Abstract
A global population of already more than seven billion people has led to an increased demand for food and water, and especially the demand for meat. Moreover, the cost of feed used in animal production has also increased dramatically, which requires animal breeders to find alternatives to reduce feed consumption. Understanding the biology underlying feed efficiency (FE) allows for a better selection of feed-efficient animals. Non-coding RNAs (ncRNAs), especially micro RNAs (miRNAs) and long non-coding RNAs (lncRNAs), play important roles in the regulation of bio-logical processes and disease development. The functions of ncRNAs in the biology of FE have emerged as they participate in the regulation of many genes and pathways related to the major FE indicators, such as residual feed intake and feed conversion ratio. This review provides the state of the art studies related to the ncRNAs associated with FE in livestock species. The contribution of ncRNAs to FE in the liver, muscle, and adipose tissues were summarized. The research gap of the function of ncRNAs in key processes for improved FE, such as the nutrition, heat stress, and gut–brain axis, was examined. Finally, the potential uses of ncRNAs for the improvement of FE were discussed.
Keywords:
feed efficiency; miRNAs; lncRNAs; residual feed intake; food conversion ratio; pigs; chicken; sheep and cattle 1. Introduction
The world’s population increased substantially in the last century and is expected to increase by 2 billion people in the next 30 years. While the need to increase animal production to meet global needs seems evident, the limited availability of resources is an important challenge in improving meat and milk production. Moreover, the cost of feed used in animal production has also increased dramatically, which requires animal breeders to find alternatives to reduce feed consumption while maintaining animal productivity. One available alternative to alleviate these problems is to emphasize feed efficiency in animal production. Genetic selection of feed-efficient animals via animal breeding approaches will provide a sustainable way of improving production while reducing feed intake. Thus, less pressure will be placed on the number of resources that are required for animal production. In addition, improving the feed efficiency of livestock has the potential to create a more environmentally friendly farming environment and provide better animal welfare for farmed animals.
In growing animals, ratio or residual (regression) traits can be used to describe feed efficiency [1]. Currently, many breeding companies use a feed conversion ratio (FCR) or feed:gain ratio as an indicator of feed efficiency. The FCR is computed as the total amount of feed intake divided into the total amount of gained weight. Koch et al. [2] introduced the concept of residual feed intake (RFI) and defined it as the difference between an animal’s actual feed intake and its predicted feed intake, the prediction usually being estimated based on energy requirements for maintenance, production, and body condition change. Therefore, a low RFI animal is more feed efficient than an animal with high RFI. Selection based on RFI has been proposed to improve feed efficiency because of its phenotypic independence on growth and maintenance requirements [3]. Some indirect measures of feed efficiency have also been proposed, such as relative growth rate (growth relative to instantaneous body size) [4] and the Kleiber ratio (ADG per unit metabolic BW) [5]. These two measures are rarely used in a breeding program, and the current breeding programs in livestock production mainly use either FCR or RFI as an indicator for improving feed efficiency.
Non-coding RNAs (ncRNAs) are transcribed RNA molecules that do not encode a protein [6], but play numerous regulatory roles in diverse biological processes, including epigenetic modification of DNA and regulation of transcriptional and post-transcriptional gene expression [7]. Amongst the most characterized ncRNAs are long non-coding RNAs (lncRNAs) and microRNAs (miRNAs). lncRNAs, which are at least 200 bases in length, can regulate gene expression through different mechanisms, including DNA methylation [8], histone modification [9], alteration of promoter activities by nucleosome repositioning [10], and epigenetic silencing and repression [11]. In the last few years, the increasing amount of evidence supports that lncRNAs are related to metabolic and immunological regulation and phenotypic variation of complex traits in domestic animals [12,13]. MicroRNAs (miRNAs) are small non-coding RNAs of 18 to 23 nucleotides and highly conserved between species [14]. Through the post-transcriptional regulation of gene expression technology, the miRNAs were found in all bodily fluids and tissues and most cell types [15] and were detected to be associated with different essential biological processes. They can regulate the gene expression through binding to target messenger RNA (mRNA), which ultimately leads to the degradation or inhibition of the targeted transcript. Recent studies have indicated that miRNAs have critical roles in regulating cellular and physiological processes, including neurogenesis, insulin secretion, cellular proliferation, differentiation, and apoptosis [16].
As the whole-genome sequencing of all livestock was completed, a tremendous success in the characterization of ncRNAs in farm animals was also observed. The studies associated with ncRNA in livestock have increased significantly in the last ten years (Table 1). More and more ncRNAs in livestock have been detected over the years. Not only the growth of the number of ncRNAs, but also more functions of ncRNAs in livestock have been described. Several reviews have examined the roles of ncRNAs in livestock phenotypes [6,12,13,17,18,19]. Given the importance of feed efficiency, it is essential to explore the miRNAs roles in the regulation of feed efficiency. This review provides the state-of-the-art studies related to the ncRNAs that underlie feeding efficiency in different livestock species. In this review, the contribution of ncRNAs to feed efficiency and related traits in the liver, muscle, and adipose tissues are summarized. Meanwhile, the research gap of the function of ncRNAs in related processes of feed efficiency, such as the nutrition, heat stress, and the gut–brain axis, are examined. Finally, the potential application of miRNAs in the improvement of feed efficiency are discussed. This review assists the elucidation of ncRNA mechanisms that could improve feed efficiency in livestock species.

Table 1.
Number of miRNAs and lncRNAs in livestock species.
2. The miRNAs Functions in Feed Efficiency
Pigs, cattle, sheep, and poultry are the most important farm animals for human society and provide the primary resource of milk, meat, egg, and wool. Some miRNAs in different tissues, including skeletal muscle, liver, and adipose tissues, have been discovered to have important roles in regulating feed efficiency in those important farm animals (Table 2). The detected feed-efficiency-related miRNAs help to better understand the molecular mechanisms and biological processes associated with feed efficiency in livestock.

Table 2.
The miRNAs involved in the feed efficiency of different livestock species.
2.1. The miRNA in Pig Feed Efficiency
Pig is an essential agricultural animal providing a cost-effective source of meat for human consumption. Over 40% of the world’s meat intake is pork [48]. More than 60% of the total costs of pig production is spent on feeding, and therefore, the critical approach to reduce costs in pig farming is improving feed efficiency. Feeding efficiency in pigs has been reported to be associated with many biological processes and pathways [20,49,50,51]. In pigs, miRNAs have been reported to play important roles in regulating feed efficiency in different tissues (Figure 1 and Table 2).
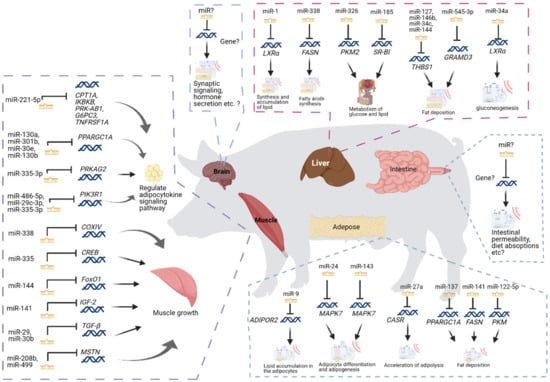
Figure 1.
The miRNAs in pig feed efficiency. This figure was created with BioRender.com (2021).
Skeletal muscle, which is the main place of carbohydrate and lipid metabolism, plays a significant role in utilizing and storing a large proportion of the energy obtained from the feed [52,53,54]. Therefore, the bioenergetic processes within the muscle can deeply influence feed efficiency. Some miRNAs detected in skeletal muscle correspond to two key pathways: one related to mitochondria and energy metabolism and the other related to skeletal muscle growth. Several miRNAs, including miR-338, miR-335, and miR-144, are related to mitochondria and energy metabolism [20,21]. More than 90% of cellular energy is produced by mitochondria through oxidative phosphorylation (OXPHOS) [55]. The miR-338 has been reported to inhibit COX-IV, one of the genes associated with OXPHOS and ATP synthesis and mitochondria transcriptional control, at mRNA and protein levels [56]. The PGC-1 protein is essential in mitochondrial biogenesis by activating cAMP response element binding protein (CREB) and nuclear respiratory factors. The miR-335, which was found to be up-regulated in low RFI pigs, has been reported to target the CREB [57]. The AMP-activated protein kinase (AMPK), which is a key regulator of cellular and whole-body energy balance, can increase mitochondrial proteins of oxidative metabolism, as well as promote the expression of Hexokinase II (HK2) through CREB in skeletal muscle [58,59,60]. The miR-144 has been reported to inhibit the phosphorylation of AMPK alpha, and therefore influence the level of energy metabolism in skeletal muscle [20,61]. The miR-221-5p has been reported to target CPT1A, IKBKB, PRKAB1, G6PC3, and TNFRSF1A genes to regulate the adipocytokine signaling pathway [22]. Several miRNAs, which include miR-208b, miR-499, miR-29, and miR-30b, were reported to be related to the growth and development of skeletal muscle [20]. Previous studies have reported that miR-29 and miR-30b are inhibitors of the transforming growth factor-beta (TGF-β) signaling pathway, which is considered the most potent negative regulator of skeletal muscle growth and development [62,63,64,65]. Both miR-208b and miR-499 have been reported to inhibit the gene expression associated with myostatin, which is an essential part of the TGF-β signaling pathway [66]. Meanwhile, miR-141 was reported to target the gene associated with insulin-like growth factor 2, which is an inhibitor of myogenesis in the myogenesis pathway, and therefore regulates the growth of skeletal muscle [20]. In addition, several miRNAs, including miR-130a, miR-301b, miR-30e, miR-130b, miR-335-3p, miR-486-5p, miR-29c-3p, and miR-335-3p, detected in skeletal muscle were reported to be involved in the adipocytokine signaling pathway [20].
The liver has been regarded as the central organ for systemic metabolism [67,68] and plays an essential role in feed efficiency, as it modulates the efficiency of converting energy obtained from macronutrients into muscle and/or adipose tissue [69]. Several miRNAs detected in pig’s liver were predicted to act as negative regulators of gene expression in fat formation. The miR-545-3p may regulate fat deposition by regulating the GRAMD3 gene, which has been reported as a candidate gene for ectopic fat [23], because of the high negative correlation between GRAMD3 and miR-545-3p [24]. The miR-338 was predicted to negatively regulate the genes associated with fatty acid synthase [24], which is a critical lipogenic enzyme and the rate-limiting step in de novo fatty acid synthesis [70]. The miR-127, miR-146b, miR-34c, and miR-144 have been predicted to inhibit the expression level of THBS1 [24], which is important for the pathogenesis of insulin resistance and adipose tissue inflammation [71]. Several miRNAs detected in the liver of pig were predicted to be associated with the metabolism of glucose, lipid, and protein. For instance, miR-34a, miR-326, miR-1, and miR-185 were identified to participate in the metabolism of glucose and lipids [25]. The lower expression of miR-34a in pig liver can increase the expression of the SIRT1 gene, which plays an important role in fat metabolism [72], and therefore increases gluconeogenesis [25]. The miR-1 was predicted to promote the synthesis and accumulation of lipids by increasing the expression of LXRα [25], that regulates the expression of genes associated with fatty acid synthesis, glucose metabolism, and sterol efflux [73].
