Y-Chromosomal Insights into Breeding History and Sire Line Genealogies of Arabian Horses
Abstract
1. Introduction
2. Materials and Methods
2.1. Creating a Refined Y-Chromosomal Haplotype Phylogeny
2.1.1. Next Generation Sequencing (NGS) Data
- Whole Genome NGS Data
- Target Enriched Sequencing Data
2.1.2. Data Analysis
- Variant Ascertainment
- MSY Haplotype Tree
2.2. MSY Haplotyping
2.2.1. Creating the Backbone Structure
2.2.2. Haplotype Determination
2.2.3. Datasets
- Globally Active Arabian Sire Lines
- Sampling Local Arabian Lines and Other Populations
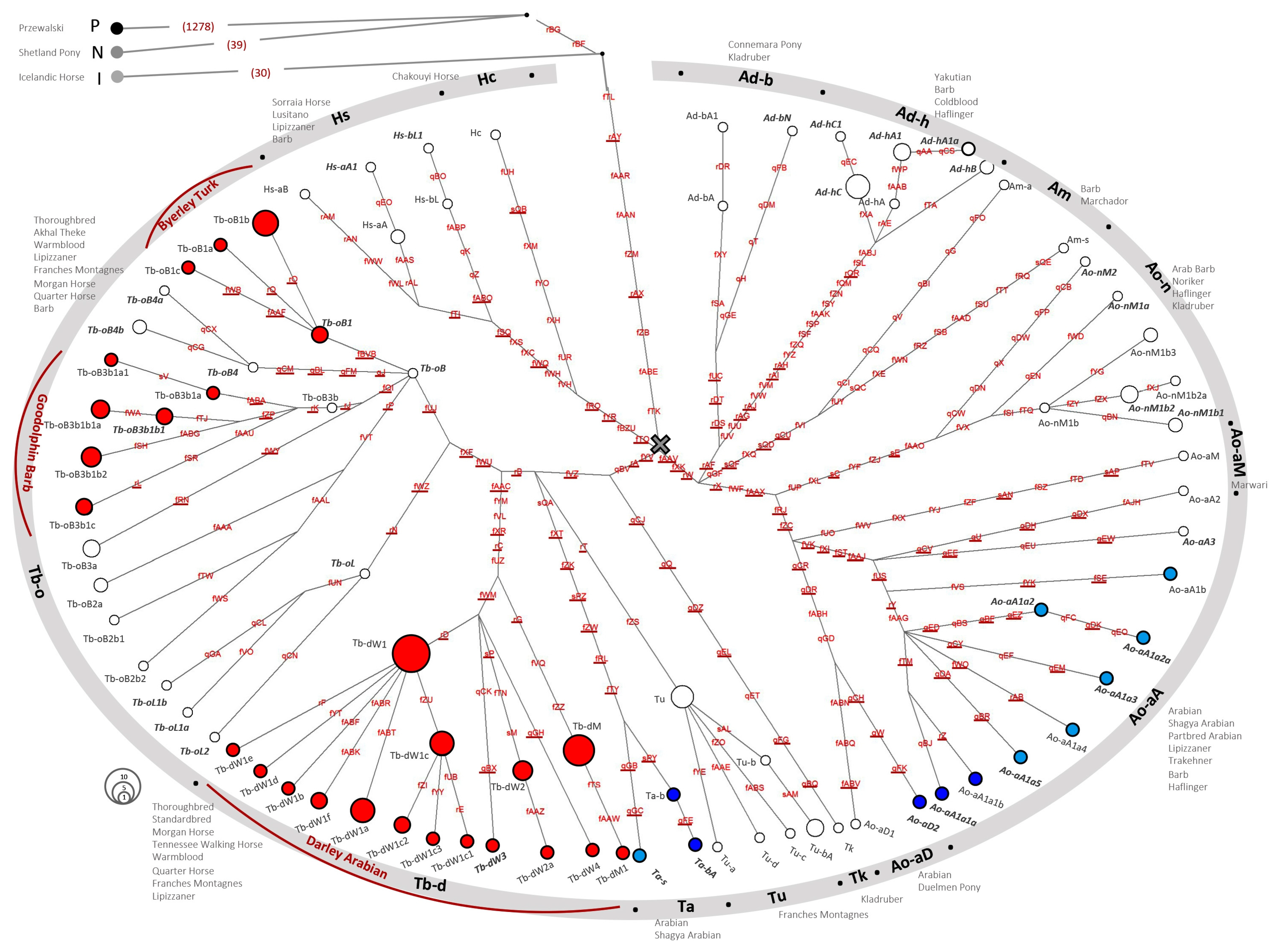
3. Results and Discussion
3.1. A Refinement of the Crown HT Structure from NGS Data
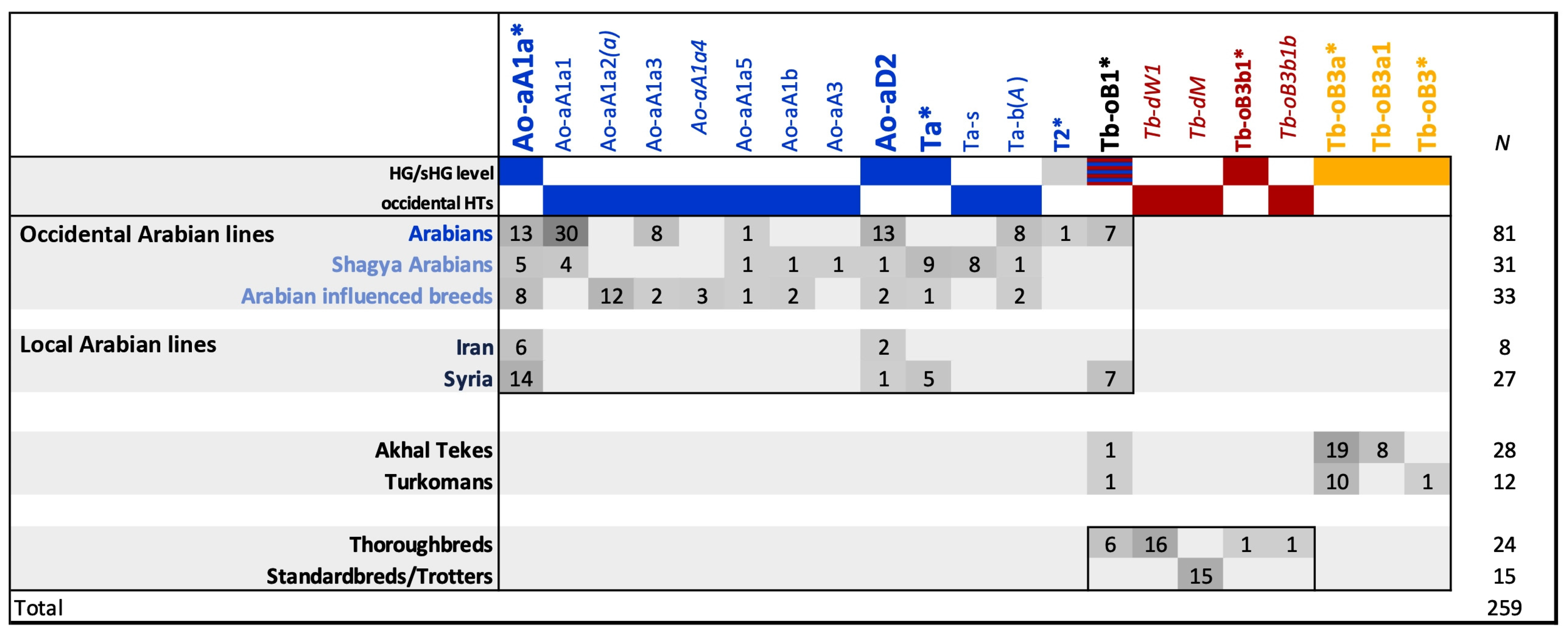
3.2. Allocation of the MSY Signature in Arabian Sire Lines
3.3. Horse MSY Haplotyping in Arabian Horses on the Genealogical Scale
3.4. On the History of Arabian Horse Breeding beyond Pedigrees
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Allele | Variant or Alternative form of the DNA Sequence at a Given Locus |
|---|---|
| clade | a branch on a phylogenetic tree that is formed from individuals of common descent; synonymous to ‘monophyletic group’ |
| crown Haplogroup | very recently expanded horse MSY HG, predominant in modern breeds |
| founder/foundation sire | in this article this term is used to mean the earliest recorded paternal ancestor of an established sire line |
| genetic marker | a genetic marker is a DNA sequence with a known physical location on a chromosome that is polymorphic and thus informative for differentiating individuals |
| genotype | alleles possessed by an individual at one locus |
| haplotype (HT) | a set of DNA polymorphisms that are inherited together due to linkage; a Y-chromosomal haplotype is characterized by the allelic state at several markers |
| haplotyping | the laboratory process of determining the haplotype of an individual via the genotyping of appropriate markers |
| haplogroup (HG) | a monophyletic group of MSY HTs |
| indel | an insertion or deletion of bases in the genome that occurs at a specific genomic position |
| key variant | in this article, this term entitles the marker selected for haplotyping among markers with tautological readout |
| locus | a location in the genome; references to any sequence or genomic region, including non-coding regions |
| mitochondrial DNA (mtDNA) | circular, double-stranded DNA located in the matrix of a mitochondrion; the mtDNA is inherited uniparentally from mother to offspring |
