Selection of Candidate Genes Conferring Blast Resistance and Heat Tolerance in Rice through Integration of Meta-QTLs and RNA-Seq
Abstract
1. Introduction
2. Materials and Methods
2.1. Bibliographic Collection and Significant Data Summary
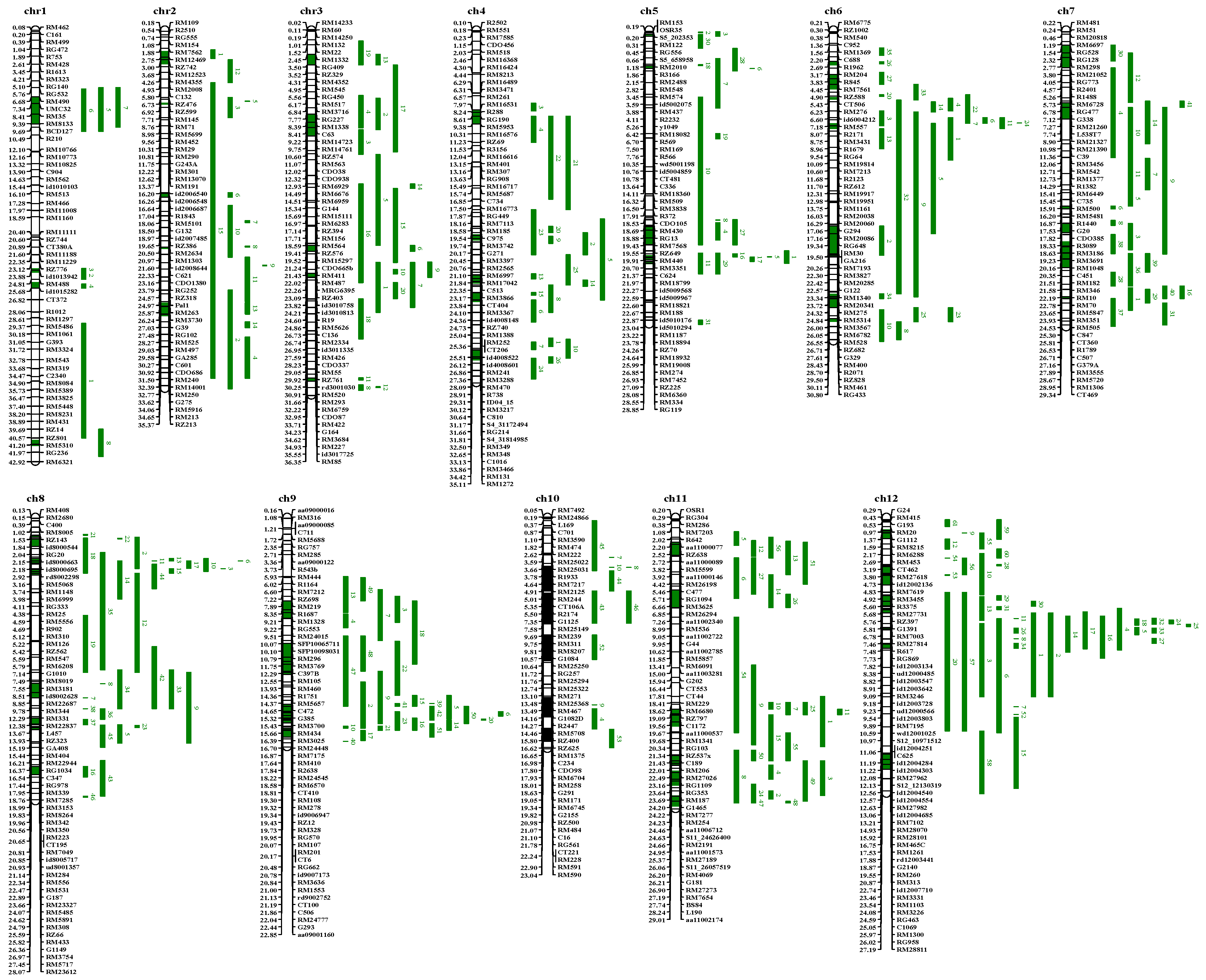
2.2. Meta-QTL Analysis
2.3. Difference Analysis of RNA-Seq Data
2.4. Integration Analysis of Meta-QTLs and RNA-Seq
3. Results
3.1. Compilation and Characterization of QTL Studies Regarding Blast Resistance in Rice
3.2. Meta-Analysis Results for Blast-Resistant QTLs in Rice
3.3. Compilation and Characterization of QTL Studies Regarding Heat Tolerance in Rice
3.4. Meta-Analysis Results for Heat-Tolerant QTLs in Rice
3.5. Integration Analysis Results for Meta-QTLs and RNA-Seq
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Confalonieri, R.; Rosenmund, A.S.; Baruth, B. An improved model to simulate rice yield. Agron. Sustain. Dev. 2009, 29, 463–474. [Google Scholar] [CrossRef]
- Carriger, S.; Vallée, D. More Crop per Drop; IWA Publishing: London, UK, 2007; Volume 6, pp. 10–13. [Google Scholar]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the feld. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef]
- Onaga, G.; Wydra, K.; Koopmann, B.; Chebotarov, D.; Séré, Y.; Von Tiedemann, A. High temperature effects on Pi54 conferred resistance to Magnaporthe oryzae in two genetic backgrounds of Oryza sativa. J. Plant Physiol. 2017, 212, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Schwarczinger, I.; Kolozsváriné Nagy, J.; Király, L.; Mészáros, K.; Bányai, J.; Kunos, V.; Fodor, J.; Künstler, A. Heat stress pre-exposure may differentially modulate plant defense to powdery mildew in a resistant and susceptible barley genotype. Genes (Basel) 2021, 12, 776. [Google Scholar] [CrossRef] [PubMed]
- Dossa, G.S.; Quibod, I.; Atienza-Grande, G.; Oliva, R.; Maiss, E.; Vera Cruz, C.; Wydra, K. Rice pyramided line IRBB67 (Xa4/Xa7) homeostasis under combined stress of high temperature and bacterial blight. Sci. Rep. 2020, 10, 683. [Google Scholar] [CrossRef] [PubMed]
- Aoun, N.; Tauleigne, L.; Lonjon, F.; Deslandes, L.; Vailleau, F.; Roux, F.; Berthomé, R. Quantitative disease resistance under elevated temperature: Genetic basis of new resistance mechanisms to Ralstonia solanacearum. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- E, G.D. Azoxystrobin rate and timing effects on rice head blast incidence and rice grain and milling yields. Plant Dis. 2006, 90, 1055–1058. [Google Scholar]
- Noguchi, M.T.; Yasuda, N.; Fujita, Y. Evidence of genetic exchange by parasexual recombination and genetic analysis of pathogenicity and mating type of parasexual recombinants in rice blast fungus, Magnaporthe oryzae. Phytopathology 2006, 96, 746–750. [Google Scholar] [CrossRef][Green Version]
- Zeng, J.; Feng, S.; Cai, J.; Wang, L.; Lin, F.; Pan, Q. Distribution of mating type and sexual status in Chinese rice blast populations. Plant Dis. 2009, 93, 238–242. [Google Scholar] [CrossRef]
- Devi, S.; Sharma, G.D. Blast disease of rice caused by Magnaporthe grisea: A review. Assam Univ. J. Sci. Technol. 2010, 6, 144–154. [Google Scholar]
- Zhao, C.; Piao, S.; Wang, X.; Huang, Y.; Ciais, P.; Elliott, J.; Huang, M.; Janssens, I.; Li, T.; Lian, X.; et al. Plausible rice yield losses under future climate warming. Nat. Plants 2016, 3, 16202. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasulu, N.; Butardo, V.M.J.; Misra, G.; Cuevas, R.P.; Anacleto, R.; Kavi Kishor, P.B. Designing climate-resilient rice with ideal grain quality suited for high-temperature stress. J. Exp. Bot. 2015, 66, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.R.; Madhav, M.S.; Singh, B.K.; Shanker, P.; Jana, T.K.; Dalal, V. High resolution mapping, cloning and molecular characterization of the Pi-k(h) gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genom. 2005, 274, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Y.; He, J.B.; Li, A.H.; Fang, N.Y.; He, W.W.; Dang, L.L.; Zeng, G.Y.; Huang, J.; Bao, Y.M.; Zhang, H.S. Population structure analysis and association mapping of blast resistance in indica rice (Oryza sativa L.) landraces. Genet. Mol. Res. 2016, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Yang, W.T.; Kim, D.H.; Kim, K.M. Identification of a novel gene, Osbht, in response to high temperature tolerance at booting stage in rice. Int. J. Mol. Sci. 2020, 21, 5862. [Google Scholar] [CrossRef] [PubMed]
- Tabata, M.; Hirabayashi, H.; Takeuchi, Y.; Ando, I.; Iida, Y.; Ohsawa, R. Mapping of quantitative trait loci for the occurrence of white-back kernels associated with high temperatures during the ripening period of rice (Oyza sativa L.). Breed. Sci. 2007, 57, 47–52. [Google Scholar] [CrossRef][Green Version]
- Stermer, B.A.; Hammerschmidt, R. Heat shock induces resistance to Cladosporium cucumerinum and enhances peroxidase activity in cucumbers. Physiol. Plant Pathol. 1984, 25, 239–249. [Google Scholar] [CrossRef]
- Vallelian-Bindschedler, L.; Schweizer, P.; Mösinger, E.; Metraux, J.P. Heat-induced resistance in barley to powdery mildew (Blumeria graminisf. sp. hordei) is associated with a burst of active oxygen species. Physiol. Mol. Plant Pathol. 1998, 52, 185–199. [Google Scholar] [CrossRef]
- Widiastuti, A.; Yoshino, M.; Saito, H.; Maejima, K.; Zhou, S.; Odani, H.; Sato, T. Induction of disease resistance against Botrytis cinerea by heat shock treatment in melon (Cucumis melo L.). Physiol. Mol. Plant Pathol. 2011, 75, 157–162. [Google Scholar] [CrossRef]