The adipose tissue has been suspected of playing a vital role in feed efficiency, as it is the master regulator of systemic lipid storage and an active endocrine organ [74,75]. Several adipocytokines secreted by adipose tissue have been reported to communicate with skeletal muscle, liver and brain and influence various processes, including appetite, lipid, and glucose metabolism and energy homeostasis [74,76,77]. Several miRNAs detected in the adipose tissue were defined to be associated with adipogenesis. The overexpression of miR-9 in the adipose tissue may contribute to the lipid accumulation in the adipocytes [26]. The miR-24-3p has been reported to target the genes associated with adipocyte differentiation in MAPK7-signaling pathways and contribute to adipogenesis [27,78]. Several miRNAs detected in the adipose tissue were reported to be associated with adipocyte lipid metabolism. The miR-27a and miR-143 were reported to regulate the porcine adipocyte lipid metabolism [28]. The over-expression of miR-27a in adipose tissue could accelerate adipolysis to release more glycerol and free fatty acids. The over-expression of miR-143 could accumulate more triglycerides in the adipocytes and therefore promote adipogenesis [28]. Three miRNA, miR-137, miR-144, and miR-122-5p, were defined as the candidate key regulators of fat deposition [31].
In addition to liver, muscle, and adipose tissues, feed efficiency in pigs is also known to be involved in appetite and hormone regulation in the brain [79] as well as the microbiota [80,81] and nutrition digestion and absorption [82,83] in the gut tract. Although various genes have been reported to be involved in these processes, the information associated with how miRNAs can contribute to gene regulation in these tissues is still missing (Figure 1). Therefore, a comprehensive picture of feed efficiency regulations by miRNAs requires the exploration of their functions in the brain and gut.
Although many miRNAs have been reported to be related to feed efficiency in pigs, most studies focused on one tissue and the functions of some miRNAs. Comprehensive studies for miRNAs in multiple tissues for feed efficiency are still lacking. The discoveries of feed-efficiency-related miRNAs in pig’s liver, muscle, and adipose tissues would enhance our understanding of molecular mechanisms of the control of miRNAs in biological processes associated with feed efficiency.
2.2. The miRNA in Cattle Feed Efficiency
The unique ability of cattle to convert lignocellulosic biomass into valuable protein makes cattle one of the most crucial farm animals for human society. Approximately 45% of the global protein supply for humans is provided by meat and milk from cattle and bison [84]. Feed efficiency has some different aspects in the beef cattle and cows. Feed, which is one of the most important factors influencing the profitability of beef cattle farming, represents up to three-quarters of total beef production costs [85]. Compared with other livestock, such as pigs, chickens, and sheep, beef cattle have the lowest production efficiency [84]. Consequently, the cattle industry has shown great interest in improving the feed efficiency of beef production systems. In beef cattle, the miRNAs in the liver and skeletal muscle have been related to feeding efficiency [32,33,34,86,87]. In cows, in addition to the major tissues involved in feed efficiency, the milk production or lactation stages are also important since nutrition is used in the processing of producing milk [88,89]. In fact, the lactation stages could impact miRNAs’ expression and functions [90,91]. However, there is no research devoted to identifying miRNAs in both feed efficiency and lactation in cows. Several miRNAs have been related to regulating feed efficiency in different tissues (Table 2, Figure 2).
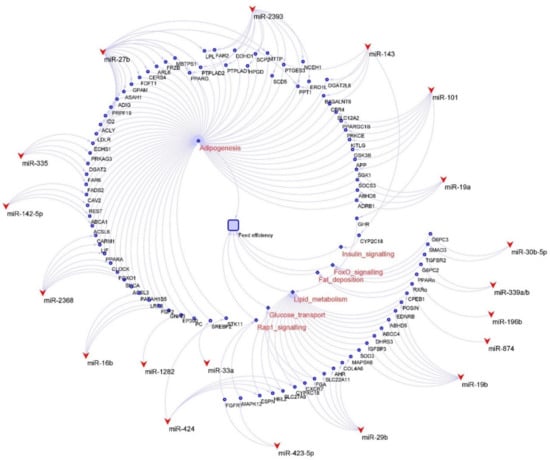
Figure 2.
Candidate miRNAs and related genes and pathways for feed efficiency in cattle. The round shapes indicate genes, the diamond shapes show the pathways, and the V shapes indicate miRNA.
The liver, which is a central controller of metabolism and a significant driver of whole-animal oxygen consumption, plays a crucial role in the feed efficiency of cattle [32]. The main functions of miRNAs in the liver have been reported to regulate the energy metabolism and hepatic metabolism of nutrients, including lipids, carbohydrates, vitamins and minerals, and proteins and amino acids [92]. The feed efficiency is influenced by insulin and energy metabolism, and higher insulin and glucagon levels could reduce feed intake [50,93,94]. Several miRNAs in high RFI cattle have been reported to play important roles in metabolic homeostasis, including glucose and lipid metabolism. For example, miR-143, the most expressed miRNA in the bovine liver, was up-regulated in high RFI cattle [32] and reported to target insulin signaling and its regulation, and therefore inhibit the activation of insulin-stimulated AKT and the homeostasis of glucose homeostasis [95]. The miR-122-3p, which is linked to metabolic control and affects hepatic cholesterol and lipid metabolism [96], was also reported to be highly expressed in bovine liver and up-regulated in high RFI cattle [32]. The miR-29b, which has functions on regulating glucose transport in the liver, muscle, and adipose [97], was also reported to be up-regulated in the liver of high RFI cattle [32]. The miR-30b-5p and miR-339a/b were reported to target the genes associated with the FoxO signaling pathway, which regulates glucose metabolism and resistance to oxidative stress [98], and therefore contribute to higher feed efficiency in Nellore cattle [33]. In addition to those miRNAs, other cattle hepatic miRNA, such as miR-19b, miR-101, miR-106b, and miR-142-3p, were also reported to be up-regulated in high RFI cattle and correlated with lipid metabolism [32,38].
The metabolism of skeletal muscle greatly contributes to the variations in feed efficiency, as 30% of energy expenditure in cattle maintenance is used for turnover of body proteins, and approximately two-thirds of the whole-body protein turnover in mammals is associated with skeletal muscle and liver [33,34,99]. Therefore, the energy metabolism and growth of skeletal muscle have been suggested as a potential strategy for improving feed efficiency in bovines [33,34]. The miRNAs detected in the skeletal muscle of cattle have been reported to be related to the growth of skeletal muscle and energy metabolism in skeletal muscle in previous studies. The miR-423-5p in the skeletal muscle of cattle was found to be differentially expressed between low and high RFI beef cattle [33]. The author also reported that the miR-423-5p targets the genes associated with the Rap1 signaling pathway [33], which is associated with controlling mitogen-activated protein kinase activity [100] and regulating the storage of nutrients in the white adipose tissue and skeletal muscle [94]. The greater expression of miR-34a and miR-2899 was reported in the skeletal muscle of higher RFI cattle [34]. Both miRNAs were predicted to regulate the mRNA expression of heat shock protein beta 1 [34], which influences the degradation of muscle proteins and the rate of protein turnover in skeletal muscle [101]. The miR-148a-3p was found to be highly expressed in skeletal muscle and predicted to target the gene KLF6, which is very important for the development of skeletal muscles in bovines [35]. The skeletal-muscle-derived satellite cells (MDSCs) were reported to regulate postnatal skeletal muscle growth and regeneration. The growth of muscle tissue is highly correlated with the differentiation of MDSCs in cattle [102]. The expressions of 564 known and 53 novel miRNAs in hindlimb muscle were reported to be associated with the differentiation of MDSC in cattle [103]. The miR-224 and miR-130 were reported to impact on the differentiation of adipocytes [36,37].
The adipose tissue, the predominant anatomic site for lipogenesis in ruminants, modulates a large variety of processes related to feed intake, energy homeostasis, and whole-body physiology through its endocrinological activity [104]. The main functions of miRNAs in the adipose tissue have been reported to relate to adipose tissue metabolism and adipogenesis, which are highly associated with feed efficiency traits [39]. Several miRNAs including miR-16b, miR-19a, miR92a/b, miR-101, miR-103, miR106, miR-142-5p, miR-196a, miR-296, miR-2368, and miR-2454, were predicted to target genes associated with functions related to lipid metabolism and/or adipogenesis [38]. The miR-33a and miR-1281 in bovine adipose tissue were reported to, respectively, regulate the genes SREBF2 and EP300, which are involved in lipid metabolism [39]. The miRNAs, including miR-143, miR-27, miR-335, and miR-378, were predicted to have important regulatory functions in adipose tissue and during adipogenesis [40]. MiRNAs were identified and predicted to regulate adipogenesis through their targets and related pathways [41]. Among them, miR-196b and miR-874 were predicted to influence the signal translation of the PPAR pathway, and therefore regulate fat deposition [41]. The miR-424 was reported to promote bovine adipogenesis through an unconventional post-transcriptional regulation of STK11 gene [42]. In addition, 131 bovine miRNAs were predicted to upregulate the bovine adipocytes, and 119 bovine miRNAs were reported to downregulate bovine adipocytes [105].
While genomics has played an important role in improving feed efficiency in cattle [106,107,108], the biology underlying feed efficiency is still not completely understood. In beef cattle, protein turnover, tissue metabolism and stress, digestibility, heat increment, fermentation, and physical activity are the major processes contributing to the variation of residual feed intake [109], but very few studies focus on the roles of miRNAs in these processes. Similarly, some factors, such as feeding behaviors, physical activity, digestibility, and methane emissions, significantly contribute to the variation of feed efficiency in cows. The biological and physical aspects of these effects are not fully addressed. Therefore, it is worthwhile to investigate whether miRNAs are involved in the regulation of the process. Evidently, further studies are required to characterize the link between host miRNAs in regulating the rumen metabolites, the microbiome in divergent feed efficient cattle. Meanwhile, several miRNAs, such as miR-424 or miR-101, could impact the feed efficiency via regulating genes in different processes (Figure 2); therefore, these miRNAs might be prioritized for functional characterization. Nevertheless, the identified differentially expressed feed-efficiency-associated miRNAs in cattle not only help people to understand their potential molecular regulatory mechanisms relating to feed efficiency in cattle, but also provide potential candidate molecular targets for the selection of cattle with improved feed efficiency.
2.3. The miRNA in Sheep Feed Efficiency
Sheep is one of the most crucial farm animals worldwide and provides high-quality meat and milk to human society. Increasing feed efficiency is important for the sheep farming industry to keep a stable output and improve the overall profitability from farming sheep because feed represents 65–70% of the total cost of sheep production systems [110,111,112]. The energy metabolism and growth of skeletal muscle play important roles in the feed efficiency of sheep, as skeletal muscle is a major tissue for energy utilization and maintenance of metabolic health as well as provides lean tissues for meat animals [113,114]. Several miRNAs in skeletal muscle and adipose tissue have been found to play important roles in regulating feed efficiency (Figure 3 and Table 2).