| modern breed | in this article, this term is used for breeds that were developed or created within the westernized industry of horse breeding, spanning the recent 200 years; here it applies to Arabians, Thoroughbreds, Warmbloods, Central European and British Coldbloods, Central European and British Ponies, Baroque Type, Iberian, and New World breeds |
| most recent common ancestor (MRCA) | the sequence status from which all HT variations in a clade descend |
| male-specific region of the Y chromosome (MSY) | the non-recombining region of the Y chromosome which is inherited as a single linkage group from father to sons |
| patriline | the line of descent traced through the paternal side of the pedigree; synonymous with ‘male-tail line’ |
| parsimony | a principle when drawing phylogenetic trees where the most likely tree is the one with the fewest evolutionary changes |
| pedigree | the record of descent of a horse |
| phylogeny | a branching tree showing the evolutionary relationships among a set of DNA sequences |
| sequence assembly | recreation of the original genome from the sequenced reads |
| short tandem repeats (STRs) | a tract of repetitive DNA in which short DNA motifs (ranging in length from one to six or more base pairs) are repeated; synonymous to ‘microsatellites’ |
| single copy Y (scY) regions | well-explored regions of the MSY that are screenable with short-read data |
| single nucleotide variant (SNVs) | a variant of a single nucleotide that occurs at a specific genomic position |
| sire line | members of a sire line descend from the same foundation sire in their patriline (male-tail line); hence the breeding influence of a foundation sire is apparent from the sire line distribution range |
| subline HT | those HTs which emerged recently, within the pedigree supported timeframe, from a new mutation and are therefore unique markers for a particular stallion |
| target enriched sequencing | NGS sequencing of a specific (desired) regions of the genome |
| variant calling | identification of variation, mostly SNVs and Indels, present in sequence data by comparing NGS data from individuals with a reference sequence |
| Y chromosome | one of the two sex chromosomes; determines male sex in horses |
Appendix B
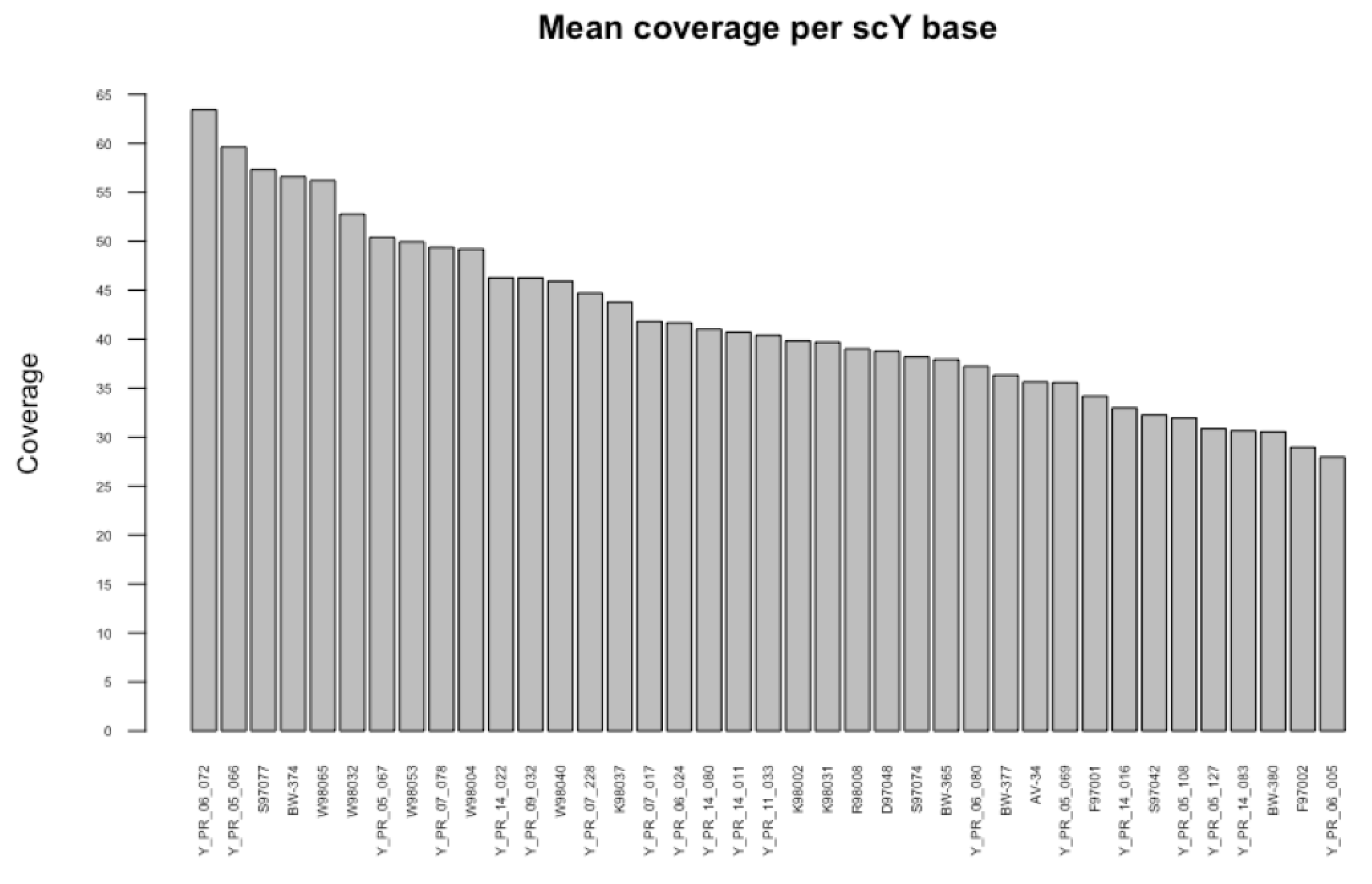
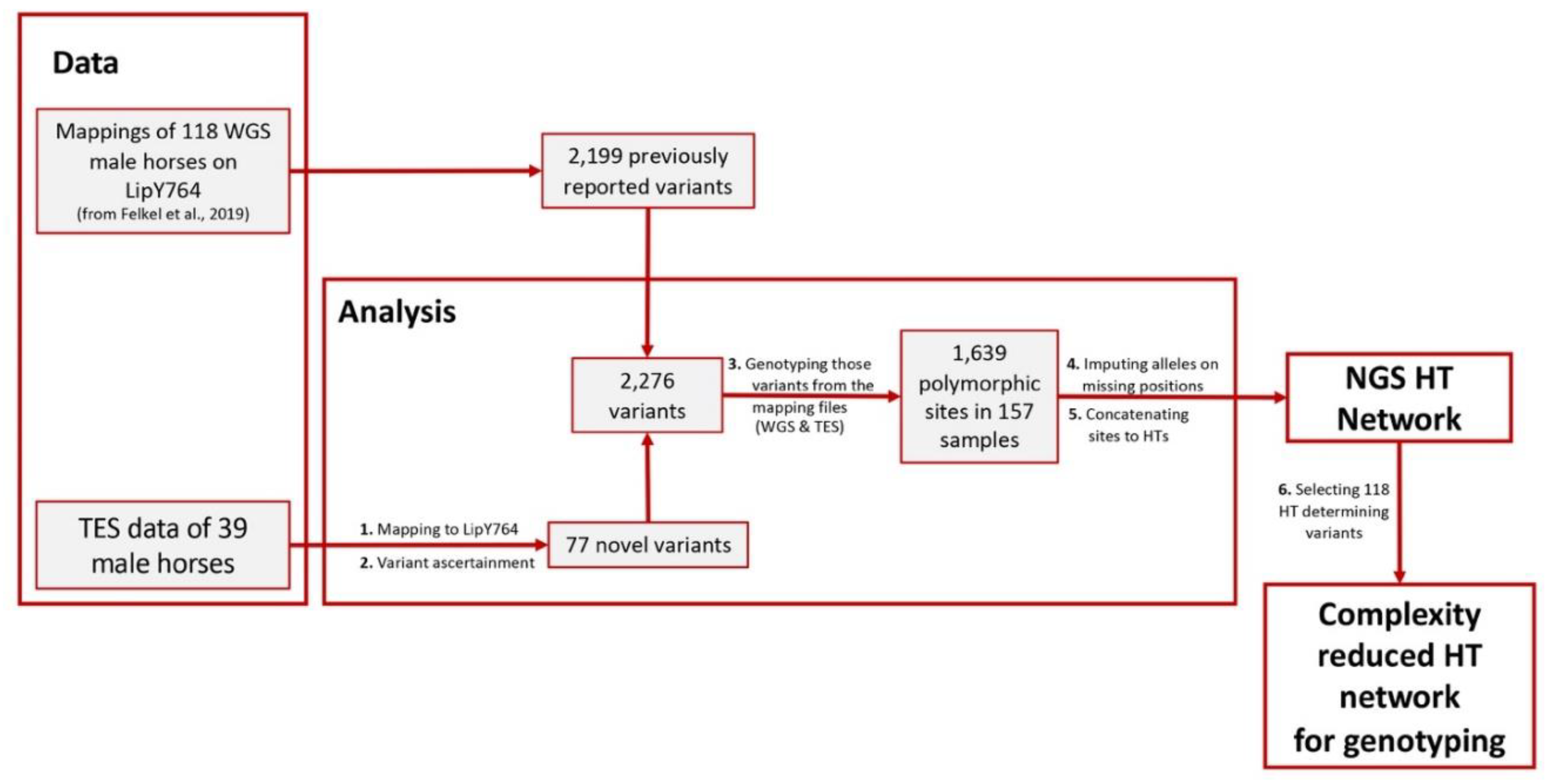
Appendix C
| Major Crown Clades (n = 3—A, H, and T) | Haplogroups | Subhaplogroups and Haplotypes Detectable with Genotyping | Comments Regarding Nomenclature |
|---|---|---|---|
| Clade A was first described in an Arabian horse ‘A’ | |||
| Ad-b | A draft: british | ||
| Ad-h | A draft: heavy | ||
| Am | A marchador | ||
| Ao-aA | A original: arabian Autochthonous | ||