- Fu, D.; Uauy, C.; Distelfeld, A.; Blechl, A.; Epstein, L.; Chen, X.; Sela, H.; Fahima, T.; Dubcovsky, J. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 2009, 323, 1357–1360. [Google Scholar] [CrossRef] [PubMed]
- Webb, K.M.; Oña, I.; Bai, J.; Garrett, K.A.; Mew, T.; Vera Cruz, C.M.; Leach, J.E. A benefit of high temperature: Increased effectiveness of a rice bacterial blight disease resistance gene. New Phytol. 2010, 185, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Negeri, A.; Wang, G.F.; Benavente, L.; Kibiti, C.M.; Chaikam, V.; Johal, G.; Balint-Kurti, P. Characterization of temperature and light effects on the defense response phenotypes associated with the maize Rp1-D21 autoactive resistance gene. BMC Plant Biol. 2013, 13, 106. [Google Scholar] [CrossRef] [PubMed]
- Squeglia, F.; Berisio, R.; Shibuya, N.; Kaku, H. Defense against pathogens: Structural insights into the mechanism of chitin induced activation of innate immunity. Curr. Med. Chem. 2017, 24, 3980–3986. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Abuqamar, S.; Luo, H.; Laluk, K.; Mickelbart, M.V.; Mengiste, T. Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J. 2009, 58, 347–360. [Google Scholar] [CrossRef]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.; Qiu, A.L.; Zhang, T.X.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Qi, H.X.; Yin, D.S.; Zeng, F.S.; Zhang, S.; Yu, D.Z. Mapping QTLs for rice blast resistance in DH line derived from Muwanggu and E’wan8. J. Plant Pathol. 2012, 42, 600–607. [Google Scholar]
- Kongprakhon, P.; Cuestamarcos, A.; Hayes, P.M.; Hongtrakul, V.; Sirithunya, P.; Toojinda, T.; Sangduen, N. Four QTL in rice associated with broad spectrum resistance to blast isolates from rice and barley. J. Phytopathol. 2010, 158, 125–131. [Google Scholar] [CrossRef]
- Fang, N.Y.; Wang, R.S.; He, W.W.; Yin, C.F.; Guan, C.H.; Chen, H.; Huang, J.; Wang, J.F.; Bao, Y.M.; Zhang, H.S. QTL mapping of panicle blast resistance in japonica landrace heikezijing and its application in rice breeding. Mol. Breeding 2016, 12, 18–29. [Google Scholar] [CrossRef]
- Chen, X.L.; Jia, Y.L.; Jia, M.H.; Shannon, R.M.; Pinson, S.; Wang, X.Y.; Wu, B.M. Functional interactions between major rice blast resistance genes, Pi-ta and Pi-b, and minor blast resistance QTLs. Phytopathology 2018, 108, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- He, W.W.; Fang, N.Y.; Wang, R.S.; Wu, Y.Y.; Zeng, G.Y.; Guan, C.H.; Chen, H.; Huang, J.; Wang, J.F.; Bao, Y.M. Fine mapping of a new race-specific blast resistance gene Pi-hk2 in Japonica Heikezijing from Taihu region of China. Phytopathology 2017, 107, 84–91. [Google Scholar] [CrossRef]
- Nagaoka, I.; Sasahara, H.; Tabuchi, H.; Shigemune, A.; Matsushita, K.; Maeda, H.; Goto, A.; Fukuoka, S.; Ando, T.; Miura, K. Quantitative trait loci analysis of blast resistance in Oryza sativa L. ‘Hokuriku 193’. Breed. Sci. 2017, 67, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Urso, S.; Desiderio, F.; Biselli, C.; Bagnaresi, P.; Crispino, L.; Piffanelli, P.; Abbruscato, P.; Assenza, F.; Guarnieri, G.; Cattivelli, L. Genetic analysis of durable resistance to Magnaporthe oryzae in the rice accession Gigante Vercelli identified two blast resistance loci. Mol. Genet. Genom. 2016, 291, 17–32. [Google Scholar] [CrossRef]
- Sato, H.; Takeuchi, Y.; Hirabayashi, H.; Nemoto, H.; Hirayama, M.; Kato, H.; Imbe, T.; Ando, I. Mapping QTLs for field resistance to rice blast in the Japanese upland rice Variety Norin12. Breed. Sci. 2006, 56, 415–418. [Google Scholar] [CrossRef][Green Version]
- Lestari, P.; Trijatmiko, K.R.; Reflinur; Warsun, A.; Tasliah; Ona, I.; Cruz, C.V.; Bustamam, M. Mapping quantitative trait loci conferring blast resistance in upland indica rice (Oryza sativa L.). J. Crop Sci. Biotechnol. 2011, 14, 57–63. [Google Scholar] [CrossRef]
- Rahim, H.A.; Bhuiyan, M.A.; Lim, L.S.; Sabu, K.K.; Saad, A.; Azhar, M.; R, W. Identification of quantitative trait loci for blast resistance in BC2F3 and BC2F5 advanced backcross families of rice. Genet. Mol. Res. 2012, 11, 3277–3289. [Google Scholar] [CrossRef]
- Jiang, H.C.; Feng, Y.T.; Qiu, L.; Gao, G.J.; Zhang, Q.L.; He, Y.Q. Identification of blast resistance QTLs based on two advanced backcross populations in rice. Rice 2020, 13, 31–32. [Google Scholar] [CrossRef]
- Angeles-Shim, R.B.; Reyes, V.P.; del Valle, M.M.; Lapis, R.S.; Shim, J.; Sunohara, H.; Jena, K.K.; Ashikari, M.; Doi, K. Marker-assisted introgression of quantitative resistance gene pi21 confers broad spectrum resistance to rice blast. Rice Sci. 2020, 27, 113–123. [Google Scholar] [CrossRef]
- Cao, L.; Zhu, J.; Zhao, S.; He, L.; Yan, Q. Mapping QTLs for heat tolerance in a DH population from indica-japonica cross of rice (Oryza sative). J. Agric. Biotechnol. 2002, 10, 210–214. [Google Scholar]
- Zhu, C.L.; Xiao, Y.H.; Wang, C.M.; Jiang, L.; Zhai, H.Q.; Wan, J.M. Mapping QTL for heat-tolerance at grain filling stage in rice. Rice Sci. 2005, 12, 33–38. [Google Scholar]
- Ye, C.; Argayoso, M.A.; Redoña, E.D.; Sierra, S.N.; Laza, M.A.; Dilla, C.J.; Mo, Y.; Thomson, M.J.; Chin, J.; Delaviña, C.B.; et al. Mapping QTL for heat tolerance at flowering stage in rice using SNP markers. Plant Breeding 2012, 131, 33–41. [Google Scholar] [CrossRef]
- Li, M.M.; Li, X.; Yu, L.Q.; Wu, J.; Li, H.; Liu, J.; Ma, X.; Jo, S.; Park, D.S.; Song, Y.; et al. Identification of QTLs associated with heat tolerance at the heading and flowering stage in rice (Oryza sativa L.). Euphytica 2018, 214, 70–81. [Google Scholar] [CrossRef]
- Cao, Z.; Li, Y.; Zeng, B.; Mao, L.; Cai, Y.; Wu, X.; Yuan, L. QTL mapping for heat tolerance of chalky grain rate of Oryza glaberrima steud. Chin. J. Rice Sci. 2020, 34, 135–142. [Google Scholar]