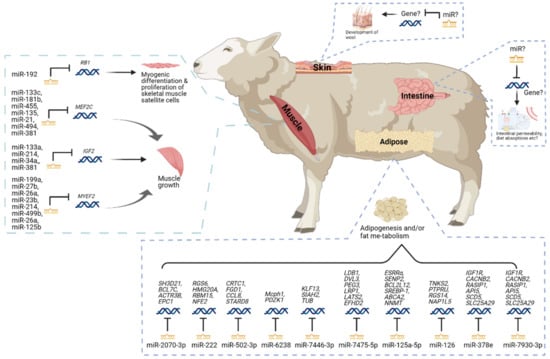
Figure 3.
The miRNAs in sheep feed efficiency. This figure was created with BioRender.com (2021).
The miRNAs detected in the skeletal muscle of sheep have been related to the growth of skeletal muscle in previous studies. A total of 345 miRNAs, including 151 up-regulated and 94 down-regulated miRNAs, were differentially expressed in sheep’s skeletal muscle [43]. Among those miRNAs, some were predicted to be involved in the signaling transduction pathways associated with muscle development, such as the Wnt signaling pathway, hippo signaling pathway, and thyroid hormone signaling pathway [43]. Meanwhile, many of them were associated with regulating the growth of skeletal muscle. For example, miR-133c, miR-181b, miR-455, miR-135, miR-21, miR-494, and miR-381 were predicted to regulate the genes associated with myocyte enhancer factor 2 proteins [43], which are key transcriptional regulators of skeletal muscle development [115]. The miR-133a, miR-214, miR-34a and miR-381 were predicted to regulate the genes associated with insulin-like growth factor I [43], which plays a critical role in muscle regeneration [116]. The miR-199a, miR-27b, miR-26a, miR-23b, miR-214, miR-499b, miR-26a, and miR-125b were predicted to regulate the genes associated with myelin expression factor 2 [43], which contributes to the skeletal muscle differentiation [117]. A miRNA, miR-192, was reported to regulate the myogenic differentiation and proliferation of skeletal muscle sheep satellite cells [44], which are responsible for producing myoblasts in skeletal muscle [118], through regulating the repression of retinoblastoma 1, which is a known regulator of myogenesis [119]. Additionally, miR-1, miR-133b, miR-206, and miR-486 were also reported to be differential expressed in the skeletal muscle of sheep and predicted to correlate with the development of skeletal muscle [120].
The adipose tissue is not only one of the main sites for lipogenesis in ruminants, but also modulates and participates in a large variety of processes related to feed intake, energy homeostasis, and whole-body physiology through its endocrinological activity [104,121]. Therefore, the adipose tissue is highly correlated with feed efficiency in ruminants. Several miRNAs in sheep’s adipose tissue were reported to be associated with adipose tissue development and metabolism. Eight hundred fifteen miRNAs were found to be differentially expressed between fat-tailed (Kazakhstan) and thin-tailed (Tibetan) sheep breeds [45]. Among these miRNAs, several miRNAs, including miR-2070-3p, miR-222, miR-502-3p, miR-6238, miR-7446-3p, miR-7475-5p, miR-125a-5p, miR-126, miR-378e, and miR-7930-3p, were related to adipogenesis and/or fat metabolism in sheep. Meanwhile, a proportion of those 815 miRNAs were predicted to play roles in adiposity, adipocyte development and differentiation, and other metabolic disturbances in other species. For example, miR-378 was reported to target MAPK1 and PPAR genes, which are associated with fat deposition and fatty acid metabolism, and promote bovine adipogenesis in white adipose tissue [122]; miR-103, miR-30, miR-27, and miR-138 have been reported to regulate adipogenesis [123,124,125,126,127,128]; miR-122, miR-370, and miR-378 have been reported to play important roles in lipid metabolism [129,130,131]; and miR-148a was reported to target MAPKAPK5, MAPK3, and MAP2K2 genes and modulate fat deposition [132,133]. Additionally, 54 miRNAs were reported to be differentially expressed in 2 sheep breeds (Han and Dorset), including 35 down-regulated and 19 up-regulated miRNAs in the Han sheep. Among them, ten up-regulated miRNAs in the Han sheep were predicted to target 12 genes associated with enriching the lipid metabolic process [134].
One of the interesting aspects of feed efficiency in sheep is that the trait depends on the production needs as the feed efficiency for meat sheep might differ from the one for wool sheep. In addition to the muscle and adipose tissue (Figure 3), the skin might also be an important tissue for regulating miRNA in feed efficiency in sheep, as a part of energy might be used for the development of wool [135]. Meanwhile, the gastrointestinal tract has also been related to the feed efficiency in sheep [136,137], but little is known about the involvement of miRNAs in the gastrointestinal tract. In addition, compared to cattle or pigs, the miRNAs studies in feed-efficiency-related tissues, such as liver or digestive systems in sheep, are fewer. Further studies in these tissues will provide insight into the biological pathways and regulatory molecules related to the feed efficiency of sheep and form putative regulatory candidates for future research on feed efficiency traits in sheep.
2.4. The miRNA in Chicken Feed Efficiency
Increasing feed efficiency and breast yield is the major focus of the poultry industry to meet the growing consumer demand for white meat. In addition to particular success in genetic/genomic selection for larger and leaner chicken [138], genetic and genomic studies have significantly saved feeding costs and resources while increasing productivity and reducing greenhouse gas emissions [139,140]. In fact, current efforts to increase feed efficiency in broilers are primarily related to genetic selection. Genetic selection contributes about 85–90% to the increased feed efficiency in broiler, while feeding strategies and management are responsible for 10–15% of increased feed efficiency [141]. An important contributing factor for improving feed efficiency is the better understanding of genetics and biology underlying feed efficiency traits, thanks to many studies in gene mapping, transcriptomics, and other omics techniques [142,143,144]. To date, many QTLs linked to feed efficiency and its related traits have been deposited in the chicken QTL database (http://www.animalgenome.org/cgi-bin/QTLdb/GG/index (accessed on 8 July 2021). Although 882 precursor miRNAs, which generate 1232 mature miRNAs for Gallus gallus in the miRNA database miRBase (Release 22.1; www.miRbase.org (accessed on 10 August 2021), have been identified, limited numbers of miRNAs studies have been devoted to understanding the mechanism of feed efficiency [46,145] and related traits, such as growth [146] and skeletal muscle development [47,147,148,149], in chicken. In a genome-wide association study, Yuan et al. [46] identified three significant SNPs for feed efficiency traits located in the vicinity of the miR-15a. The authors performed enrichment analyses of the genes targeted by miR-15a and suggested this miRNA could play important roles in feed efficiency via controlling genes in the insulin signaling pathway, known for the regulation of appetite and feed intakes. Luo et al. [145] also identified a SNP (g.5678784A>T) in the miR-1596, which is important for RFI. No functional studies have been devoted to characterizing the roles of miRNAs in chicken feed efficiency. Using the systematic transcriptomics analyses of mRNAs and miRNA data, Li et al. [47] suggested that miR-142-5p can regulate FOXO3 in the regulation of the skeletal muscle growth in chickens. As mentioned above, the liver is a key tissue for feed efficiency and miRNAs, and Li et al. [150] identified 67 miRNAs higher at 20 weeks (pre-egg laying) and 13 miRNAs higher at 30 weeks (egg-laying) in this tissue of the Lushi hens. Hicks et al. [151] identified 40 differential expressed miRNAs when comparing the liver transcriptomics profiling of E18 and D3 chicken. Fat deposit is also an important process related to feed efficiency, and miRNAs have been indicated to play important roles in this process [152,153,154]. Some notable pathways related to the differentiation (in vitro) of primary chicken pre-adipocytes are MAPK signaling, insulin signaling, and fatty acid metabolism [154]. In fact, several miRNAs are suggested to play roles in liver function. For example, Ma et al. [155] reported miR-101-2-5p could target the ApoB gene in the liver of chicken (Gallus Gallus), and Tian et al. [156] showed miR-34a-5p targeted ACSL1 protein expression to increases hepatic triglycerides and total cholesterol levels in laying hens. In a recent study, Marchesi et al. [157] identified two miRNAs, miR-1730 and miR-1744, that were associated with the FCR trait in broilers. The authors also reported miRNAs related to the feed-efficiency-related traits, such as mir-1641, for feed intake or mir-1759 for body weight gain [158]. Notably, mir-1759 has been involved in triglyceride synthesis and adipocyte differentiation via regulating LPIN1 [158].
Considering that feed accounts for approximately 70% of the total production cost in the poultry industry, it is important for the continued development of functional studies of miRNAs to improve feed efficiency in poultry. Several miRNAs (miR-1730, mir-1759, and miR-1744) could be used as biomarkers for the characterization of high and low feed efficiency animals. Given the fact that many mRNAs transcriptomic studies have been performed for feed efficiency in chicken [159,160,161,162], the integration of miRNAs sequencing with transcriptomics analyses might enhance the identification of miRNAs’ roles in feed efficiency in chicken.
3. Long Non-Coding RNA
Increasing feed efficiency is continuously gaining importance for ecological and economic reasons, as it has the potential to contribute to both increased productivity and reduced environmental impact in livestock production systems. In the past decade, more and more evidence has shown that lncRNAs play roles in regulating feed efficiency and related traits, including energy metabolism and the development of skeletal muscle in livestock species (Table 3).

Table 3.
Long non-coding RNAs in feed efficiency.
In pigs, 811 lncRNAs were detected to be differentially expressed in the muscle and adipose tissue of piglets, which may result in differences in muscle and fat development [174]. Several lncRNAs were detected and predicted to regulate lipid metabolism and adipogenesis in pigs. Seventeen lncRNAs were reported to regulate eight genes associated with the PPAR signaling pathway [164], which is highly related to fatty acid and sterol metabolism as well as adipogenic differentiation [163]. The lncRNA, XLOC_014379, was reported to target enzyme SCD, which plays a critical role in transforming saturated fatty acid to endogenous oleic acid in food, regulating unsaturated fatty acid biosynthesis, and promoting lipid deposition [175], and thus regulating fatty acid metabolism [164]. Meanwhile, nine and eleven key lncRNAs were detected in the fatty acid metabolism and adipocyte differentiation networks, respectively [164]. In addition, the porcine PU.1 antisense lncRNA, which can form a sense–antisense RNA duplex with PU.1 mRNA to inhibit its translation, was reported to promote adipogenesis during the pre-adipocyte differentiation process [165].
In cattle, 126 lncRNA were reported to be associated with feed efficiency [166]. Among them, 71 detected lncRNAs (21 were identified in the adrenal gland, 8 in liver, 10 in muscle, 15 in the hypothalamus, and 17 in the pituitary gland) were identified as the key lncRNAs, which have the potential to regulate the expression of mRNA associated with feed efficiency in cattle [166]. Meanwhile, several of them were detected within the genomic regions of QTL for traits related to feed efficiency, feed intake, and fat deposition [166]. For example, TCONS_00119451 and TCONS_00119463 were reported to overlap seven QTLs, which include QTL:56461, QTL:20842, QTL:20843, QTL:20844, QTL:20845, QTL:20846, and QTL:20847, associated with RFI; TCONS_00032445, TCONS_00062811, and TCONS_00149966 were reported to overlap the QTLs related to dry matter intake; TCONS_00188391 and TCONS_00190543 were reported to coincide the QTLs associated with feed conversion ratio; and TCONS_00119451 and TCONS_00119463 were reported to overlap eleven QTLs related to fat deposition related traits [166]. Several other lncRNAs were predicted to be associated with energy metabolism in cattle. For example, MSTRG.4390 and MSTRG.5042 were reported to participate in the pathway enrichments for fatty acid β-oxidation and the TCA-cycle, respectively [167], which play important roles in mitochondrial function and energy metabolism for feed efficiency; MSTRG.4390 and MSTRG.5042 were also correlated with the expression of PCK1 gene and FBP1 gene [167], which occupy key roles in the biological pathway for energy balance in cattle through influencing gluconeogenesis [176]; and MSTRG.4802 was predicted to be involved in oxidative phosphorylation and mitochondrial dysfunction [167].