| Ao-aA1a*, Ao-aA1a1, Ao-aA1a2, Ao-aA1a2a, Ao-aA1a3, Ao-aA1a4, Ao-aA1a5, Ao-aA1b, Ao-aA2, Ao-aA3 | |||
| Ao-aD | A original: arabian Duelmener | ||
| Ao-aD1, Ao-aD2 | |||
| Ao-aM | A original: arabian Marwari | ||
| Ao-n | A original: noriker | ||
| Clade H was first described in a Spanish (H) Sorraia horse | |||
| Hc | H china | ||
| Hs | H spanish | ||
| Hs-a | H spanish: autochthonous | ||
| Hs-b | H spanish: barb | ||
| Clade T was first described in Thoroughbreds (T) | |||
| Ta | T arabian | ||
| Ta* | |||
| Ta-s | T arabian: shagya | ||
| Ta-b | T arabian: bairactar | ||
| Ta-bA | |||
| Tb-d | T(horough)bred: darley | ||
| Tb-dM | T(horough)bred: darley Mambrino | ||
| Tb-dW | T(horough)bred: darley Whalebone | ||
| Tb-dW1, Tb-dW2, Tb-dW3, Tb-dW4 | |||
| Tb-o | T(horough)bred: other | ||
| Tb-oB | T(horough)bred: other Byerley/Godolphin | ||
| Tb-oB1*, Tb-oB1a, Tb-oB1b, Tb-oB1c, Tb-oB2, Tb-oB3*, Tb-oB3a*, Tb-oB3a1, Tb-oB3b*, Tb-oB3b1*, Tb-oB3b1a, Tb-oB3b1b, Tb-oB3b1c, Tb-oB4 | |||
| Tb-oL | T(horough)bred: other Lipizzan | ||
| Tk | T kladruber | ||
| Tu | T ubiquitous | ||
| Non-crown Haplogroups | |||
| I | Icelandic horse | ||
| N | Northern Europe | ||
| P | Przewalski |
References
- Kelekna, P. The Horse in Human History; Cambridge University Press: New York, NY, USA, 2009; ISBN 978-0521736299. [Google Scholar]
- Librado, P.; Khan, N.; Fages, A.; Kusliy, M.A.; Suchan, T.; Tonasso-Calvière, L.; Schiavinato, S.; Alioglu, D.; Fromentier, A.; Perdereau, A.; et al. The origins and spread of domestic horses from the Western Eurasian steppes. Nature 2021, 598, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Fages, A.; Gaunitz, C.; Leonardi, M.; Wagner, S.; Khan, N.; Hanghøj, K.; Alquraishi, S.A.; Alfarhan, A.H.; Al-Rasheid, K.A.; et al. The evolutionary origin and genetic makeup of domestic horses. Genetics 2016, 204, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.L.; Mickelson, J.R.; Cothran, E.G.; Andersson, L.S.; Axelsson, J.; Bailey, E.; Bannasch, D.; Binns, M.M.; Borges, A.S.; Brama, P.; et al. Genetic Diversity in the Modern Horse Illustrated from Genome-Wide SNP Data. PLoS ONE 2013, 8, e54997. [Google Scholar] [CrossRef] [PubMed]
- Bower, M.A.; Campana, M.G.; Nisbet, R.; Weller, R.; Whitten, M.; Edwards, C.J.; Stock, F.; Barrett, E.; O’Connell, T.; Hill, E.W.; et al. Truth in the bones: Resolving the identity of the founding elite thoroughbred racehorses. Archaeometry 2012, 54, 916–925. [Google Scholar] [CrossRef]