- Zhang, T.; Yang, L.; Jang, K.F.; Huang, M.; Huang, M.; Chen, W.F.; Zheng, J.K. QTL mapping for heat tolerance of the tassel period of rice. Mol. Plant Breed. 2008, 6, 867–873. [Google Scholar]
- Kobayashi, A.; Sonoda, J.; Sugimoto, K.; Kondo, M.; Iwasawa, N.; Hayashi, T.; Tomita, K.; Yano, M.; Shimizu, T. Detection and verification of QTLs associated with heat-induced quality decline of rice (Oryza sativa L.) using recombinant inbred lines and near-isogenic lines. Breed. Sci. 2013, 63, 339–346. [Google Scholar] [CrossRef]
- Islam, M.S.; Ontoy, J.; Subudhi, P.K. Meta-analysis of quantitative trait loci associated with seedling-stage salt tolerance in rice (Oryza sativa L.). Plants 2019, 8, 33. [Google Scholar] [CrossRef]
- Swamy, B.P.M.; Vikram, P.; Dixit, S.; Ahmed, H.U.; Kumar, A. Meta-analysis of grain yield QTL identified during agricultural drought in grasses showed consensus. BMC Genet. 2011, 12, 319–337. [Google Scholar] [CrossRef]
- Qi, Z.M.; Zhang, Z.G.; Wang, Z.Y.; Yu, J.Y.; Qin, H.T.; Mao, X.R.; Jiang, H.W.; Xin, D.W.; Yin, Z.G.; Zhu, R.S. Meta-analysis and transcriptome profiling reveal hub genes for soybean seed storage composition during seed development. Plant Cell Environ. 2018, 41, 2109–2127. [Google Scholar] [CrossRef]
- Goffinet, B.; Gerber, S. Quantitative trait loci: A meta-analysis. Genetics 2000, 155, 463–473. [Google Scholar] [CrossRef]
- Wang, Y.J.; Xu, J.; Deng, D.X.; Ding, H.D.; Bian, Y.L.; Yin, Z.T.; Wu, Y.R.; Zhou, B.; Zhao, Y. A comprehensive meta-analysis of plant morphology, yield, stay-green, and virus disease resistance QTL in maize (Zea mays L.). Planta 2016, 243, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.G.; Qi, H.D.; Chen, Q.S.; Zhang, Z.G.; Jiang, H.W.; Zhu, R.S.; Hu, Z.B.; Wu, X.X.; Li, C.D.; Zhang, Y. Soybean plant height QTL mapping and meta-analysis for mining candidate genes. Plant Breeding 2017, 136, 688–698. [Google Scholar] [CrossRef]
- Priyanka, J.; Singh, P.K.; Kapoor, R.; Khanna, A.; Solanke, A.U.; Krishnan, S.G.; Singh, A.K.; Sharma, V.; Sharma, T.R. Understanding host-pathogen interactions with expression profiling of NILs carrying rice-blast resistance Pi9 gene. Front. Plant Sci. 2017, 8, 93–113. [Google Scholar]
- Kong, W.; Zhang, C.; Qiang, Y.; Zhong, H.; Zhao, G.; Li, Y. Integrated RNA-Seq analysis and Meta-QTLs mapping provide insights into cold stress response in rice seedling roots. Int. J. Mol. Sci. 2020, 21, 4615. [Google Scholar] [CrossRef]
- Delfino, P.; Zenoni, S.; Imanifard, Z.; Tornielli, G.B.; Bellin, D. Selection of candidate genes controlling veraison time in grapevine through integration of meta-QTL and transcriptomic data. BMC Genet. 2019, 20, 739–757. [Google Scholar] [CrossRef]
- Darvasi, A.; Soller, M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav. Genet. 1997, 27, 125–132. [Google Scholar] [CrossRef]
- Guo, B.; Sleper, D.A.; Lu, P.; Shannon, J.G.; Nguyen, H.; Arelli, P.R. QTLs associated with resistance to soybean cyst nematode in soybean: Meta-analysis of QTL locations. Crop Sci. 2006, 46, 595–602. [Google Scholar] [CrossRef]
- Courtois, B.; Ahmadi, N.; Khowaja, F.S.; Price, A.H.; Rami, J.; Frouin, J.; Hamelin, C.; Ruiz, M. Rice root genetic architecture: Meta-analysis from a drought QTL database. Rice 2009, 2, 115–128. [Google Scholar] [CrossRef]
- Arcade, A.; Labourdette, A.; Falque, M.; Mangin, B.; Chardon, F.; Charcosset, A.; Joets, J. BioMercator: Integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 2004, 20, 2324–2326. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Walter, W.; Sánchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Chen, L.; He, H. Identification of rice blast resistance candidate genes based on meta-QTL and RNA-aeq analysis. Acta Agron. Sin. 2021, 1–18. [Google Scholar]
- Zhu, M.; Wang, L.; Pan, Q. Identification and characterization of a new blast resistance gene located on rice chromosome 1 through linkage and differential analyses. Phytopathology 2004, 94, 515–519. [Google Scholar] [CrossRef]
- Shi, B.H.; Zhang, J.H.; Zheng, Y.M.; Liu, Y.Q.; Vera Cruz, C.M.; Zheng, T.Q.; Zhao, M.F. Identification of a new resistance gene Pi-Da(t) from Dacca6 against rice blast fungus (Magnaporthe oryzae) in Jin23B background. Mol. Breed. 2012, 30, 1089–1096. [Google Scholar] [CrossRef]
- Pan, Q.H.; Wang, L.; Ikehashi, H.; Yamagata, H.; Tanisaka, T. Identification of two new genes conferring resistance to rice blast in the Chinese native cultivar ‘Maowangu’. Plant Breeding 1998, 117, 27–31. [Google Scholar] [CrossRef]
- Pan, Q.H.; Wang, L.; Tanisaka, T. A new blast resistance gene identified in the Indian native rice cultivar Aus373 through allelism and linkage tests. Plant Pathol. 1999, 48, 288–293. [Google Scholar] [CrossRef]
- Chen, X.W.; Li, S.G.; Xu, J.C.; Zhai, W.X.; Ling, Z.Z.; Ma, B.T.; Wang, Y.P.; Wang, W.M.; Cao, G.; Ma, Y.Q.; et al. Identification of two blast resistance genes in a rice variety, Digu. J. Phytopathology 2004, 152, 77–85. [Google Scholar] [CrossRef]
- Ahn, S.N.; Kim, D.M.; Ju, H.G.; Kang, J.W.; Han, S.S. A new rice variety’Hwaweon 5’with durable resistance to rice blast. Korean J. Breed. Sci. 2013, 45, 142–147. [Google Scholar]
- Wong, H.L.; Sakamoto, T.; Kawasaki, T.; Umemura, K.; Shimamoto, K. Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 2004, 135, 1447–1456. [Google Scholar] [CrossRef]