In sheep, some lncRNAs were found to be associated with feed efficiency and its related traits. Ten lncRNAs were identified as being differentially expressed between high and low RFI sheep [168]. Among them, LNC_000890 was predicted to regulate the metabolic efficiency of liver tissue metabolic efficiency and is co-expressed with the ADRA2A gene, which is significantly associated with feed efficiency [177], and therefore represents a crucial regulator for feed efficiency in sheep [168]. The lincRNA.16164 was predicted to target the TSHZ1 gene [169], which is linked to body weight and lipid metabolism [178]. In addition, six lncRNAs were found to overlap with QTLs associated with tail fat deposition [169].
In chicken, there are some reports about some lncRNAs that regulate the development of skeletal muscle [170,171,179]. The lncRNA gga-lnc-0181 was found to be highly expressed in skeletal muscle and predicted to play a functional role in muscle development [170]. The lncRNA-Six1 was reported to regulate the Six1 gene, which regulates the skeletal muscle development and transformation of muscle fiber types [180,181,182], thus promoting cell proliferation and being involved in muscle growth [171]. Several lncRNAs were detected and reported to be associated with the regulation of lipid metabolism. For example, the lncRNA, lnc_DHCR24, was repeated to be involved in lipid metabolism [172]. In addition, seven lncRNAs were found to be differentially expressed in the entire differentiation process of intramuscular preadipocytes, and therefore play an important role in the intramuscular preadipocytes [173].
Although the number of detected feed-efficiency-related lncRNAs is limited, the lncRNAs mentioned above in pig, cattle, sheep, and chicken demonstrate that lncRNAs also play important roles in regulating feed efficiency in livestock species. The poor accuracy of transcript detection caused by the limitations of high-throughput technologies and the low-level and extremely tissue-specific expression of lncRNAs is the major challenge associated with lncRNA analysis [12]. Only a small fraction of lncRNAs have been detected to be associated with feed efficiency in livestock species, and this knowledge gap highlights the need to explore the mechanisms of lncRNA in controlling the gene expressions related to feed efficiency.
4. Conclusions and Future Perspectives
In this paper, we have reviewed the role of ncRNAs in feed efficiency by emphasizing the state of the art of the current studies. Although a significant increase in the number of miRNAs related to feed efficiency in each livestock species was found, many research gaps should be filled in order to obtain a holistic picture of the ncRNAs for the traits. In cattle and pigs, many ncRNAs related to FE have already been identified, but information is still lacking for poultry. Since the major studies only focus on exploring or profiling ncRNAs related to the traits, the apparent need will be the functional characterization of their ncRNAs in vitro and in vivo. Moreover, many studies only look at one or some organs related to feed efficiency, and comprehensive approaches should be made. Although livers and muscles are major organs related to feed efficiency, the roles of GIT and brain in the regulation should not be ignored. For instance, miRNAs have been mentioned to play essential roles in the gut microbiota and brain signals in the diet response [183]. Therefore, it is expected that miRNAs could also mediate the regulation of feed efficiency-related genes in the gut–brain axis. The future of livestock farming will rely on precision nutrition [184,185]. Many ncRNAs have been linked to different specific diets and feeding formulas in livestock species. The question of using ncRNAs for a better animal diet, which could lead to improved feed efficiency, still needs to be answered. Heat stress is also important for feed efficiency, and several studies have indicated that miRNAs can play roles in the regulation of heat stress [186]. Developing some miRNAs as biomarkers for the selection of animals for both heat tolerance and improved tolerance should be prioritized in future research. Additionally, the ncRNAs have been shown to work together in the competitive endogenous networks [187]; therefore, the comprehensive understanding of ncRNA function needs to be put in the network perspectives. Consequently, future experiments should include several types of ncRNAs characterization at the same time.
From the livestock industry perspective, the most challenging question to be answered is how to use the knowledge of ncRNAs for improving feed efficiency. The development of ncRNA-based biomarkers is one of the options that can be considered. The combination of extracellular miRNAs with other phenotypic measurements will assess feed efficiency and related traits more accurately, allowing livestock producers to monitor feeding management and nutritional needs to optimize the use of feed. Another fascinating option is to use the genome-editing or RNA interfering tools to alter the miRNA expressions, consequently changing the downstream impact on the feed efficiency traits.
Finally, it is also worth mentioning the possible use of artificial intelligence, sensors, and big data to better understand the roles of ncRNAs on feed efficiency. Firstly, these methods and tools will help to better characterize the phenotype, which is the most important step before functional investigation. Secondly, deeper sequencing and better quality data will hopefully give a better capture of ncRNAs expression, especially the novel ncRNAs, and their link to the traits. Artificial intelligence, especially machine learning approaches, has been significantly involved in feed efficiency improvement. Their methods can also be used to accurately characterize ncRNAs functions in FE, such as the classification of ncRNAs involved in high and low feed efficient groups.
In conclusion, ncRNAs have a promising role in improving feed efficiency for major livestock species through regulating the expression of genes and pathways on FE-related organs. Although much needs to be performed to see the practical application of ncRNAs in reducing feed intake in the livestock industry, the rapid change in technologies and methods might help to shorten these research and application gaps.
Author Contributions
Conceptualization, Y.M., G.H., D.N.D. and P.D.; writing—original draft preparation, G.H., D.N.D. and P.D.; writing—review and editing, G.H., D.N.D., P.D. and Y.M.; supervision, Y.M. and D.N.D. All authors have read and agreed to the published version of the manuscript.
Funding
The authors gratefully acknowledge financial support from Natural Sciences and Engineering Research Council (NSERC) of Canada, Mitacs, Canada Mink Breeders Association, Nova Scotia Mink Breeders Association, Nova Scotia Department of Agriculture, and Mink Veterinary Consulting & Research Service Ltd.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Authors are thankful to the anonymous reviewers whose critiques and comments greatly improved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Berry, D.P.; Crowley, J.J. Cell Biology Symposium: Genetics of feed efficiency in dairy and beef cattle. J. Anim. Sci. 2013, 91, 1594–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, R.M.; Swiger, L.A.; Chambers, D.; Gregory, K.E. Efficiency of Feed Use in Beef Cattle. J. Anim. Sci. 1963, 22, 486–494. [Google Scholar] [CrossRef]
- Berry, D.; Crowley, J. Residual intake and body weight gain: A new measure of efficiency in growing cattle. J. Anim. Sci. 2012, 90, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Fitzhugh, H., Jr.; Taylor, S.C. Genetic analysis of degree of maturity. J. Anim. Sci. 1971, 33, 717–725. [Google Scholar] [CrossRef]
- Kleiber, M. The Fire of Life: An Introduction to Animal Energetics; John Wiley and Sons Inc.: New York, NY, USA; London, UK, 1961. [Google Scholar]
- Do, D.N.; Ibeagha-Awemu, E.M. Non-Coding RNA Roles in Ruminant Mammary Gland Development and Lactation; InTech: London, UK, 2017. [Google Scholar]
- Gomes, A.Q.; Nolasco, S.; Soares, H. Non-coding RNAs: Multi-tasking molecules in the cell. Int. J. Mol. Sci. 2013, 14, 16010–16039. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.; Shiekhattar, R. Where long noncoding RNAs meet DNA methylation. Cell Res. 2014, 24, 263–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667. [Google Scholar] [CrossRef] [Green Version]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosinska-Selbi, B.; Mielczarek, M.; Szyda, J. Review: Long non-coding RNA in livestock. Animal 2020, 14, 2003–2013. [Google Scholar] [CrossRef]
- Weikard, R.; Demasius, W.; Kuehn, C. Mining long noncoding RNA in livestock. Anim. Genet. 2017, 48, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, L.L.; Matukumalli, L.K.; Sonstegard, T.S.; Van Tassell, C.P.; Gasbarre, L.C.; Capuco, A.V.; Smith, T.P.L. Discovery and profiling of bovine microRNAs from immune-related and embryonic tissues. Physiol. Genom. 2007, 29, 35–43. [Google Scholar] [CrossRef]
- Halushka, M.K.; Fromm, B.; Peterson, K.J.; McCall, M.N. Big Strides in Cellular MicroRNA Expression. Trends Genet. 2018, 34, 165–167. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Do, D.N.; Dudemaine, P.-L.; Fomenky, B.; Ibeagha-Awemu, E.M. Transcriptome Analysis of Non-Coding RNAs in Livestock Species: Elucidating the Ambiguity; InTech: Rijeka, Croatia, 2017; Chapter 5. [Google Scholar]
- Do, D.N.; Dudemaine, P.-L.; Mathur, M.; Suravajhala, P.; Zhao, X.; Ibeagha-Awemu, E.M. MiRNA Regulatory Functions in Farm Animal Diseases, and Biomarker Potentials for Effective Therapies. Int. J. Mol. Sci. 2021, 22, 3080. [Google Scholar] [CrossRef] [PubMed]