- Raudsepp, T.; Finno, C.J.; Bellone, R.R.; Petersen, J.L. Ten years of the horse reference genome: Insights into equine biology, domestication and population dynamics in the post-genome era. Anim. Genet. 2019, 50, 569–597. [Google Scholar] [CrossRef]
- Fages, A.; Hanghøj, K.; Khan, N.; Gaunitz, C.; Seguin-Orlando, A.; Leonardi, M.; McCrory Constantz, C.; Gamba, C.; Al-Rasheid, K.A.; Albizuri, S.; et al. Tracking Five Millennia of Horse Management with Extensive Ancient Genome Time Series. Cell 2019, 177, 1419–1435.e31. [Google Scholar] [CrossRef]
- Hendricks, B. International Encyclopedia of Horse Breeds; University of Oklahoma Press: Norman, OK, USA, 2007; ISBN 9780806138848. [Google Scholar]
- Arnason, T.; van Vleck, L.D. Genetic Improvement of the Horse. In The Genetics of the Horse; Bowling, A., Ruvinsky, A., Eds.; CABI Publishing: Wallingford, UK, 2000; p. 527. ISBN 9780851999258. [Google Scholar]
- Grilz-Seger, G.; Druml, T. Lipizzaner Hengststämme; Vehling Medienservice und Verlag GmbH: Graz, Austria, 2011. [Google Scholar]
- Petersen, J.L.; Mickelson, J.R.; Cleary, K.D.; McCue, M.E. The american quarter horse: Population structure and relationship to the thoroughbred. J. Hered. 2014, 105, 148–162. [Google Scholar] [CrossRef][Green Version]
- Weatherby, J. An Introduction to a General Stud Book; H. Reynell: London, UK, 1791. [Google Scholar]
- Cunningham, E.P.; Dooley, J.J.; Splan, R.K.; Bradley, D.G. Microsatellite diversity, pedigree relatedness and the contributions of founder lineages to thoroughbred horses. Anim. Genet. 2001, 32, 360–364. [Google Scholar] [CrossRef]
- McGivney, B.A.; Han, H.; Corduff, L.R.; Katz, L.M.; Tozaki, T.; MacHugh, D.E.; Hill, E.W. Genomic inbreeding trends, influential sire lines and selection in the global Thoroughbred horse population. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Zechner, P.; Sölkner, J.; Bodo, I.; Druml, T.; Baumung, R.; Achmann, R.; Marti, E.; Habe, F.; Brem, G. Analysis of diversity and population structure in the Lipizzan horse breed based on pedigree information. Livest. Prod. Sci. 2002, 77, 137–146. [Google Scholar] [CrossRef]
- Pjontek, J.; Kadlečík, O.; Kasarda, R.; Horný, M. Pedigree analysis in four Slovak endangered horse breeds. Czech J. Anim. Sci. 2012, 57, 54–64. [Google Scholar] [CrossRef]
- Calafell, F.; Larmuseau, M.H. The Y chromosome as the most popular marker in genetic genealogy benefits interdisciplinary research. Hum. Genet. 2017, 136, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Jobling, M.A.; Tyler-Smith, C. Fathers and sons: The Y chromosome and human evolution. Trends Genet. 1995, 11, 449–456. [Google Scholar] [CrossRef]