- Naqvi, N.I.; Chattoo, B.B. Development of a sequence characterized amplified region (SCAR) based indirect selection method for a dominant blast-resistance gene in rice. Genome 1996, 39, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Wang, L.; Ikehashi, H.; Tanisaka, T. Identification of a new blast resistance gene in the indica rice cultivar Kasalath using Japanese differential cultivars and isozyme markers. Phytopathology 1996, 86, 1071–1075. [Google Scholar] [CrossRef]
- Jeung, J.U.; Kim, B.R.; Cho, Y.C.; Han, S.S.; Moon, H.P.; Lee, Y.T.; Jena, K.K. A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor. Appl. Genet. 2007, 115, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Li, Z.; Wu, J.; Wang, Y.; Wu, L.; Wang, S.; Wang, D.; Wen, T.; Liang, Y.; Sun, P.; et al. Molecular mapping of the Pi2/9 allelic gene Pi2-2 conferring broad-spectrum resistance to Magnaporthe oryzae in the rice cultivar Jefferson. Rice (N. Y.) 2012, 5, 29. [Google Scholar] [CrossRef]
- Tabien, R.E.; Li, Z.; Paterson, A.H.; Marchetti, M.A.; Stansel, J.W.; Pinson, S.R.M.; Park, W.D. Mapping of four major rice blast resistance genes from ’Lemont’ and ’Teqing’ and evaluation of their combinatorial effect for field resistance. Theor. Appl. Genet. 2000, 101, 1215–1225. [Google Scholar] [CrossRef]
- Liu, X.; Lin, F.; Wang, L.; Pan, Q. The in silico map-based cloning of Pi36, a rice coiled-coil nucleotide-binding site leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics 2007, 176, 2541–2549. [Google Scholar] [CrossRef]
- Lee, S.; Wamishe, Y.; Jia, Y.; Liu, G.; Jia, M.H. Identification of two major resistance genes against race IE-1k of Magnaporthe oryzae in the indica rice cultivar Zhe733. Mol. Breeding 2009, 24, 127–134. [Google Scholar] [CrossRef]
- Zhu, L.; Xu, J.; Chen, Y.; Ling, Z.; Lu, C.; Xu, Y. Identification of an unknown rice blast resistance gene by molecular markers. Sci. China 1994, 1048–1052. [Google Scholar]
- He, X.; Liu, X.; Wang, L.; Wang, L.; Lin, F.; Cheng, Y.; Chen, Z.; Liao, Y.; Pan, Q. Identification of the novel recessive gene pi55(t) conferring resistance to Magnaporthe oryzae. Sci. China Life Sci. 2012, 55, 141–149. [Google Scholar] [CrossRef][Green Version]
- Lee, S.K.; Song, M.Y.; Seo, Y.S.; Kim, H.K.; Ko, S.; Cao, P.J.; Suh, J.P.; Yi, G.; Roh, J.H.; Lee, S.; et al. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 2009, 181, 1627–1638. [Google Scholar] [CrossRef]
- Lin, F.; Liu, Y.; Wang, L.; Liu, X.; Pan, Q. A high-resolution map of the rice blast resistance gene Pi15 constructed by sequence-ready markers. Plant Breeding 2007, 126, 287–290. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, B.; Zhu, X.; Yang, J.; Bordeos, A.; Wang, G.; Leach, J.E.; Leung, H. Fine-mapping and molecular marker development for Pi56(t), a NBS-LRR gene conferring broad-spectrum resistance to Magnaporthe oryzae in rice. Theor. Appl. Genet. 2013, 126, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, G.; Xie, H.; Ling, Z. Genetic analysis and mapping of rice blast resistance genes induced by cloud in japonica rice. J. Agric. Biotechnol. 2003, 11, 241–244. [Google Scholar]
- Liu, M.; Zhang, S.; Hu, J.; Sun, W.; Padilla, J.; He, Y.; Li, Y.; Yin, Z.; Liu, X.; Wang, W.; et al. Phosphorylation-guarded light-harvesting complex II contributes to broad-spectrum blast resistance in rice. Proc. Natl. Acad. Sci. USA 2019, 116, 17572–17577. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L.; Mackill, D.J.; Bonman, J.M.; Mccouch, S.R.; Champoux, M.C.; Nelson, R.J. RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 1994, 136, 1421–1434. [Google Scholar] [CrossRef] [PubMed]
- Zenbayashi-Sawata, K.; Ashizawa, T.; Koizumi, S. Pi34-AVRPi34: a new gene-for-gene interaction for partial resistance in rice to blast caused by Magnaporthe grisea. J. Gene. Plant Pathol. 2005, 71, 395–401. [Google Scholar] [CrossRef]
- Gowda, M.; Roy-Barman, S.; Chattoo, B.B. Molecular mapping of a novel blast resistance gene Pi38 in rice using SSLP and AFLP markers. Plant Breed. 2006, 125, 596–599. [Google Scholar] [CrossRef]
- Chen, D.H.; Viña, d.; Inukai, M.; Mackill, T.D.J.; Ronald, P.C.; Nelson, R.J. Molecular mapping of the blast resistance gene, Pi44(t), in a line derived from a durably resistant rice cultivar. Theor. Appl. Genet. 1999, 98, 1046–1053. [Google Scholar] [CrossRef]
- Das, A.; Soubam, D.; Singh, P.K.; Thakur, S.; Singh, N.K.; Sharma, T.R. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct. Integr. Genom. 2012, 12, 215–228. [Google Scholar] [CrossRef]
- Huang, H.; Huang, L.; Feng, G.; Wang, S.; Wang, Y.; Liu, J.; Jiang, N.; Yan, W.; Xu, L.; Sun, P.; et al. Molecular mapping of the new blast resistance genes Pi47 and Pi48 in the durably resistant local rice cultivar Xiangzi 3150. Phytopathology 2011, 101, 620–626. [Google Scholar] [CrossRef][Green Version]
- Ashikawa, I.; Hayashi, N.; Yamane, H.; Kanamori, H.; Wu, J.; Matsumoto, T.; Ono, K.; Yano, M. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 2008, 180, 2267–2276. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Yang, Q.; Wang, H.; Guo, T.; Liu, Y.; Zhu, X.; Chen, Z. Identification and fine mapping of a resistance gene to Magnaporthe oryzae in a space-induced rice mutant. Mol. Breed. 2011, 28, 303–312. [Google Scholar] [CrossRef]
- Li, P.; Shi, X.; Wang, J.; Liu, C.; Zhang, H. Molecular mapping of blast resistance genes in japonica rice cultivars Heibaizijing in Taihu. Chin. J. Rice Sci. 2007, 21, 579–584. [Google Scholar]
- Fjellstrom, R.G.; Conaway-Bormans, C.A.; Mcclung, A.M.; Marchetti, M.A.; Park, W.D. Development of DNA markers suitable for marker assisted selection of three pi genes conferring resistance to multiple Pyricularia grisea pathotypes. Crop Sci. 2003, 44, 1790–1798. [Google Scholar] [CrossRef]