- Fatima, A.; Morris, D.G. MicroRNAs in domestic livestock. Physiol. Genom. 2013, 45, 685–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, L.; Hou, Y.; Wu, H.; Miao, Y.; Li, X.; Cao, J.; Brameld, J.M.; Parr, T.; Zhao, S. Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential Residual Feed Intake in pigs. Sci. Rep. 2015, 5, 11953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, W.; Tang, Y.; Zhao, Y.; Zhao, J.; Zhang, L.; Wei, W.; Chen, J. MiR-144-3p Targets FoxO1 to Reduce Its Regulation of Adiponectin and Promote Adipogenesis. Front. Genet. 2020, 11, 603144. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wen, S.; Wu, J.; Zeng, B.; Chen, T.; Luo, J.; Shu, G.; Wang, S.-B.; Zhang, Y.; Xi, Q. Comparative Analysis of MicroRNA Expression Profiles Between Skeletal Muscle- and Adipose-Derived Exosomes in Pig. Front. Genet. 2021, 12, 631230. [Google Scholar] [CrossRef] [PubMed]
- Chu, A.Y.; Deng, X.; Fisher, V.A.; Drong, A.; Zhang, Y.; Feitosa, M.F.; Liu, C.-T.; Weeks, O.; Choh, A.C.; Duan, Q. Multiethnic genome-wide meta-analysis of ectopic fat depots identifies loci associated with adipocyte development and differentiation. Nat. Genet. 2017, 49, 125–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, K.; Zhao, X.; Ao, H.; Chen, S.; Yang, T.; Tan, Z.; Wang, Y.; Zhang, F.; Liu, Y.; Ni, H. Transcriptome analysis of miRNA and mRNA in the livers of pigs with highly diverged backfat thickness. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Sun, W.-k.; Cheng, C.; Chen, Y.-h.; Zeng, K.; Chen, X.; Gu, Y.; Gao, R.; Liu, R. Comparison of liver microRNA transcriptomes of Tibetan and Yorkshire pigs by deep sequencing. Genes 2016, 577, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Mentzel, C.M.J.; Anthon, C.; Jacobsen, M.J.; Karlskov-Mortensen, P.; Bruun, C.S.; Jørgensen, C.B.; Gorodkin, J.; Cirera, S.; Fredholm, M. Gender and obesity specific microRNA expression in adipose tissue from lean and obese pigs. PLoS ONE 2015, 10, e0131650. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Wu, Y.; Wang, J.; Chen, J.; Huang, Y.; Rao, J.; Feng, C. MicroRNA-24 promotes 3T3-L1 adipocyte differentiation by directly targeting the MAPK7 signaling. Biochem. Biophys. Res. Commun. 2016, 474, 76–82. [Google Scholar] [CrossRef]
- Wang, T.; Li, M.; Guan, J.; Li, P.; Wang, H.; Guo, Y.; Shuai, S.; Li, X. MicroRNAs miR-27a and miR-143 regulate porcine adipocyte lipid metabolism. Int. J. Mol. Sci. 2011, 12, 7950–7959. [Google Scholar] [CrossRef]
- Yang, W.; Tang, K.; Wang, Y.; Zan, L. MiR-27a-5p Increases Steer Fat Deposition Partly by Targeting Calcium-sensing Receptor (CASR). Sci. Rep. 2018, 8, 3012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Du, J.; Wang, L.; Niu, L.; Zhao, Y.; Tang, G.; Jiang, Y.; Shuai, S.; Bai, L.; Li, X.; et al. MicroRNA-143a-3p modulates preadipocyte proliferation and differentiation by targeting MAPK7. Biomed. Pharmacother. 2018, 108, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Xing, K.; Zhao, X.; Liu, Y.; Zhang, F.; Tan, Z.; Qi, X.; Wang, X.; Ni, H.; Guo, Y.; Sheng, X.; et al. Identification of Differentially Expressed MicroRNAs and Their Potential Target Genes in Adipose Tissue from Pigs with Highly Divergent Backfat Thickness. Animals 2020, 10, 624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Husseini, W.; Chen, Y.; Gondro, C.; Herd, R.M.; Gibson, J.P.; Arthur, P.F. Characterization and Profiling of Liver microRNAs by RNA-sequencing in Cattle Divergently Selected for Residual Feed Intake. Asian-Australas. J. Anim. Sci. 2016, 29, 1371–1382. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, P.S.N.; Coutinho, L.L.; Tizioto, P.C.; Cesar, A.S.M.; de Oliveira, G.B.; Diniz, W.J.d.S.; De Lima, A.O.; Reecy, J.M.; Mourão, G.B.; Zerlotini, A.; et al. An integrative transcriptome analysis indicates regulatory mRNA-miRNA networks for residual feed intake in Nelore cattle. Sci. Rep. 2018, 8, 17072. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, E.B.; Gionbelli, M.P.; Rodrigues, R.T.S.; Bonilha, S.F.M.; Newbold, C.J.; Guimarães, S.E.F.; Silva, W.; Verardo, L.L.; Silva, F.F.; Detmann, E.; et al. Differentially expressed mRNAs, proteins and miRNAs associated to energy metabolism in skeletal muscle of beef cattle identified for low and high residual feed intake. BMC Genom. 2019, 20, 501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.; Yang, J.; Jiang, R.; Yang, Z.; Li, H.; Huang, Y.; Lan, X.; Lei, C.; Ma, Y.; Qi, X.; et al. miR-148a-3p regulates proliferation and apoptosis of bovine muscle cells by targeting KLF6. J. Cell. Physiol. 2019, 234, 15742–15750. [Google Scholar] [CrossRef]
- Zhang, Y.-Y.; Wang, H.-B.; Wang, Y.-N.; Wang, H.-C.; Zhang, S.; Hong, J.-Y.; Guo, H.-F.; Chen, D.; Yang, Y.; Zan, L.-S. Transcriptome analysis of mRNA and microRNAs in intramuscular fat tissues of castrated and intact male Chinese Qinchuan cattle. PLoS ONE 2017, 12, e0185961. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Lee, M.J.; Abdelmohsen, K.; Kim, W.; Kim, M.M.; Srikantan, S.; Martindale, J.L.; Hutchison, E.R.; Kim, H.H.; Marasa, B.S.; et al. miR-130 Suppresses Adipogenesis by Inhibiting Peroxisome Proliferator-Activated Receptor γ Expression. Mol. Cell. Biol. 2011, 31, 626–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romao, J.M.; Jin, W.; He, M.; McAllister, T.; Guan, L.L. Altered MicroRNA Expression in Bovine Subcutaneous and Visceral Adipose Tissues from Cattle under Different Diet. PLoS ONE 2012, 7, e40605. [Google Scholar] [CrossRef]
- Romao, J.M.; Jin, W.; He, M.; McAllister, T.; Guan, L.L. MicroRNAs in bovine adipogenesis: Genomic context, expression and function. BMC Genom. 2014, 15, 137. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zheng, Y.; Wang, G.; Li, H. Identification of microRNA and bioinformatics target gene analysis in beef cattle intramuscular fat and subcutaneous fat. Mol. Biosyst. 2013, 9, 2154–2162. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhang, X.; Huang, W.; Miao, X. Identification and characterization of differentially expressed miRNAs in subcutaneous adipose between Wagyu and Holstein cattle. Sci. Rep. 2017, 7, 44026. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, S.; Zhang, W.; Cheng, G.; Khan, R.; Junjvlieke, Z.; Li, S.; Zan, L. miR-424 Promotes Bovine Adipogenesis Through an Unconventional Post-Transcriptional Regulation of STK11. Front. Genet. 2020, 11, 145. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Li, C.; Li, X.; Yao, Y.; Ni, W.; Zhang, X.; Cao, Y.; Hazi, W.; Wang, D.; Quan, R.; et al. Expression profiles of microRNAs in skeletal muscle of sheep by deep sequencing. Asian-Australas. J. Anim. Sci. 2019, 32, 757–766. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Kang, Y.; Wang, H.-Y.; Guan, W.-J.; Li, X.-C.; Jiang, L.; He, X.-H.; Pu, Y.-B.; Han, J.-L.; Ma, Y.-H. Expression profiling and functional characterization of miR-192 throughout sheep skeletal muscle development. Sci. Rep. 2016, 6, 30281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Wang, X.; Yuan, C.; Kang, D.; Xu, X.; Zhou, J.; Geng, R.; Yang, Y.; Yang, Z.; Chen, Y. Integrating miRNA and mRNA Expression Profiling Uncovers miRNAs Underlying Fat Deposition in Sheep. Biomed Res. Int. 2017, 2017, 1857580. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Chen, S.; Shi, F.; Wu, G.; Liu, A.; Yang, N.; Sun, C. Genome-wide association study reveals putative role of gga-miR-15a in controlling feed conversion ratio in layer chickens. BMC Genom. 2017, 18, 699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Abdalla, B.A.; Zheng, M.; He, X.; Cai, B.; Han, P.; Ouyang, H.; Chen, B.; Nie, Q.; Zhang, X. Systematic transcriptome-wide analysis of mRNA–miRNA interactions reveals the involvement of miR-142-5p and its target (FOXO3) in skeletal muscle growth in chickens. Mol. Genet. Genom. 2018, 293, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, D.; Hernandez-Sanchez, J.; Chen, J.; Liu, C.; Wu, Z.; Fang, M.; Li, N. Genome wide association analysis reveals new production trait genes in a male Duroc population. PLoS ONE 2015, 10, e0139207. [Google Scholar] [CrossRef] [Green Version]
- Do, D.N.; Ostersen, T.; Strathe, A.B.; Mark, T.; Jensen, J.; Kadarmideen, H.N. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC Genet. 2014, 15, 27. [Google Scholar] [CrossRef] [Green Version]
- Do, D.N.; Strathe, A.B.; Ostersen, T.; Pant, S.D.; Kadarmideen, H.N. Genome-wide association and pathway analysis of feed efficiency in pigs reveal candidate genes and pathways for residual feed intake. Front. Genet. 2014, 5, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gondret, F.; Vincent, A.; Houée-Bigot, M.; Siegel, A.; Lagarrigue, S.; Causeur, D.; Gilbert, H.; Louveau, I. A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs. BMC Genom. 2017, 18, 244. [Google Scholar] [CrossRef] [Green Version]
- Morales, P.E.; Bucarey, J.L.; Espinosa, A. Muscle lipid metabolism: Role of lipid droplets and perilipins. J. Diabetes Res. 2017, 2017, 1789395. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.K. Muscle as a secretory organ. Compr. Physiol. 2013, 3, 1337–1362. [Google Scholar] [PubMed]
- Turner, N.; Cooney, G.J.; Kraegen, E.W.; Bruce, C.R. Fatty acid metabolism, energy expenditure and insulin resistance in muscle. J. Endocrinol. 2014, 220, T61–T79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuo, K.; Shigeo, O. Regulation of mitochondrial ATP synthesis in mammalian cells by transcriptional control. Int. J. Biochem. 1990, 22, 219–229. [Google Scholar] [CrossRef]
- Aschrafi, A.; Schwechter, A.D.; Mameza, M.G.; Natera-Naranjo, O.; Gioio, A.E.; Kaplan, B.B. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J. Neurosci. 2008, 28, 12581–12590. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.T.; Nakamura, K.; Davies, R.; Nahas, S.A.; Brown, C.; Tunuguntla, R.; Gatti, R.A.; Hu, H. ATM–dependent miR-335 targets CtIP and modulates the DNA damage response. PLoS Genet. 2013, 9, e1003505. [Google Scholar] [CrossRef] [Green Version]
- Bijland, S.; Mancini, S.J.; Salt, I.P. Role of AMP-activated protein kinase in adipose tissue metabolism and inflammation. Clin. Sci. 2013, 124, 491–507. [Google Scholar] [CrossRef] [Green Version]
- Miranda, N.; Tovar, A.R.; Palacios, B.; Torres, N. AMPK as a cellular energy sensor and its function in the organism. Rev. Investig. Clin. 2007, 59, 458–469. [Google Scholar] [PubMed]