- Radovic, L.; Remer, V.; Reiter, S.; Bozlak, E.; Felkel, S.; Grilz-Seger, G.; Brem, G.; Wallner, B. Y chromosome genetic variation and deep genealogies provide new insights on Lipizzan sire lines. J. Equine Vet. Sci. 2021, 100, 103501. [Google Scholar] [CrossRef]
- Wallner, B.; Palmieri, N.; Vogl, C.; Rigler, D.; Bozlak, E.; Druml, T.; Jagannathan, V.; Leeb, T.; Fries, R.; Tetens, J.; et al. Y Chromosome Uncovers the Recent Oriental Origin of Modern Stallions. Curr. Biol. 2017, 27, 2029–2035.e5. [Google Scholar] [CrossRef]
- Felkel, S.; Vogl, C.; Rigler, D.; Jagannathan, V.; Leeb, T.; Fries, R.; Neuditschko, M.; Rieder, S.; Velie, B.D.; Lindgren, G.; et al. Asian horses deepen the MSY phylogeny. Anim. Genet. 2018, 49, 90–93. [Google Scholar] [CrossRef]
- Felkel, S.; Vogl, C.; Rigler, D.; Dobretsberger, V.; Chowdhary, B.P.; Distl, O.; Fries, R.; Jagannathan, V.; Janečka, J.E.; Leeb, T.; et al. The horse Y chromosome as an informative marker for tracing sire lines. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Han, H.; Wallner, B.; Rigler, D.; MacHugh, D.E.; Manglai, D.; Hill, E.W. Chinese Mongolian horses may retain early domestic male genetic lineages yet to be discovered. Anim. Genet. 2019, 50, 399–402. [Google Scholar] [CrossRef]
- Liu, S.; Yang, Y.; Pan, Q.; Sun, Y.; Ma, H.; Liu, Y.; Wang, M.; Zhao, C.; Wu, C. Ancient Patrilineal Lines and Relatively High ECAY Diversity Preserved in Indigenous Horses Revealed with Novel Y-Chromosome Markers. Front. Genet. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Castaneda, C.; Juras, R.; Khanshour, A.M.; Randlaht, I.; Wallner, B.; Rigler, D.; Lindgren, G.; Raudsepp, T.; Cothran, E.G. Population genetic analysis of the Estonian native horse suggests diverse and distinct genetics, ancient origin and contribution from unique patrilines. Genes 2019, 10, 629. [Google Scholar] [CrossRef]
- Wallner, B.; Vogl, C.; Shukla, P.; Burgstaller, J.P.; Druml, T.; Brem, G. Identification of Genetic Variation on the Horse Y Chromosome and the Tracing of Male Founder Lineages in Modern Breeds. PLoS ONE 2013, 8, e60015. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, E.J.; Sadeghi, R.; Schlamp, F.; Holl, H.M.; Moradi-Shahrbabak, M.; Miraei-Ashtiani, S.R.; Abdalla, S.; Shykind, B.; Troedsson, M.; Stefaniuk-Szmukier, M.; et al. Genome Diversity and the Origin of the Arabian Horse. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Glazewska, I. Speculations on the origin of the Arabian horse breed. Livest. Sci. 2010, 129, 49–55. [Google Scholar] [CrossRef]
- About the World Arabian Horse Organization (WAHO). Available online: http://www.waho.org (accessed on 27 October 2021).