- Hua, L.; Wu, J.; Chen, C.; Wu, W.; He, X.; Lin, F.; Wang, L.; Ashikawa, I.; Matsumoto, T.; Wang, L.; et al. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor. Appl. Genet. 2012, 125, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.H.; Mackill, D.J.; Bonman, J.M.; McCouch, S.R.; Guiderdoni, E.; Notteghem, J.L.; Tanksley, S.D. Molecular mapping of genes for resistance to rice blast (Pyricularia grisea Sacc.). Theor. Appl. Genet. 1996, 93, 859–863. [Google Scholar] [CrossRef]
- Cao, W.; Chu, R.; Zhang, Y.; Luo, J.; Su, Y.; Liu, J.; Zhang, H.; Wang, J.; Bao, Y. OsJAMyb, a R2R3-type MYB transcription factor enhanced blast resistance in transgenic rice. Physiol. Mol. Plant Pathol. 2015, 92, 154–160. [Google Scholar] [CrossRef]
- Li, H.; Zhou, S.Y.; Zhao, W.S.; Su, S.C.; Peng, Y.L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009, 69, 337–346. [Google Scholar] [CrossRef]
- Causse, M.A.; Fulton, T.M.; Cho, Y.G.; Ahn, S.N.; Chunwongse, J.; Wu, K.; Xiao, J.; Yu, Z.; Ronald, P.C.; Harrington, S.E. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 1994, 138, 1251–1274. [Google Scholar] [CrossRef]
- Zheng, K.; Qian, H.; Zhuang, J.; Lu, J.; Lin, H. Identification of blast resistance genes in rice by DNA markers. Chin. J. Plant Pathol. 1995, 25, 307–313. [Google Scholar]
- Imbe, T.; Oba, S.; Yanoria, M.J.T.; Tsunematsu, H. A new gene for blast resistance in rice cultivar, IR24. Rice Genet. Newsl. 1997, 14, 60–62. [Google Scholar]
- Hayashi, N.; Ando, I.; Imbe, T. Identification of a new resistance gene to a Chinese blast fungus isolate in the Japanese rice cultivar Aichi Asahi. Phytopathology 1998, 88, 822–887. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mittal, D.; Chakrabarti, S.; Sarkar, A.; Singh, A.; Grover, A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 2009, 47, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liang, X.; Lu, L.; Xu, Z.; Huang, J.; He, H.; Peng, X. Two glyoxylate reductase isoforms are functionally redundant but required under high photorespiration conditions in rice. BMC Plant Biol. 2020, 20, 357. [Google Scholar] [CrossRef]
- Bheemanahalli, R.; Knight, M.; Quinones, C.; Doherty, C.J.; Jagadish, S.V.K. Genome-wide association study and gene network analyses reveal potential candidate genes for high night temperature tolerance in rice. Sci. Rep. 2021, 11, 6747. [Google Scholar] [CrossRef]
- Vitoriano, C.B.; Calixto, C.P.G. Reading between the lines: RNA-seq data mining reveals the alternative message of the rice leaf transcriptome in response to heat stress. Plants (Basel) 2021, 10, 1647. [Google Scholar] [CrossRef]
- Guo, M.; Wang, R.; Wang, J.; Hua, K.; Wang, Y.; Liu, X.; Yao, S. ALT1, a Snf2 family chromatin remodeling ATPase, negatively regulates alkaline tolerance through enhanced defense against oxidative stress in rice. PLoS ONE 2014, 9, e112515. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Wei, C.; Liu, X.; Wang, M.; Yu, F.; Xie, Q.; Tu, J. The ring finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol. 2016, 170, 429–443. [Google Scholar] [CrossRef]
- Singh, A.; Singh, U.; Mittal, D.; Grover, A. Genome-wide analysis of rice ClpB/HSP100, ClpC and ClpD genes. BMC Genet. 2010, 11, 95. [Google Scholar] [CrossRef]
- Zou, J.; Liu, A.; Chen, X.; Zhou, X.; Gao, G.; Wang, W.; Zhang, X. Expression analysis of nine rice heat shock protein genes under abiotic stresses and ABA treatment. J. Plant Physiol. 2009, 166, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, L.; Ou, S.; Wang, R.; Wang, Y.; Chu, C.; Yao, S. Natural variations of SLG1 confer high-temperature tolerance in indica rice. Nat. Commun. 2020, 11, 5441. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 2008, 227, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Dong, G.; Wang, F.; Shi, Y.; Zhu, J.; Zhang, Y.; Ruan, B.; Wu, Y.; Feng, X.; Zhao, C.; et al. A β-ketoacyl carrier protein reductase confers heat tolerance via the regulation of fatty acid biosynthesis and stress signaling in rice. New Phytol. 2021, 232, 655–672. [Google Scholar] [CrossRef]
- Chen, K.; Guo, T.; Li, X.M.; Zhang, Y.M.; Yang, Y.B.; Ye, W.W.; Dong, N.Q.; Shi, C.; Kan, Y.; Xiang, Y.H.; et al. Translational regulation of plant response to high temperature by a dual-function tRNAHis guanylyltransferase in rice. Mol. Plant 2019, 12, 1123–1142. [Google Scholar] [CrossRef]
- Guo, X.; Wu, Y.; Wang, Y.; Chen, Y.; Chu, C. OsMSRA4.1 and OsMSRB1.1, two rice plastidial methionine sulfoxide reductases, are involved in abiotic stress responses. Planta 2009, 230, 227–238. [Google Scholar] [CrossRef]
- Van Breusegem, F.; Dekeyser, R.; Garcia, A.B.; Claes, B.; Gielen, J.; Van Montagu, M.; Caplan, A.B. Heat-inducible rice hsp82 and hsp70 are not always co-regulated. Planta 1994, 193, 57–66. [Google Scholar] [CrossRef]
- Wei, H.; Liu, J.; Wang, Y.; Huang, N.; Zhang, X.; Wang, L.; Zhang, J.; Tu, J.; Zhong, X. A dominant major locus in chromosome 9 of rice (Oryza sativa L.) confers tolerance to 48 °C high temperature at seedling stage. J. Hered. 2013, 104, 287–294. [Google Scholar] [CrossRef]
- Reyes, V.P.; Angeles-Shim, R.B.; Mendioro, M.S.; Manuel, M.C.C.; Lapis, R.S.; Shim, J.; Sunohara, H.; Nishiuchi, S.; Kikuta, M.; Makihara, D.; et al. Marker-assisted introgression and stacking of major QTLs controlling grain number (Gn1a) and number of primary branching (WFP) to NERICA cultivars. Plants (Basel) 2021, 10, 844. [Google Scholar] [CrossRef]
- Fukuoka, S.; Saka, N.; Koga, H.; Ono, K.; Shimizu, T.; Ebana, K.; Hayashi, N.; Takahashi, A.; Hirochika, H.; Okuno, K.; et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 2009, 325, 998–1001. [Google Scholar] [CrossRef]