- Thomson, D.M.; Herway, S.T.; Fillmore, N.; Kim, H.; Brown, J.D.; Barrow, J.R.; Winder, W.W. AMP-activated protein kinase phosphorylates transcription factors of the CREB family. J. Appl. Physiol. 2008, 104, 429–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turczyńska, K.M.; Bhattachariya, A.; Säll, J.; Göransson, O.; Swärd, K.; Hellstrand, P.; Albinsson, S. Stretch-sensitive down-regulation of the miR-144/451 cluster in vascular smooth muscle and its role in AMP-activated protein kinase signaling. PLoS ONE 2013, 8, e65135. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Qiu, W.; Zhou, Y.; Wen, P.; Fang, L.; Cao, H.; Zen, K.; He, W.; Zhang, C.; Dai, C. A microRNA-30e/mitochondrial uncoupling protein 2 axis mediates TGF-β1-induced tubular epithelial cell extracellular matrix production and kidney fibrosis. Kidney Int. 2013, 84, 285–296. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-J.; Lee, Y.-S.; Zimmers, T.A.; Soleimani, A.; Matzuk, M.M.; Tsuchida, K.; Cohn, R.D.; Barton, E.R. Regulation of muscle mass by follistatin and activins. Mol. Endocrinol. 2010, 24, 1998–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandit, K.V.; Milosevic, J.; Kaminski, N. MicroRNAs in idiopathic pulmonary fibrosis. Transl. Res. 2011, 157, 191–199. [Google Scholar] [CrossRef]
- Patel, V.; Noureddine, L. MicroRNAs and fibrosis. Curr. Opin. Nephrol. Hypertens. 2012, 21, 410. [Google Scholar] [CrossRef] [PubMed]
- Allen, D.L.; Loh, A.S. Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am. J. Physiol. Cell Physiol. 2011, 300, C124–C137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rui, L. Energy metabolism in the liver. Compr. Physiol. 2011, 4, 177–197. [Google Scholar]
- Shimizu, N.; Maruyama, T.; Yoshikawa, N.; Matsumiya, R.; Ma, Y.; Ito, N.; Tasaka, Y.; Kuribara-Souta, A.; Miyata, K.; Oike, Y. A muscle-liver-fat signalling axis is essential for central control of adaptive adipose remodelling. Nat. Commun. 2015, 6, 6693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horodyska, J.; Hamill, R.M.; Reyer, H.; Trakooljul, N.; Lawlor, P.G.; McCormack, U.M.; Wimmers, K. RNA-seq of liver from pigs divergent in feed efficiency highlights shifts in macronutrient metabolism, hepatic growth and immune response. Front. Genet. 2019, 10, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakil, S.J. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry 1989, 28, 4523–4530. [Google Scholar] [CrossRef]
- Matsuo, Y.; Tanaka, M.; Yamakage, H.; Sasaki, Y.; Muranaka, K.; Hata, H.; Ikai, I.; Shimatsu, A.; Inoue, M.; Chun, T.-H. Thrombospondin 1 as a novel biological marker of obesity and metabolic syndrome. Metabolism 2015, 64, 1490–1499. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.; Islam, R. Mammalian Sirt1: Insights on its biological functions. Cell Commun. Signal. 2011, 9, 11. [Google Scholar] [CrossRef] [Green Version]
- Pawar, A.; Botolin, D.; Mangelsdorf, D.J.; Jump, D.B. The role of liver X receptor-alpha in the fatty acid regulation of hepatic gene expression. J. Biol. Chem. 2003, 278, 40736–40743. [Google Scholar] [CrossRef] [Green Version]
- Konige, M.; Wang, H.; Sztalryd, C. Role of adipose specific lipid droplet proteins in maintaining whole body energy homeostasis. Biochim. Biophys. Acta 2014, 1842, 393–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed-Ali, V.; Pinkney, J.; Coppack, S. Adipose tissue as an endocrine and paracrine organ. Int. J. Obes. 1998, 22, 1145–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerre-Millo, M. Adipose tissue hormones. J. Endocrinol. Investig. 2002, 25, 855–861. [Google Scholar] [CrossRef]
- Trayhurn, P.; Bing, C. Appetite and energy balance signals from adipocytes. Philos. Trans. R. Soc. B 2006, 361, 1237–1249. [Google Scholar] [CrossRef]
- Matoušková, P.; Hanousková, B.; Skálová, L. MicroRNAs as Potential Regulators of Glutathione Peroxidases Expression and Their Role in Obesity and Related Pathologies. Int. J. Mol. Sci. 2018, 19, 1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Wang, X.; Zhou, S.; Wu, J.; Geng, Q.; Ruan, D.; Qiu, Y.; Quan, J.; Ding, R.; Cai, G. Brain Transcriptome Analysis Reveals Potential Transcription Factors and Biological Pathways Associated with Feed Efficiency in Commercial DLY Pigs. DNA Cell Biol. 2020, 40, 272–282. [Google Scholar] [CrossRef]
- Bergamaschi, M.; Tiezzi, F.; Howard, J.; Huang, Y.J.; Gray, K.A.; Schillebeeckx, C.; McNulty, N.P.; Maltecca, C. Gut microbiome composition differences among breeds impact feed efficiency in swine. Microbiome 2020, 8, 110. [Google Scholar] [CrossRef]
- Yang, H.; Huang, X.; Fang, S.; He, M.; Zhao, Y.; Wu, Z.; Yang, M.; Zhang, Z.; Chen, C.; Huang, L. Unraveling the fecal microbiota and metagenomic functional capacity associated with feed efficiency in pigs. Front. Microbiol. 2017, 8, 1555. [Google Scholar] [CrossRef] [Green Version]
- De Lange, C.; Levesque, C.; Kerr, B. Amino acid nutrition and feed efficiency. In Feed Efficiency in Swine; Springer: Berlin/Heidelberg, Germany, 2012; pp. 81–100. [Google Scholar]
- Metzler-Zebeli, B.U.; Lawlor, P.G.; Magowan, E.; Zebeli, Q. Interactions between metabolically active bacteria and host gene expression at the cecal mucosa in pigs of diverging feed efficiency. J. Anim. Sci. 2018, 96, 2249–2264. [Google Scholar] [CrossRef]
- Mottet, A.; de Haan, C.; Falcucci, A.; Tempio, G.; Opio, C.; Gerber, P. Livestock: On our plates or eating at our table? A new analysis of the feed/food debate. Glob. Food Sec. 2017, 14, 1–8. [Google Scholar] [CrossRef]
- Nielsen, M.K.; MacNeil, M.D.; Dekkers, J.C.M.; Crews, D.H.; Rathje, T.A.; Enns, R.M.; Weaber, R.L. Review: Life-cycle, total-industry genetic improvement of feed efficiency in beef cattle: Blueprint for the Beef Improvement Federation11The development of this commentary was supported by the Beef Improvement Federation. Prof. Anim. Sci. 2013, 29, 559–565. [Google Scholar] [CrossRef]
- Barendse, W.; Reverter, A.; Bunch, R.J.; Harrison, B.E.; Barris, W.; Thomas, M.B. A Validated Whole-Genome Association Study of Efficient Food Conversion in Cattle. Genetics 2007, 176, 1893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukiibi, R.; Johnston, D.; Vinsky, M.; Fitzsimmons, C.; Stothard, P.; Waters, S.M.; Li, C. Bovine hepatic miRNAome profiling and differential miRNA expression analyses between beef steers with divergent feed efficiency phenotypes. Sci. Rep. 2020, 10, 19309. [Google Scholar] [CrossRef]
- Hardie, L.; VandeHaar, M.; Tempelman, R.; Weigel, K.; Armentano, L.; Wiggans, G.; Veerkamp, R.; de Haas, Y.; Coffey, M.; Connor, E. The genetic and biological basis of feed efficiency in mid-lactation Holstein dairy cows. J. Dairy Sci. 2017, 100, 9061–9075. [Google Scholar] [CrossRef] [Green Version]
- Hurley, A.; Lopez-Villalobos, N.; McParland, S.; Lewis, E.; Kennedy, E.; O’Donovan, M.; Burke, J.L.; Berry, D. Characteristics of feed efficiency within and across lactation in dairy cows and the effect of genetic selection. J. Dairy Sci. 2018, 101, 1267–1280. [Google Scholar] [CrossRef]
- Do, D.N.; Li, R.; Dudemaine, P.-L.; Ibeagha-Awemu, E.M. MicroRNA roles in signalling during lactation: An insight from differential expression, time course and pathway analyses of deep sequence data. Sci. Rep. 2017, 7, 44605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Moisá, S.; Khan, M.; Wang, J.; Bu, D.; Loor, J. MicroRNA expression patterns in the bovine mammary gland are affected by stage of lactation. J. Dairy Sci. 2012, 95, 6529–6535. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Verfaillie, C.M. MicroRNAs: The fine modulators of liver development and function. Liver Int. 2014, 34, 976–990. [Google Scholar] [CrossRef] [Green Version]
- Kogelman, L.J.A.; Cirera, S.; Zhernakova, D.V.; Fredholm, M.; Franke, L.; Kadarmideen, H.N. Identification of co-expression gene networks, regulatory genes and pathways for obesity based on adipose tissue RNA Sequencing in a porcine model. BMC Med. Genet. 2014, 7, 57. [Google Scholar] [CrossRef] [Green Version]
- Seabury, C.M.; Oldeschulte, D.L.; Saatchi, M.; Beever, J.E.; Decker, J.E.; Halley, Y.A.; Bhattarai, E.K.; Molaei, M.; Freetly, H.C.; Hansen, S.L.; et al. Genome-wide association study for feed efficiency and growth traits in U.S. beef cattle. BMC Genom. 2017, 18, 386. [Google Scholar] [CrossRef] [Green Version]
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef]
- Lewis, A.P.; Jopling, C.L. Regulation and biological function of the liver-specific miR-122. Biochem. Soc. Trans. 2010, 38, 1553–1557. [Google Scholar] [CrossRef]
- Pandey, A.K.; Verma, G.; Vig, S.; Srivastava, S.; Srivastava, A.K.; Datta, M. miR-29a levels are elevated in the db/db mice liver and its overexpression leads to attenuation of insulin action on PEPCK gene expression in HepG2 cells. Mol. Cell. Endocrinol. 2011, 332, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Eijkelenboom, A.; Mokry, M.; de Wit, E.; Smits, L.M.; Polderman, P.E.; van Triest, M.H.; van Boxtel, R.; Schulze, A.; de Laat, W.; Cuppen, E.; et al. Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol. Syst. Biol. 2013, 9, 638. [Google Scholar] [CrossRef] [PubMed]
- Frayn, K.N. Metabolic Regulation: A Human Perspective, 3rd ed.; Wiley-Blackwell: Oxford, UK, 2010. [Google Scholar]