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sánchez, D.; Schlötterer, C. ReadTools: A universal toolkit for handling sequence data from different sequencing platforms. Mol. Ecol. Resour. 2018, 18, 676–680. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.T.; Abecasis, G.R.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Lippold, S.; Xu, H.; Ko, A.; Li, M.; Renaud, G.; Butthof, A.; Schröder, R.; Stoneking, M. Human paternal and maternal demographic histories: Insights from high-resolution Y chromosome and mtDNA sequences. Investig. Genet. 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Hammer, M.F. A nomenclature system for the tree of human Y-Chromosomal binary haplogroups. Genome Res. 2002, 12, 339–348. [Google Scholar] [CrossRef]
- Achilli, A.; Olivieri, A.; Soares, P.; Lancioni, H.; Kashani, B.H.; Perego, U.A.; Nergadze, S.G.; Carossa, V.; Santagostino, M.; Capomaccio, S.; et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. USA 2012, 109, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Raswan, C.R.; Seydel, H. Der Araber und Sein Pferd; Dritte Nac.; Olms Georg: Hildesheim, Germany, 1998. [Google Scholar]
- Forbis, J. Das klassische arabische Pferd; Paul Parey: Hamburg, Germany, 1980; ISBN 978-3489609322. [Google Scholar]
- Arab Families by Grace Dashiell (Western Horseman June, 1950). Available online: https://wiwfarm.com/GDArabianFamilies.html (accessed on 23 November 2021).
- Wells, M.L. The Illustrated Guide to The Morab Horse, 1st ed.; Holladay Publishing: Manning, NC, USA, 2009; ISBN 0578004658. [Google Scholar]
- The two Siglavys with world influence by Furrer, B in Arabische Pferde in the Focus (1/2019). Available online: https://in-the-focus.com/2019/04/ap-1-2019-ist-online/ (accessed on 21 January 2022).
- Pruski, W. Organizowanie państwowych stadnin arabskich. In Dwa Wieki Polskiej Hodowli Koni Arabskich (1778–1978) i Jej Sukcesy na Świecie; Państwowe Wydawnictwo Rolnicze i Leśne: Warszawa, Poland, 1983; pp. 278–322. [Google Scholar]
- Jobling, M.A.; Tyler-Smith, C. Human Y-chromosome variation in the genome-sequencing era. Nat. Publ. Gr. 2017, 18, 485–497. [Google Scholar] [CrossRef]
- Almarzook, S.; Reissmann, M.; Arends, D.; Brockmann, G.A. Genetic diversity of Syrian Arabian horses. Anim. Genet. 2017, 48, 486–489. [Google Scholar] [CrossRef]
- Khanshour, A.M.; Cothran, E.G. Maternal phylogenetic relationships and genetic variation among Arabian horse populations using whole mitochondrial DNA D-loop sequencing. BMC Genet. 2013, 14, 1–12. [Google Scholar] [CrossRef]
- Khanshour, A.; Conant, E.; Juras, R.; Cothran, E.G. Microsatellite analysis of genetic diversity and population structure of Arabian horse populations. J. Hered. 2013, 104, 386–398. [Google Scholar] [CrossRef]
- Machmoum, M.; Boujenane, I.; Azelhak, R.; Badaoui, B.; Petit, D.; Piro, M. Genetic Diversity and Population Structure of Arabian Horse Populations Using Microsatellite Markers. J. Equine Vet. Sci. 2020, 93, 103200. [Google Scholar] [CrossRef]
- Flade, J.E. Das Araberpferd, 3rd ed.; A. Ziemsen Verlag: Wittenberg Lutherstadt, Germany, 1977; p. 144. [Google Scholar]
- Wrangel, G.C.G. Die Rassen des Pferdes. Ihre Entstehung, Geschichtliche Entwicklung und Charakteristische Kennzeichen. Erster Band; Verlag von Schickhardt Ebner (Konrad Witter): Stuttgart, Germany, 1908. [Google Scholar]