- Neeraja, C.N.; Maghirang-Rodriguez, R.; Pamplona, A.; Heuer, S.; Collard, B.C.; Septiningsih, E.M.; Vergara, G.; Sanchez, D.; Xu, K.; Ismail, A.M.; et al. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor. Appl. Genet. 2007, 115, 767–776. [Google Scholar] [CrossRef]
- Muthu, V.; Abbai, R.; Nallathambi, J.; Rahman, H.; Ramasamy, S.; Kambale, R.; Thulasinathan, T.; Ayyenar, B.; Muthurajan, R. Pyramiding QTLs controlling tolerance against drought, salinity, and submergence in rice through marker assisted breeding. PLoS ONE 2020, 15, e0227421. [Google Scholar] [CrossRef] [PubMed]
- Ballini, E.; Morel, J.; Droc, G.; Price, A.H.; Courtois, B.; Notteghem, J.; Tharreau, D. A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant Microbe Interact. 2008, 21, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.S.; Presting, G.G.; Barbazuk, W.B.; Goicoechea, J.L.; Blackmon, B.P.; Fang, G.C.; Kim, H.; Frisch, D.; Yu, Y.; Sun, S. An integrated physical and genetic map of the rice genome. Plant Cell. 2002, 14, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Raza, Q.; Riaz, A.; Bashir, K.; Sabar, M. Reproductive tissues-specific meta-QTLs and candidate genes for development of heat-tolerant rice cultivars. Plant Mol. Biol. 2020, 104, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Richa, K.; Tiwari, I.M.; Devanna, B.N.; Botella, J.R.; Sharma, V.; Sharma, T.R. Novel chitinase gene LOC_Os11g47510 from Indica rice Tetep provides enhanced resistance against sheath blight pathogen Rhizoctonia solani in rice. Front. Plant Sci. 2017, 8, 596. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Takenaka, Y.; Nakano, S.; Tamoi, M.; Sakuda, S.; Fukamizo, T. Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: Chitinase inhibitor allosamidin enhances stress tolerance. Biosci. Biotechnol. Biochem. 2009, 73, 1066–1071. [Google Scholar] [CrossRef]
- Mir, Z.A.; Ali, S.; Shivaraj, S.M.; Bhat, J.A.; Singh, A.; Yadav, P.; Rawat, S.; Paplao, P.K.; Grover, A. Genome-wide identification and characterization of chitinase gene family in Brassica juncea and Camelina sativa in response to Alternaria brassicae. Genomics 2020, 112, 749–763. [Google Scholar] [CrossRef]
- Miyamoto, K.; Shimizu, T.; Lin, F.; Sainsbury, F.; Thuenemann, E.; Lomonossoff, G.; Nojiri, H.; Yamane, H.; Okada, K. Identification of an E-box motif responsible for the expression of jasmonic acid-induced chitinase gene OsChia4a in rice. J. Plant Physiol. 2012, 169, 621–627. [Google Scholar] [CrossRef]
- Fu, C.; Wang, F.; Liu, W.; Liu, D.; Li, J.; Zhu, M.; Liao, Y.; Liu, Z.; Huang, H.; Zeng, X.; et al. Transcriptomic analysis reveals new insights into high-temperature-dependent glume-unclosing in an elite rice male sterile line. Front. Plant. Sci. 2017, 8, 112. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jeong, J.S.; Kim, Y.S.; Redillas, M.C.; Jang, G.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013, 11, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Ngernmuen, A.; Suktrakul, W.; Kate-Ngam, S.; Jantasuriyarat, C. Transcriptome comparison of defense responses in the rice variety ’Jao Hom Nin’ regarding two blast resistant genes, pish and pik. Plants (Basel) 2020, 9, 694. [Google Scholar] [CrossRef] [PubMed]
- Richa, K.; Tiwari, I.M.; Kumari, M.; Devanna, B.N.; Sonah, H.; Kumari, A.; Nagar, R.; Sharma, V.; Botella, J.R.; Sharma, T.R. Functional characterization of novel chitinase genes present in the sheath blight resistance QTL: qSBR11-1 in rice line Tetep. Front. Plant Sci. 2016, 7, 244. [Google Scholar] [CrossRef] [PubMed]
- Bouktila, D.; Khalfallah, Y.; Habachi-Houimli, Y.; Mezghani-Khemakhem, M.; Makni, M.; Makni, H. Full-genome identification and characterization of NBS-encoding disease resistance genes in wheat. Mol. Genet. Genom. 2015, 290, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tao, F.; An, F.; Zou, Y.; Tian, W.; Chen, X.; Xu, X.; Hu, X. Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2017, 18, 649–661. [Google Scholar] [CrossRef]
- Wang, J.; Tian, W.; Tao, F.; Wang, J.; Shang, H.; Chen, X.; Xu, X.; Hu, X. TaRPM1 positively regulates wheat high-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. Front. Plant Sci. 2020, 10, 1679. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yang, S.; Yan, Y.; Xiao, Z.; Cheng, J.; Wu, J.; Qiu, A.; Lai, Y.; Mou, S.; Guan, D.; et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 2015, 66, 3163–3174. [Google Scholar] [CrossRef]
- Wang, J.; Tao, F.; Tian, W.; Guo, Z.; Chen, X.; Xu, X.; Shang, H.; Hu, X. The wheat WRKY transcription factors TaWRKY49 and TaWRKY62 confer differential high-temperature seedling-plant resistance to Puccinia striiformis f. sp. tritici. PLoS ONE 2017, 12, e0181963. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Suzuki, N.; Rizhsky, L.; Hegie, A.; Koussevitzky, S.; Mittler, R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 2007, 144, 1777–1785. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Qi, J.; Ren, N.; Cheng, J.; Erb, M.; Mao, B.; Lou, Y. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 2009, 60, 638–648. [Google Scholar] [CrossRef]
- Cheng, A.X.; Xiang, C.Y.; Li, J.X.; Yang, C.Q.; Hu, W.L.; Wang, L.J.; Lou, Y.G.; Chen, X.Y. The rice (E)-beta-caryophyllene synthase (OsTPS3) accounts for the major inducible volatile sesquiterpenes. Phytochemistry 2007, 68, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Ruan, B.; Hua, Z.; Zhao, J.; Zhang, B.; Ren, D.; Liu, C.; Yang, S.; Zhang, A.; Jiang, H.; Yu, H.; et al. OsACL-A2 negatively regulates cell death and disease resistance in rice. Plant Biotechnol. J. 2019, 17, 1344–1356. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, F.; Li, J.; Chen, J.P.; Zhang, H.M. Integrative analysis of the micrornaome and transcriptome illuminates the response of susceptible rice plants to rice stripe virus. PLoS ONE 2016, 11, e0146946. [Google Scholar] [CrossRef]