- Kooistra, M.R.H.; Dubé, N.; Bos, J.L. Rap1: A key regulator in cell-cell junction formation. J. Cell Biol. 2007, 120, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, N.-K.; Lim, D.; Lee, S.-H.; Cho, Y.-M.; Park, E.-W.; Lee, C.-S.; Shin, B.-S.; Kim, T.-H.; Yoon, D. Heat Shock Protein B1 and Its Regulator Genes Are Negatively Correlated with Intramuscular Fat Content in the Longissimus Thoracis Muscle of Hanwoo (Korean Cattle) Steers. J. Agric. Food Chem. 2011, 59, 5657–5664. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.H.A.; Kaster, N.; Khan, R.; Abdelnour, S.A.; El-Hack, M.E.A.; Khafaga, A.F.; Taha, A.; Ohran, H.; Swelum, A.A.; Schreurs, N.M.; et al. The Role of MicroRNAs in Muscle Tissue Development in Beef Cattle. Genes 2020, 11, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.W.; Sun, X.F.; Tong, H.L.; Wang, Y.H.; Li, S.F.; Yan, Y.Q.; Li, G.P. Effect of differentiation on microRNA expression in bovine skeletal muscle satellite cells by deep sequencing. Cell. Mol. Biol. 2016, 21, 8. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Fang, X.; Gao, M.; Mi, J.; Zhang, X.; Xia, L.; Zhao, Z.; Albrecht, E.; Maak, S.; Yang, R. Isolation and Identification of Bovine Preadipocytes and Screening of MicroRNAs Associated with Adipogenesis. Animals 2020, 10, 818. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Oliveira, H.R.; Houlahan, K.; Fonseca, P.A.; Lam, S.; Butty, A.M.; Seymour, D.J.; Vargas, G.; Chud, T.C.; Silva, F.F. Genetic mechanisms underlying feed utilization and implementation of genomic selection for improved feed efficiency in dairy cattle. Can. J. Anim. Sci. 2020, 100, 587–604. [Google Scholar] [CrossRef]
- Kenny, D.; Fitzsimons, C.; Waters, S.; McGee, M. Invited review: Improving feed efficiency of beef cattle–the current state of the art and future challenges. Animal 2018, 12, 1815–1826. [Google Scholar] [CrossRef] [Green Version]
- VandeHaar, M.J.; Armentano, L.E.; Weigel, K.; Spurlock, D.M.; Tempelman, R.J.; Veerkamp, R. Harnessing the genetics of the modern dairy cow to continue improvements in feed efficiency. J. Dairy Sci. 2016, 99, 4941–4954. [Google Scholar] [CrossRef] [Green Version]
- Herd, R.; Arthur, P. Physiological basis for residual feed intake. J. Anim. Sci. 2009, 87, E64–E71. [Google Scholar] [CrossRef]
- Cammack, K.M.; Leymaster, K.A.; Jenkins, T.G.; Nielsen, M.K. Estimates of genetic parameters for feed intake, feeding behavior, and daily gain in composite ram lambs1,2. J. Anim. Sci. 2005, 83, 777–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, T.; Heard, J.; Malcolm, B. System changes to a lamb farm in south-west Victoria: Some pre-experimental modelling. AFBM J. 2014, 11, 1–18. [Google Scholar]
- Zhang, X.; Wang, W.; Mo, F.; La, Y.; Li, C.; Li, F. Association of residual feed intake with growth and slaughtering performance, blood metabolism, and body composition in growing lambs. Sci. Rep. 2017, 7, 12681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Huang, Y.; Du, M. Farm animals for studying muscle development and metabolism: Dual purposes for animal production and human health. Anim. Front. 2019, 9, 21–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picard, B.; Lefaucheur, L.; Berri, C.; Duclos, M.J. Muscle fibre ontogenesis in farm animal species. Reprod. Nutr. Dev. 2002, 42, 415–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, C.M.; Hu, J.; Barnes, R.M.; Heidt, A.B.; Cornelissen, I.; Black, B.L. Myocyte enhancer factor 2C function in skeletal muscle is required for normal growth and glucose metabolism in mice. Skelet. Muscle 2015, 5, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, E.R.; Morris, L.; Musaro, A.; Rosenthal, N.; Sweeney, H.L. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J. Cell Biol. 2002, 157, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Zetser, A.; Gredinger, E.; Bengal, E. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation participation of the MEF2C transcription factor. J. Biol. Chem. 1999, 274, 5193–5200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Relaix, F.; Zammit, P.S. Satellite cells are essential for skeletal muscle regeneration: The cell on the edge returns centre stage. Development 2012, 139, 2845. [Google Scholar] [CrossRef] [Green Version]
- Huh, M.S.; Parker, M.H.; Scimeè, A.; Parks, R.; Rudnicki, M.A. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 2004, 166, 865–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, M.; Kumar, A.; Siddaraju, N.K.; Fairoze, M.N.; Chhabra, P.; Ahlawat, S.; Vijh, R.K.; Yadav, A.; Arora, R. Differential expression of miRNAs in skeletal muscles of Indian sheep with diverse carcass and muscle traits. Sci. Rep. 2020, 10, 16332. [Google Scholar] [CrossRef] [PubMed]
- Greathead, H.M.R.; Dawson, J.M.; Scollan, N.D.; Buttery, P.J. In vivo measurement of lipogenesis in ruminants using [1-14C]acetate. Br. J. Nutr. 2001, 86, 37–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, W.; Dodson, M.V.; Moore, S.S.; Basarab, J.A. Characterization of microRNA expression in bovine adipose tissues: A potential regulatory mechanism of subcutaneous adipose tissue development. BMC Mol. Biol. 2010, 11, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, H.; Lim, B.; Lodish, H.F. MicroRNAs Induced During Adipogenesis that Accelerate Fat Cell Development Are Downregulated in Obesity. Diabetes 2009, 58, 1050. [Google Scholar] [CrossRef] [Green Version]
- Enomoto, H.; Furuichi, T.; Zanma, A.; Yamana, K.; Yoshida, C.; Sumitani, S.; Yamamoto, H.; Enomoto-Iwamoto, M.; Iwamoto, M.; Komori, T. Runx2 deficiency in chondrocytes causes adipogenic changes in vitro. J. Cell Biol. 2004, 117, 417. [Google Scholar]
- Karbiener, M.; Neuhold, C.; Opriessnig, P.; Prokesch, A.; Bogner-Strauss, J.G.; Scheideler, M. MicroRNA-30c promotes human adipocyte differentiation and co-represses PAI-1 and ALK2. RNA Biol. 2011, 8, 850–860. [Google Scholar] [CrossRef] [Green Version]
- Zaragosi, L.-E.; Wdziekonski, B.; Brigand, K.L.; Villageois, P.; Mari, B.; Waldmann, R.; Dani, C.; Barbry, P. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 2011, 12, R64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Q.; Gao, Z.; Alarcon, R.M.; Ye, J.; Yun, Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009, 276, 2348–2358. [Google Scholar] [CrossRef] [PubMed]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Hernando, C.; Suárez, Y.; Rayner, K.J.; Moore, K.J. MicroRNAs in lipid metabolism. Curr. Opin. Lipidol. 2011, 22, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Gerin, I.; Bommer, G.T.; McCoin, C.S.; Sousa, K.M.; Krishnan, V.; MacDougald, O.A. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E198–E206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilton, C.; Neville, M.J.; Karpe, F. MicroRNAs in adipose tissue: Their role in adipogenesis and obesity. Int. J. Obes. 2013, 37, 325–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, C.; Zhang, M.; Tong, M.; Yang, L.; Pang, L.; Chen, L.; Xu, G.; Chi, X.; Hong, Q.; Ni, Y.; et al. miR-148a is Associated with Obesity and Modulates Adipocyte Differentiation of Mesenchymal Stem Cells through Wnt Signaling. Sci. Rep. 2015, 5, 9930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, D.; Mao, C.; Quattrochi, B.; Friedline, R.H.; Zhu, L.J.; Jung, D.Y.; Kim, J.K.; Lewis, B.; Wang, Y.-X. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 2014, 5, 4725. [Google Scholar] [CrossRef] [Green Version]
- Miao, X.; Luo, Q.; Qin, X.; Guo, Y. Genome-wide analysis of microRNAs identifies the lipid metabolism pathway to be a defining factor in adipose tissue from different sheep. Sci. Rep. 2015, 5, 18470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunch, T.D.; Evans, R.; Wang, S.; Brennand, C.; Whittier, D.; Taylor, B. Feed efficiency, growth rates, carcass evaluation, cholesterol level and sensory evaluation of lambs of various hair and wool sheep and their crosses. Small Rumin. Res. 2004, 52, 239–245. [Google Scholar] [CrossRef]
- McLoughlin, S.; Spillane, C.; Claffey, N.; Smith, P.E.; O’Rourke, T.; Diskin, M.G.; Waters, S.M. Rumen microbiome composition is altered in sheep divergent in feed efficiency. Front. Microbiol. 2020, 11, 1981. [Google Scholar] [CrossRef] [PubMed]
- Patil, R.D.; Ellison, M.J.; Wolff, S.M.; Shearer, C.; Wright, A.M.; Cockrum, R.R.; Austin, K.J.; Lamberson, W.R.; Cammack, K.M.; Conant, G.C. Poor feed efficiency in sheep is associated with several structural abnormalities in the community metabolic network of their ruminal microbes. J. Anim. Sci. 2018, 96, 2113–2124. [Google Scholar] [CrossRef] [PubMed]
- Zuidhof, M.; Schneider, B.; Carney, V.; Korver, D.; Robinson, F. Growth, efficiency, and yield of commercial broilers from 1957, 1978, and 2005. Poult. Sci. 2014, 93, 2970–2982. [Google Scholar] [CrossRef] [PubMed]
- Sell-Kubiak, E.; Wimmers, K.; Reyer, H.; Szwaczkowski, T. Genetic aspects of feed efficiency and reduction of environmental footprint in broilers: A review. J. Appl. Genet. 2017, 58, 487–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tallentire, C.W.; Leinonen, I.; Kyriazakis, I. Breeding for efficiency in the broiler chicken: A review. Agron. Sustain. Dev. 2016, 36, 66. [Google Scholar] [CrossRef] [Green Version]
- Reyer, H.; Hawken, R.; Murani, E.; Ponsuksili, S.; Wimmers, K. The genetics of feed conversion efficiency traits in a commercial broiler line. Sci. Rep. 2015, 5, 16387. [Google Scholar] [CrossRef] [Green Version]
- Metzler-Zebeli, B.; Magowan, E.; Hollmann, M.; Ball, M.; Molnár, A.; Lawlor, P.; Hawken, R.; O’Connell, N.; Zebeli, Q. Assessing serum metabolite profiles as predictors for feed efficiency in broiler chickens reared at geographically distant locations. Br. Poult. Sci. 2017, 58, 729–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, M.; Proszkowiec-Weglarz, M. Mechanisms regulating feed intake, energy expenditure, and body weight in poultry. Poult. Sci. 2007, 86, 1478–1490. [Google Scholar] [CrossRef]
- Yang, L.; He, T.; Xiong, F.; Chen, X.; Fan, X.; Jin, S.; Geng, Z. Identification of key genes and pathways associated with feed efficiency of native chickens based on transcriptome data via bioinformatics analysis. BMC Genom. 2020, 21, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, C.; Sun, L.; Ma, J.; Wang, J.; Qu, H.; Shu, D. Association of single nucleotide polymorphisms in the micro RNA miR-1596 locus with residual feed intake in chickens. Anim. Genet. 2015, 46, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Nie, Q.; Zhang, X. Overview of genomic insights into chicken growth traits based on genome-wide association study and microRNA regulation. Curr. Genom. 2013, 14, 137–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glazov, E.A.; Cottee, P.A.; Barris, W.C.; Moore, R.J.; Dalrymple, B.P.; Tizard, M.L. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008, 18, 957–964. [Google Scholar] [CrossRef] [Green Version]