- Nissen, J. Enzyklopädie der Pferderassen, Europa Band 2; Franck-Kosmos Verlags-GmbH & Co: Stuttgart, Germany, 1998. [Google Scholar]
- Orlando, L. Ancient Genomes Reveal Unexpected Horse Domestication and Management Dynamics. BioEssays 2020, 42, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bowling, A.; Del Valle, A.; Bowling, M. A pedigree-based study of mitochondrial D-loop DNA sequence variation among Arabian horses. Anim. Genet. 2000, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Warmuth, V.; Eriksson, A.; Bower, M.A.; Barker, G.; Barrett, E.; Hanks, B.K.; Li, S.; Lomitashvili, D.; Ochir-Goryaeva, M.; Sizonovg, G.V.; et al. Reconstructing the origin and spread of horse domestication in the Eurasian steppe. Proc. Natl. Acad. Sci. USA 2012, 109, 8202–8206. [Google Scholar] [CrossRef] [PubMed]

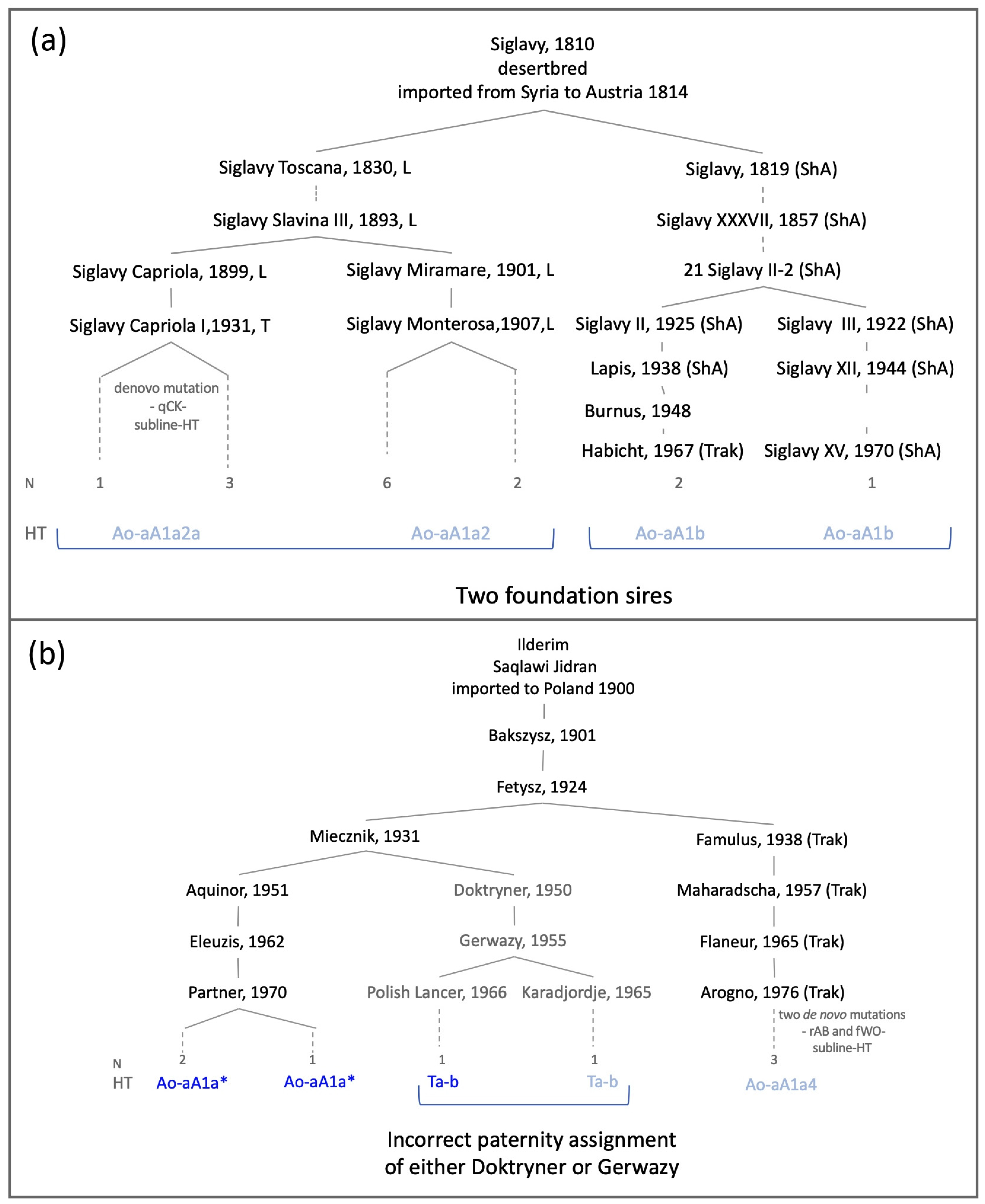
| Foundation Sire 1 | Imported | Line Represented in Dataset via | Registered Arabians Sampled From 2 | Breed Sampled 3 | Y-Chromosomal Haplotype | Remarks |
|---|---|---|---|---|---|---|
| Koheilan Adjuze db Sbaa Anazeh | 1885 Hungary | Piolun, 1934, Poland Jaszmak, 1928, Poland | Austria(1), Russia(5) | Trak(2) ShA(3) | Ao-aA1a* Ao-aA1a* | |
| Mahmoud Mirza db Mukhalladiyah Asad (Iraq) | India, Great Britain, Hungary | Jussuf I, 1962, Shagya Arabian | ShA(2) | Ao-aA1a* | ||
| Old Jellabi Speckled (Bahrain) | Bahrain Foundation horse | Dhahmaan Alawwal, 1938, Bahrain | Austria(2), Poland(1) | Ao-aA1a* | ||
| Zarif Muniqi Shammar Bedouins | 1840 Germany | Rex II 372, 1941, Fredriksborg Horse Hermolin, 1937, Knabstrupper | FH(4) Ks(2) | Ao-aA1a* Ao-aA1a* | ||
| Zobeyni db Seglawi Jedran Ibn Sbeini F’daan | Egyptian Foundation horse | Mahruss II, 1893, Egypt | Germany(1) | Ao-aA1a* | ||
| Ilderim db Saqlawi Jidran | 1900 Poland | Aquinor, 1951, Poland Maharadscha, 1957, Trakehner Doktryner, 1950, Poland | Poland(3) USA(1) | Trak(1), Wb(2) Le(1) | Ao-aA1a* Ao-aA1a4 Ta-b | subline-HT incongruence 4 |
| Saklawi I Saklawi Jidran Ibn Sudan | Egyptian Foundation horse | Ansata Ibn Halima, 1958, Egypt Aswan, 1958, Egypt Galal, 1959, Egypt Habdan Enzahi, 1952, Egypt Morafic, 1952, Egypt | Egypt(1), Qatar(3), Poland(2), Syria(1) Iran(1), Poland(1), Qatar(1), Russia(1) Egypt(1), Germany(1) Austria(1), Egypt(3), Iran(2), Poland(5), Qatar(4) | ShA(1) ShA(1) ShA(2) | Ao-aA1a1 Ao-aA1a1 Ao-aA1a1 Ao-aA1a1 Ao-aA1a1 | |
| El Deree db Saqlawi Shaifi Baqqara (Syria/Iraq) | Egyptian Foundation horse | Akhtal, 1968, Egypt | Qatar(2) | Ao-aA1a1 | ||
| Siglavy db ‚Schwarzenberg‘ Saklawi | 1814 Austria | Siglavy Monterosa, 1907, Lipizzaner Siglavy Capriola III, 1940, Lipizzaner 21 Siglavy II, 1909, Shagya Arabian | Lip(7), Kl(1) Lip(4) ShA(1), Trak(2) | Ao-aA1a2 Ao-aA1a2a Ao-aA1b | subline-HT private HT | |
| Ibrahim db Saklawi Faliti Banu Sakhr | 1907 Poland | Negatiw, 1945, Russia Ferseyn, 1937, USA | Austria(1), Iran(2), Poland(2), Russia(2) United Arab Emirates(1) | PA(1), Pi(1) | Ao-aA1a3 Ao-aA1a3 | |
| Siglavy Bagdady db Saklawi Ruala | 1902 Hungary | Siglavy Bagdady VI, 1949, Babolna | Germany(1) | ShA(1), PA(1) | Ao-aA1a5 | |
| Dahoman db Dahman Djelas Anazeh | 1852 Hungary | Dahoman XVI, 1904, Shagya Arabian | ShA(1) | Ao-aA3 | ||
| Jamil El Kebir db Saklawi Jidran F‘daan | Egyptian Foundation horse | Anter, 1946, Egypt | Germany(1) | Ao-aD2 | ||