| Meta-QTL | Flanking Makers | 95% CI (Mb) | Interval Distance (Mb) | Original QTLs | Interval Genes | Published Blast-Resistance Gene |
|---|---|---|---|---|---|---|
| Metab1-1 | RM490-RM8133 | 6.68–9.39 | 2.71 | 3 | 345 | Pi27(t) [65] |
| Metab1-2 | id1013754-RM2318 | 23.73–24.14 | 0.41 | 2 | 54 | |
| Metab1-4 | RM414-RM14 | 40.76–41.36 | 0.61 | 2 | 112 | |
| Metab2-1 | RG634-RM5654 | 2.14–3.42 | 1.28 | 2 | 187 | Pi-Da(t) [66] |
| Metab2-2 | RM492-RM2468 | 7.29–7.42 | 0.14 | 2 | 11 | Pi14 [67], Pi16(t) [68] |
| Metab2-3 | id2006540-RM7426 | 16.20–16.68 | 0.48 | 2 | 34 | |
| Metab2-4 | id2008644-RG25 | 21.60–21.77 | 0.17 | 4 | 10 | Pid1(t) [69] |
| Metab2-5 | RM5470-RM6122 | 27.15–28.44 | 1.28 | 4 | 143 | |
| Metab2-6 | GA285-RM6424 | 29.59–29.63 | 0.04 | 4 | 9 | |
| Metab3-1 | RM1332-RG409 | 2.45–3.50 | 1.04 | 2 | 182 | |
| Metab3-2 | C63-RZ574 | 8.41–10.60 | 2.19 | 4 | 343 | |
| Metab3-3 | RM411-RM487 | 21.43–22.02 | 0.59 | 4 | 52 | |
| Metab3-4 | id3010813-R19 | 24.21–24.60 | 0.39 | 7 | 30 | |
| Metab3-5 | RM3684-RM227 | 34.62–34.93 | 0.31 | 3 | 59 | |
| Metab4-1 | RM16531-RM5953 | 7.98–9.39 | 1.41 | 4 | 62 | |
| Metab4-2 | G271-RM2565 | 20.34–20.93 | 0.59 | 5 | 87 | |
| Metab4-3 | id4008148-RM1388 | 24.36–25.22 | 0.87 | 4 | 138 | |
| Metab4-4 | id4008601-RM241 | 26.12–27.04 | 0.93 | 3 | 138 | |
| Metab4-5 | RG214-RM348 | 31.85–32.84 | 0.99 | 5 | 166 | Pi45(t) [70] |
| Metab5-1 | RG556-S5_658958 | 0.45–0.66 | 0.21 | 3 | 40 | OsMT2b [71] |
| Metab5-2 | id5002075-RM437 | 3.59–3.88 | 0.29 | 4 | 35 | |
| Metab5-3 | RZ649-C624 | 19.61–21.43 | 1.83 | 5 | 218 | Pi10 [72] |
| Metab5-4 | id5009818-id5010176 | 22.44–22.87 | 0.43 | 4 | 70 | |
| Metab5-5 | id5010294-RM1187 | 23.04–23.28 | 0.24 | 7 | 33 | |
| Metab6-1 | G30-C226A | 3.18–3.54 | 0.36 | 2 | 57 | |
| Metab6-2 | RM7561-RM2126 | 4.45–5.91 | 1.45 | 2 | 186 | |
| Metab6-3 | RZ144-RZ667 | 6.72–6.93 | 0.21 | 5 | 32 | Pi8 [73], Pi13(t) [67] |
| Metab6-4 | RM19779-RM527 | 9.32–9.86 | 0.55 | 11 | 31 | Pi40 [74], Pi22 [75] |
| Metab6-5 | RM541-G122 | 19.51–22.57 | 3.06 | 3 | 269 | |
| Metab6-6 | RG778-G329 | 26.24–27.61 | 1.37 | 3 | 174 | |
| Metab6-7 | R2071-RG653 | 28.70–29.03 | 0.33 | 4 | 58 | Pitq1 [76] |
| Metab7-1 | RG528-RM21052 | 1.54–3.77 | 2.23 | 2 | 256 | |
| Metab7-2 | RM21260-RM21327 | 7.27–8.90 | 1.63 | 5 | 121 | |
| Metab7-3 | G20-CDO385 | 17.53–17.82 | 0.29 | 5 | 18 | |
| Metab7-4 | RM3691-RM1048 | 19.23–20.17 | 0.94 | 3 | 104 | |
| Metab7-5 | RM346-RM5847 | 21.05–23.65 | 2.60 | 5 | 357 | |
| Metab7-6 | R1789-C507 | 26.53–26.71 | 0.18 | 7 | 26 | |
| Metab8-1 | id8000544-RM6863 | 1.84–2.01 | 0.17 | 3 | 11 | |
| Metab8-2 | id8000695-rd8002298 | 2.18–2.92 | 0.73 | 3 | 42 | Pi-36 [77] |
| Metab8-3 | RG333-RM5556 | 4.11–4.59 | 0.48 | 9 | 54 | |
| Metab8-4 | RM126-RM6208 | 5.22–5.79 | 0.57 | 10 | 88 | Pi-42(t) [78] |
| Metab8-5 | GA408-RM339 | 16.57–17.95 | 1.38 | 8 | 92 | Pi-11 [79] |
| Metab8-6 | RM342-RM223 | 19.96–20.65 | 0.69 | 4 | 65 | |
| Metab8-7 | id8005717-RM284 | 20.85–21.15 | 0.29 | 5 | 30 | |
| Metab8-8 | RM308-RZ66 | 24.79–25.59 | 0.80 | 3 | 100 | pi-55(t) [80] |
| Metab9-1 | R1687-SFP10098031 | 8.35–10.10 | 1.75 | 6 | 101 | Pi5 [81], Pi-15 [82], pi-56 [83] |
| Metab9-2 | RM105-RM434 | 12.55–15.66 | 3.12 | 6 | 301 | |
| Metab9-3 | RM6570-RM108 | 18.58–19.30 | 0.73 | 8 | 113 | |
| Metab9-4 | RZ12-RG570 | 19.43–19.95 | 0.52 | 9 | 85 | |
| Metab9-5 | CT6-RG662 | 20.17–20.48 | 0.31 | 9 | 54 | |
| Metab9-6 | RM1553-C506 | 21.00–21.86 | 0.85 | 11 | 140 | |
| Metab10-1 | RM2125-G1125 | 4.89–7.34 | 2.44 | 6 | 127 | |
| Metab10-2 | RM25149-G1084 | 7.57–10.64 | 3.07 | 2 | 155 | |
| Metab11-1 | RZ638-RM5599 | 2.52–3.83 | 1.30 | 5 | 147 | |
| Metab11-2 | aa11002340-RM536 | 7.26–8.99 | 1.74 | 4 | 125 | Pi-y(t) [84], LHCB5 [85] |
| Metab11-3 | RM6680-RG103 | 19.08–20.80 | 1.72 | 7 | 181 | Pi-7 [86], Pi-34 [87], Pi-38 [88] |
| Metab11-4 | RG1109-RM7277 | 23.62–24.68 | 1.06 | 3 | 82 | Pi-44(t) [89] |
| Metab11-5 | RM27154-RM4069 | 25.23–26.67 | 1.44 | 3 | 123 | Pi54 [90], Pi-43(t) [78], Pi-47(t) [91] |
| Metab11-6 | RM7654-L190 | 27.67–28.76 | 1.09 | 6 | 91 | Pik-m [92], Pi-46(t) [93] Pi-hk1(t) [94], Pi-k [95], Pi-1 [96], Pi-18 [97], Pi-lm2 [76], OsJAMyb [98], OsWAK1 [99] |
| Metab12-1 | G1112-RM6288 | 1.27–2.20 | 0.92 | 4 | 147 | |
| Metab12-2 | RM3455-R3375 | 4.92–5.61 | 0.69 | 5 | 49 | |
| Metab12-3 | G1391-RM7003 | 5.81–6.78 | 0.97 | 2 | 44 | Pi-6 [100], Pi-h-1(t) [101], Pi-tq6 [76] |
| Metab12-4 | id12003144-id12003547 | 7.92–8.82 | 0.90 | 2 | 41 | Pi-20 [102], Pi-21(t) [97], Pi-157(t) [72] |
| Metab12-5 | id12003728-id12003803 | 9.18–9.54 | 0.36 | 2 | 27 | |
| Metab12-6 | C625-id12004303 | 11.06–11.22 | 0.16 | 24 | 11 | |
| Metab12-7 | RM27982-id12004685 | 12.63–13.06 | 0.43 | 5 | 20 | Pi-ta2 [95], Pi-19 [103], Pi-48(t) [91] |
| Metab12-8 | RM3331-C1069 | 23.49–25.08 | 1.59 | 4 | 161 |
| Meta-QTL | Flanking Markers | 95% CI (Mb) | Map Distance (Mb) | Original QTLs | Interval Gene | Published Heat-Tolerance Gene |
|---|---|---|---|---|---|---|
| Metah1-1 | R2159-RM1232 | 21.70–27.63 | 5.93 | 2 | 671 | OsHsfA7 [104]; OsHsfC1a [104]; OsWRKY11 [105]; OsGR2 [106]; OsRb1 [107]; OsTRBF1 [108]; OsDfr [109]; OsUBC [110] |
| Metah1-2 | RM6581-RM297 | 31.50–32.10 | 0.60 | 2 | 101 | Osbht [17] |
| Metah2-2 | R1989-RM3419 | 16.10–19.34 | 3.24 | 4 | 253 | OsHsfA5 [104]; OsClpD1 [111]; OsHsfA3 [104] |