- Khatri, B.; Seo, D.; Shouse, S.; Pan, J.H.; Hudson, N.J.; Kim, J.K.; Bottje, W.; Kong, B.C. MicroRNA profiling associated with muscle growth in modern broilers compared to an unselected chicken breed. BMC Genom. 2018, 19, 683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qiu, M.; Du, H.; Li, Q.; Gan, W.; Xiong, X.; Yu, C.; Peng, H.; Xia, B.; Song, X. Small RNA sequencing of pectoral muscle tissue reveals microRNA-mediated gene modulation in chicken muscle growth. J. Anim. Physiol. Anim. Nutr. 2020, 104, 867–875. [Google Scholar] [CrossRef]
- Li, H.; Ma, Z.; Jia, L.; Li, Y.; Xu, C.; Wang, T.; Han, R.; Jiang, R.; Li, Z.; Sun, G. Systematic analysis of the regulatory functions of microRNAs in chicken hepatic lipid metabolism. Sci. Rep. 2016, 6, 31766. [Google Scholar] [CrossRef] [Green Version]
- Hicks, J.A.; Porter, T.E.; Liu, H.-C. Identification of microRNAs controlling hepatic mRNA levels for metabolic genes during the metabolic transition from embryonic to posthatch development in the chicken. BMC Genom. 2017, 18, 687. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Liu, R.; Zhao, G.; Li, Q.; Zheng, M.; Zhang, J.; Li, S.; Liang, Z.; Wen, J. Integrated analysis of microRNA and mRNA expression profiles in abdominal adipose tissues in chickens. Sci. Rep. 2015, 5, 16132. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Y.; Jin, W.; Li, Y.; Zhang, Y.; Ma, X.; Sun, G.; Han, R.; Tian, Y.; Li, H. MicroRNAs and their regulatory networks in Chinese Gushi chicken abdominal adipose tissue during postnatal late development. BMC Genom. 2019, 20, 778. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Sun, J.; Zhu, S.; Du, Z.; Li, D.; Li, W.; Li, Z.; Tian, Y.; Kang, X.; Sun, G. MiRNAs and mRNAs Analysis during Abdominal Preadipocyte Differentiation in Chickens. Animals 2020, 10, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Z.; Li, H.; Zheng, H.; Jiang, K.; Jia, L.; Yan, F.; Tian, Y.; Kang, X.; Wang, Y.; Liu, X. MicroRNA-101-2-5p targets the ApoB gene in the liver of chicken (Gallus Gallus). Genome 2017, 60, 673–678. [Google Scholar] [CrossRef] [Green Version]
- Tian, W.-H.; Wang, Z.; Yue, Y.-X.; Li, H.; Li, Z.-J.; Han, R.-L.; Tian, Y.-D.; Kang, X.-T.; Liu, X.-J. miR-34a-5p increases hepatic triglycerides and total cholesterol levels by regulating ACSL1 protein expression in laying hens. Int. J. Mol. Sci. 2019, 20, 4420. [Google Scholar] [CrossRef] [Green Version]
- Marchesi, J.A.P.; Ono, R.K.; Cantão, M.E.; Ibelli, A.M.G.; Peixoto, J.d.O.; Moreira, G.C.M.; Godoy, T.F.; Coutinho, L.L.; Munari, D.P.; Ledur, M.C. Exploring the genetic architecture of feed efficiency traits in chickens. Sci. Rep. 2021, 11, 4622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, S.; Chen, W.; Lu, W.; Huang, Y. A single-nucleotide polymorphism in the 3′ untranslated region of the LPIN1 gene and association analysis with performance traits in chicken. Br. Poult. Sci. 2013, 54, 312–318. [Google Scholar]
- Abasht, B.; Zhou, N.; Lee, W.R.; Zhuo, Z.; Peripolli, E. The metabolic characteristics of susceptibility to wooden breast disease in chickens with high feed efficiency. Poult. Sci. 2019, 98, 3246–3256. [Google Scholar] [CrossRef]
- Xiao, C.; Deng, J.; Zeng, L.; Sun, T.; Yang, Z.; Yang, X. Transcriptome Analysis Identifies Candidate Genes and Signaling Pathways Associated With Feed Efficiency in Xiayan Chicken. Front. Genet. 2021, 12, 607719. [Google Scholar] [CrossRef]
- Yi, G.; Yuan, J.; Bi, H.; Yan, W.; Yang, N.; Qu, L. In-Depth Duodenal Transcriptome Survey in Chickens with Divergent Feed Efficiency Using RNA-Seq. PLoS ONE 2015, 10, e0136765. [Google Scholar] [CrossRef] [Green Version]
- Zhuo, Z.; Lamont, S.J.; Lee, W.R.; Abasht, B. RNA-Seq Analysis of Abdominal Fat Reveals Differences between Modern Commercial Broiler Chickens with High and Low Feed Efficiencies. PLoS ONE 2015, 10, e0135810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farmer, S.R. Regulation of PPARγ activity during adipogenesis. Int. J. Obes. 2005, 29, S13–S16. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Zhang, X.; Li, A.; Xie, L.; Miao, X. Differential regulation of mRNAs and lncRNAs related to lipid metabolism in two pig breeds. Oncotarget 2017, 8, 87539–87553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, N.; Wang, Y.; Xu, R.X.; Wang, G.Q.; Xiong, Y.; Yu, T.Y.; Yang, G.S.; Pang, W.J. PU.1 antisense lncRNA against its mRNA translation promotes adipogenesis in porcine preadipocytes. Anim. Genet. 2015, 46, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, P.A.; Reverter, A.; Berezin, R.B.; Porto-Neto, L.R.; Ribeiro, G.; Santana, M.H.A.; Ferraz, J.B.S.; Fukumasu, H. Exploring the Regulatory Potential of Long Non-Coding RNA in Feed Efficiency of Indicine Cattle. Genes 2020, 11, 997. [Google Scholar] [CrossRef] [PubMed]
- Nolte, W.; Weikard, R.; Brunner, R.M.; Albrecht, E.; Hammon, H.M.; Reverter, A.; Küehn, C. Identification and Annotation of Potential Function of Regulatory Antisense Long Non-Coding RNAs Related to Feed Efficiency in Bos taurus Bulls. Int. J. Mol. Sci. 2020, 21, 3292. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Zhang, X.X.; Li, G.Z.; Li, X.L.; Zhang, Y.K.; Zhao, Y.; Song, Q.Z.; Wang, W.M. Transcriptome analysis of long noncoding RNAs ribonucleic acids from the livers of Hu sheep with different residual feed intake. Animal 2020, 15, 100098. [Google Scholar] [CrossRef]
- Bakhtiarizadeh, M.R.; Salami, S.A. Identification and Expression Analysis of Long Noncoding RNAs in Fat-Tail of Sheep Breeds. G3-Genes Genom. Genet. 2019, 9, 1263–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Wang, S.; Wu, R.; Zhou, X.; Zhu, D.; Zhang, Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics 2012, 99, 292–298. [Google Scholar] [CrossRef] [Green Version]
- Cai, B.; Li, Z.; Ma, M.; Wang, Z.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. LncRNA-Six1 Encodes a Micropeptide to Activate Six1 in Cis and Is Involved in Cell Proliferation and Muscle Growth. Front. Physiol. 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Muret, K.; Klopp, C.; Wucher, V.; Esquerré, D.; Legeai, F.; Lecerf, F.; Désert, C.; Boutin, M.; Jehl, F.; Acloque, H.; et al. Long noncoding RNA repertoire in chicken liver and adipose tissue. Genet. Sel. Evol. 2017, 49, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Zhang, X.; Han, K.; Zhang, G.; Wang, J.; Xie, K.; Xue, Q.; Fan, X. Analysis of long noncoding RNA and mRNA using RNA sequencing during the differentiation of intramuscular preadipocytes in chicken. PLoS ONE 2017, 12, e0172389. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, G.; Chen, G.; Li, J.; Li, M.; Zou, C.; Fang, C.; Li, C. Transcriptome Analysis Suggests the Roles of Long Intergenic Non-coding RNAs in the Growth Performance of Weaned Piglets. Front. Genet. 2019, 10, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampath, H.; Miyazaki, M.; Dobrzyn, A.; Ntambi, J.M. Stearoyl-CoA desaturase-1 mediates the pro-lipogenic effects of dietary saturated fat. J. Biol. Chem. 2007, 282, 2483–2493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fassah, D.M.; Jeong, J.Y.; Baik, M. Hepatic transcriptional changes in critical genes for gluconeogenesis following castration of bulls. Asian-Australas. J. Anim. Sci. 2018, 31, 537–547. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Zhang, X.; Li, F.; Li, C.; La, Y.; Mo, F.; Li, G.; Zhang, Y.; Li, X.; Song, Q.; et al. Transcriptome Analysis Identifies Candidate Genes and Pathways Associated With Feed Efficiency in Hu Sheep. Front. Genet. 2019, 10, 1183. [Google Scholar] [CrossRef]
- Berisha, S.Z.; Serre, D.; Schauer, P.; Kashyap, S.R.; Smith, J.D. Changes in Whole Blood Gene Expression in Obese Subjects with Type 2 Diabetes Following Bariatric Surgery: A Pilot Study. PLoS ONE 2011, 6, e16729. [Google Scholar] [CrossRef] [Green Version]
- Ren, T.; Li, Z.; Zhou, Y.; Liu, X.; Han, R.; Wang, Y.; Yan, F.; Sun, G.; Li, H.; Kang, X. Sequencing and characterization of lncRNAs in the breast muscle of Gushi and Arbor Acres chickens. Genome 2018, 61, 337–347. [Google Scholar] [CrossRef] [Green Version]
- Grifone, R.; Laclef, C.; Spitz, F.; Lopez, S.; Demignon, J.; Guidotti, J.-E.; Kawakami, K.; Xu, P.-X.; Kelly, R.; Petrof, B.J.; et al. Six1 and Eya1 Expression Can Reprogram Adult Muscle from the Slow-Twitch Phenotype into the Fast-Twitch Phenotype. Mol. Cell. Biol. 2004, 24, 6253. [Google Scholar] [CrossRef] [Green Version]
- Hetzler, K.L.; Collins, B.C.; Shanely, R.A.; Sue, H.; Kostek, M.C. The homoeobox gene SIX1 alters myosin heavy chain isoform expression in mouse skeletal muscle. Acta Physiol. 2014, 210, 415–428. [Google Scholar] [CrossRef]
- Brien, J.H.; Hernandez-Lagunas, L.; Artinger, K.B.; Ford, H.L. MicroRNA-30a regulates zebrafish myogenesis through targeting the transcription factor Six1. J. Cell Biol. 2014, 127, 2291. [Google Scholar]
- Moloney, G.M.; Dinan, T.G.; Clarke, G.; Cryan, J.F. Microbial regulation of microRNA expression in the brain–gut axis. Curr. Opin. Pharmacol. 2019, 48, 120–126. [Google Scholar] [CrossRef]
- González, L.; Kyriazakis, I.; Tedeschi, L. Precision nutrition of ruminants: Approaches, challenges and potential gains. Animal 2018, 12, s246–s261. [Google Scholar] [CrossRef] [Green Version]
- Vaintrub, M.; Levit, H.; Chincarini, M.; Fusaro, I.; Giammarco, M.; Vignola, G. Precision livestock farming, automats and new technologies: Possible applications in extensive dairy sheep farming. Animal 2020, 15, 100143. [Google Scholar] [CrossRef]
- Raza, S.H.A.; Abdelnour, S.A.; Dhshan, A.I.; Hassanin, A.A.; Noreldin, A.E.; Albadrani, G.M.; Abdel-Daim, M.M.; Cheng, G.; Zan, L. Potential role of specific microRNAs in the regulation of thermal stress response in livestock. J. Therm. Biol. 2021, 96, 102859. [Google Scholar] [CrossRef]
- Sanchez-Mejias, A.; Tay, Y. Competing endogenous RNA networks: Tying the essential knots for cancer biology and therapeutics. J. Hematol. Oncol. 2015, 8. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).