K.K.; Ahmad, A.B.; Siti, K.A.; Abdul Wahab, A.Z.; Shu-Chien, A.C. Gene arrangement and adaptive evolution in the mitochondrial genomes of terrestrial sesarmid crabs Geosesarma faustum and Geosesarma penangensis. Front. Ecol. Evol. 2021, 9, 778570. [Google Scholar] [CrossRef]
- Xing, Y.; Ma, X.; Wei, Y.; Pan, D.; Liu, W.; Sun, H. The complete mitochondrial genome of the semiterrestrial crab, Chiromantes neglectum (Eubrachyura: Grapsoidea: Sesarmidae). Mitochondrial DNA B 2016, 1, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.P.; Xin, Z.Z.; Liu, Y.; Zhang, D.Z.; Wang, Z.F.; Zhang, H.B.; Chai, X.Y.; Zhou, C.L.; Liu, Q.N. The complete mitochondrial genome of Sesarmops sinensis reveals gene rearrangements and phylogenetic relationships in Brachyura. PLoS ONE 2017, 12, e0179800. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.T.; Xin, Z.Z.; Tang, Y.Y.; Yang, T.T.; Tang, B.P.; Sun, Y.; Zhang, D.Z.; Zhou, C.L.; Liu, Q.N.; Yu, X.M. Comparative mitochondrial genome analyses of sesarmid and other brachyuran crabs reveal gene rearrangements and phylogeny. Front. Genet. 2020, 11, 536640. [Google Scholar] [CrossRef]
- Park, Y.J.; Park, C.E.; Lee, S.H.; Sook Ko, H.; Ullah, I.; Hwang, U.W.; Shin, J.H. The complete mitochondrial genome sequence of the intertidal crab Parasesarma tripectinis (Arthropoda, Decapoda, Sesarmidae). Mitochondrial DNA B 2018, 3, 193–194. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, Y.; Gong, L.; Lu, X.; Jiang, L.; Liu, B.; Liu, L.; Lü, Z.; Li, P. Mitochondrial Genome of Episesarma lafondii (Brachyura: Sesarmidae) and Comparison with Other Sesarmid Crabs. J. Ocean Univ. China 2021, 20, 1545–1556. [Google Scholar] [CrossRef]
- Gan, H.Y.; Gan, H.M.; Tan, M.H.; Lee, Y.P.; Austin, C.M. The complete mitogenome of the hermit crab Clibanarius infraspinatus (Hilgendorf, 1869), (Crustacea; Decapoda; Diogenidae)—A new gene order for the Decapoda. Mitochondrial DNA A 2016, 27, 4099–4100. [Google Scholar] [CrossRef]
- Yang, J.S.; Nagasawa, H.; Fujiwara, Y.; Tsuchida, S.; Yang, W.J. The complete mitogenome of the hydrothermal vent crab Gandalfus yunohana (Crustacea: Decapoda: Brachyura): A link between the Bythograeoidea and Xanthoidea. Zool. Scr. 2010, 39, 621–630. [Google Scholar] [CrossRef]
- Baeza, J.A. The complete mitochondrial genome of the Caribbean spiny lobster Panulirus argus. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chak, T.C.; Baeza, J.A.; Barden, P. Eusociality shapes convergent patterns of molecular evolution across mitochondrial genomes of snapping shrimps. Mol. Biol. Evol. 2021, 38, 1372–1383. [Google Scholar] [CrossRef] [PubMed]
- Kilpert, F.; Podsiadlowski, L. The Australian freshwater isopod (Phreatoicidea: Isopoda) allows insights into the early mitogenomic evolution of isopods. Comp. Biochem. Physiol.-D Genom. Proteom. 2010, 5, 36–44. [Google Scholar]
- Li, Q.; Xu, C.; Wang, C.; Liu, G. The complete mitochondrial genome of red-clawed crab Chiromantes haematochir (Sesarmidae: Grapsidae). Mitochondrial DNA B 2019, 4, 53–54. [Google Scholar] [CrossRef]
- Xin, Z.Z.; Liu, Y.; Zhang, D.Z.; Chai, X.Y.; Wang, Z.F.; Zhang, H.B.; Zou, C.L.; Tang, B.P.; Liu, Q.N. Complete mitochondrial genome of Clistocoeloma sinensis (Brachyura: Grapsoidea): Gene rearrangements and higher-level phylogeny of the Brachyura. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Cheng, R.; Xiang, T.; Deng, B.; Wang, Y.; Deng, D.; Zhang, H. The complete mitochondrial genome of the Chinese Daphnia pulex (Cladocera, Daphniidae). Zookeys 2016, 615, 47–60. [Google Scholar]
- Schubart, C.D.; Ng, P.K.L. Revision of the intertidal and semiterrestrial crab genera Chiromantes Gistel, 1848, and Pseudosesarma Serène & Soh, 1970 (Crustacea: Brachyura: Sesarmidae), using morphology and molecular phylogenetics, with the establishment of nine new genera and two new species. Raffles Bull. Zool. 2020, 68, 891–994. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).