| Krzyzyk db | 1876 Poland | Enwer Bey, 1923, Poland | Poland(2) | Ao-aD2 | ||
| Mirage db Seglavi Jedran Dalia Sbaa (Iraq) | 1923 Great Britain | Bey Shah, 1976, USA | Qatar(1), Poland(2) | Ao-aD2 | ||
| Kuhailan Haifi db Koheilan Haifi Ruala | 1931 Poland | Bask, 1956, Poland Celebes, 1949, Poland Wielki Szlem, 1938, POL | Poland(2), Qatar(1) Austria(1), Iran(1), Poland(1), United Arab Emirates(1) | PA(1) WB(1) | Ao-aD2 Ao-aD2 Ao-aD2 | |
| Hadban db Hadban Inzihi S’baa | 1897 Hungary | Habdan XI, 1954, Shagya Arabian | ShA(1) | Ao-aD2 | ||
| Gazlan db Kuhaylan Tamri Would Ali Anazeh (Iraq) | 1852 Lipizza | Gazal VII, 1944, Shagya Arabian | ShA(6) | Ta* | ||
| O’Bajan db Ma’anaqi Sbaili S’baa Anazeh | 1885 Hungary | O’Bajan VII-4 530, 1936, Shagya Arabian O’Bajan X, 1929, Shagya Arabian | ShA(2) ShA(2) | Ta* Ta* | ||
| Shagya db Beni Saher | 1836 Hungary | Shagya IV, 1875, Shagya Arabian Shagya VII, 1877, Shagya Arabian Shagya XI, 1886, Shagya Arabian | ShA(3) ShA(2) ShA(3) | Ta-s Ta-s Ta-s | ||
| Mersuch db Hamdani Semri Hazaim Pasha (Iraq) | 1902 Hungary | Mersuch IV, 1936, Shagya Arabian | ShA(1) | Ta-b | ||
| Bairactar db Saklawi Jidran (Syria) | 1817 Germany | Arax, 1952, Poland Gwarny,1952 | Poland(1), Qatar(1), Russia(1) Germany(2), Poland(1) | Ta-b Ta-bA | subline-HT | |
| Dahman Amir db Dahman Amir Sultan Abdulhamid II (Turkey) | 1902 Poland | Saludo, 1954, Spain | Qatar(1) | PA(1) | Ta-b | |
| Seanderich db Saklawi Jidran Shammar | 1908 Spain | Tabal, 1952, Spain | Germany(1) | T2* | ||
| Kuhailan Afas db Koheilan Afas (Bahrain) | 1931 Poland | Comet, 1953, Poland | Germany(1), Poland(1) | Tb-oB1* | ||
| Latif db Hamdani Semri Anazeh Fedan | 1909 France | Baroud II, 1927, France Kann, 1927, France | Qatar(3), Russia(1) Russia(1) | Tb-oB1* Tb-oB1* |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Remer, V.; Bozlak, E.; Felkel, S.; Radovic, L.; Rigler, D.; Grilz-Seger, G.; Stefaniuk-Szmukier, M.; Bugno-Poniewierska, M.; Brooks, S.; Miller, D.C.; et al. Y-Chromosomal Insights into Breeding History and Sire Line Genealogies of Arabian Horses. Genes 2022, 13, 229. https://doi.org/10.3390/genes13020229
Remer V, Bozlak E, Felkel S, Radovic L, Rigler D, Grilz-Seger G, Stefaniuk-Szmukier M, Bugno-Poniewierska M, Brooks S, Miller DC, et al. Y-Chromosomal Insights into Breeding History and Sire Line Genealogies of Arabian Horses. Genes. 2022; 13(2):229. https://doi.org/10.3390/genes13020229
Chicago/Turabian StyleRemer, Viktoria, Elif Bozlak, Sabine Felkel, Lara Radovic, Doris Rigler, Gertrud Grilz-Seger, Monika Stefaniuk-Szmukier, Monika Bugno-Poniewierska, Samantha Brooks, Donald C. Miller, and et al. 2022. "Y-Chromosomal Insights into Breeding History and Sire Line Genealogies of Arabian Horses" Genes 13, no. 2: 229. https://doi.org/10.3390/genes13020229
APA StyleRemer, V., Bozlak, E., Felkel, S., Radovic, L., Rigler, D., Grilz-Seger, G., Stefaniuk-Szmukier, M., Bugno-Poniewierska, M., Brooks, S., Miller, D. C., Antczak, D. F., Sadeghi, R., Cothran, G., Juras, R., Khanshour, A. M., Rieder, S., Penedo, M. C., Waiditschka, G., Kalinkova, L., ... Wallner, B. (2022). Y-Chromosomal Insights into Breeding History and Sire Line Genealogies of Arabian Horses. Genes, 13(2), 229. https://doi.org/10.3390/genes13020229