| Metah2-3 | RM221-RG256 | 27.61–33.94 | 6.33 | 4 | 887 | OsHSP24.1 [112]; RCTU1 [113] |
| Metah3-1 | RM3372-RM22 | 1.46–1.52 | 0.06 | 2 | 13 | |
| Metah3-2 | RM7365-RM338 | 11.28–13.22 | 1.94 | 2 | 237 | |
| Metah3-3 | RM15721-RM15759 | 27.70–28.31 | 0.61 | 2 | 84 | |
| Metah3-4 | RM1352-RM143 | 32.35–33.19 | 0.84 | 2 | 149 | OsHsfA2e [114] |
| Metah4-1 | RM16424-RM8213 | 4.30–4.44 | 0.14 | 3 | 15 | |
| Metah4-2 | RG449-RM185 | 17.87–18.58 | 0.71 | 4 | 59 | HTS1 [115]; eIF3h [116] |
| Metah4-3 | G235-RM348 | 31.47–32.65 | 1.18 | 2 | 191 | |
| Metah4-4 | RM2799-RM2275 | 34.14–34.32 | 0.18 | 2 | 31 | |
| Metah5-1 | RM153-RZ556 | 0.19–2.09 | 1.90 | 3 | 264 | |
| Metah5-2 | RM1366-R1838 | 2.92–3.31 | 0.39 | 4 | 48 | |
| Metah6-1 | RM4332-RM190 | 0.72–1.76 | 1.04 | 5 | 155 | |
| Metah6-2 | RM8112-RM584 | 2.17–3.42 | 1.24 | 2 | 207 | |
| Metah6-3 | RM2615-RM4128 | 5.96–6.64 | 0.69 | 2 | 73 | |
| Metah6-4 | RM3183-RM20155 | 12.45–19.61 | 7.17 | 3 | 392 | OsMSRB1.1 [117] |
| Metah7-1 | RM192-RM3831 | 0.26–1.16 | 0.91 | 4 | 132 | |
| Metah7-2 | RM21320-C39 | 8.78–11.36 | 2.58 | 3 | 131 | |
| Metah7-3 | RZ978-RM7601 | 28.41–29.04 | 0.63 | 3 | 97 | |
| Metah8-1 | RM8018-RM6999 | 2.17–3.98 | 1.82 | 5 | 177 | |
| Metah8-2 | RM547-RM6838 | 5.59–5.85 | 0.26 | 7 | 33 | |
| Metah8-3 | RM256-RZ66 | 24.27–25.67 | 1.40 | 3 | 180 | hsp82A [118] |
| Metah9-1 | RM5526-RM7364 | 7.31–9.56 | 2.25 | 4 | 93 | OsHTAS [119] |
| Metah9-2 | RM410-R2638 | 17.64–17.84 | 0.19 | 2 | 24 | |
| Metah9-3 | RM6570-RM553 | 18.58–19.32 | 0.75 | 4 | 116 | OsHSP58.7 [112] |
| Metah9-4 | OSR28-RM107 | 19.79–20.07 | 0.28 | 4 | 43 | |
| Metah10-1 | RM1126-RM25228 | 9.70–9.95 | 0.25 | 4 | 12 | |
| Metah10-2 | RM5620-RM5373 | 17.40–18.73 | 1.32 | 3 | 161 | |
| Metah10-3 | RM6132-RM6100 | 18.79–18.82 | 0.03 | 3 | 6 | |
| Metah10-4 | RM2371-C488 | 19.58–19.96 | 0.38 | 6 | 53 | |
| Metah10-5 | RM1374-RM228 | 21.57–22.24 | 0.67 | 6 | 119 | |
| Metah11-1 | R77-R642 | 0.20–2.02 | 1.82 | 3 | 290 | |
| Metah11-2 | C1350-RM5704 | 3.81–5.48 | 1.66 | 5 | 160 | |
| Metah11-3 | RM287-RM5349 | 16.77–19.18 | 2.42 | 2 | 166 | |
| Metah11-4 | RM27234-RM6293 | 26.10–28.26 | 2.17 | 4 | 237 | |
| Metah12-1 | RM3483-RM6296 | 1.61–3.20 | 1.59 | 5 | 212 | |
| Metah12-2 | RM27877-RM270 | 9.18–10.60 | 1.42 | 4 | 81 | |
| Metah12-4 | RM4585-R496 | 26.13–26.86 | 0.73 | 2 | 72 |
| RAP-ID | Gene Symbol | Locus Name | Function Annotation | Log2foldchange | |
|---|---|---|---|---|---|
| CO | LT | ||||
| Os11g0700900 | C10923 | LOC_Os11g47500.1 | Glycosyl hydrolase, putative, expressed | 6.62 | 6.98 |
| Os08g0508800 | OsHI-LOX | LOC_Os08g39840.1 | Lipoxygenase, chloroplast precursor, putative, expressed | 6.08 | 5.58 |
| Os11g0702100 | chitinases | LOC_Os11g47600.1 | Glycosyl hydrolase, putative, expressed | 5.47 | 6.35 |
| Os11g0701500 | LOC_Os11g47560.1 | Glycosyl hydrolase, putative, expressed | 5.43 | 3.30 | |
| Os11g0684000 | OsJAMyb | LOC_Os11g45740.1 | MYB family transcription factor, putative, expressed | 5.39 | 4.98 |
| Os08g0509100 | OsLOX8 | LOC_Os08g39850.1 | Lipoxygenase, chloroplast precursor, putative, expressed | 4.21 | 3.53 |
| Os11g0701400 | C10150 | LOC_Os11g47550.1 | Glycosyl hydrolase, putative, expressed | 4.17 | 2.71 |
| Os08g0141400 | OsNDB3 | LOC_Os08g04630.1 | External NADH-ubiquinone oxidoreductase 1, mitochondrial precursor, putative, expressed | 4.01 | 4.18 |
| Os11g0660500 | OsTCTP | LOC_Os11g43900.1 | Translationally-controlled tumor protein, putative, expressed | 3.80 | 2.88 |
| Os11g0701000 | OsChib3H-c | LOC_Os11g47510.1 | Glycosyl hydrolase, putative, expressed | 3.25 | 3.36 |
| Os11g0691500 | WAK3 | LOC_Os11g46900.1 | Wall-associated receptor kinase 3 precursor, putative, expressed | 3.01 | 3.80 |
| Os09g0321900 | OsUBC6 | LOC_Os09g15320.2 | Ubiquitin-conjugating enzyme, putative, expressed | 2.44 | 2.62 |
| Os11g0691100 | OsiWAK1 | LOC_Os11g46860.1 | Wall-associated receptor kinase-like 4 precursor, putative, expressed | 2.29 | 3.02 |
| Os11g0689100 | Pi-k | LOC_Os11g46210.1 | NB-ARC domain containing protein, expressed | 2.28 | 2.65 |
| Os12g0266200 | WAK124 | LOC_Os12g16540.1 | OsWAK124-OsWAK receptor-like protein OsWAK-RLP, expressed | 2.11 | 4.28 |
| Os11g0700500 | OsMYBAS1 | LOC_Os11g47460.1 | MYB family transcription factor, putative, expressed | 2.09 | 2.39 |
| Os04g0631800 | SDK6 | LOC_Os04g53994.1 | Kinase, putative, expressed | 2.08 | 3.69 |
| Os11g0684100 | OsWRKY59 | LOC_Os11g45750.2 | WRKY protein, expressed | −2.04 | −2.21 |
| Os06g0165500 | OsRLCK198 | LOC_Os06g06960.1 | S-locus-like receptor protein kinase, putative, expressed | −2.06 | −2.41 |
| Os11g0693800 | ACLA3 | LOC_Os11g47120.1 | DEFL48-Defensin and Defensin-like DEFL family, expressed | −2.24 | −3.01 |
| Os11g0696200 | EDT1; OsACLA-2 | LOC_Os11g47330.1 | ATP-grasp domain containing protein, expressed | −2.32 | −2.87 |
| Os08g0139700 | OsTPS3 | LOC_Os08g04500.1 | Terpene synthase, putative, expressed | −2.51 | −5.75 |
| Os05g0111300 | OsMT2b | LOC_Os05g02070.2 | Metallothionein, expressed | −4.32 | −2.22 |
| Os12g0263000 | OsGS2 | LOC_Os12g16200.1 | Glutathione synthetase, chloroplast precursor, putative, expressed | -4.96 | -3.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, T.; Chen, L.; Ai, Y.; He, H. Selection of Candidate Genes Conferring Blast Resistance and Heat Tolerance in Rice through Integration of Meta-QTLs and RNA-Seq. Genes 2022, 13, 224. https://doi.org/10.3390/genes13020224
Tian T, Chen L, Ai Y, He H. Selection of Candidate Genes Conferring Blast Resistance and Heat Tolerance in Rice through Integration of Meta-QTLs and RNA-Seq. Genes. 2022; 13(2):224. https://doi.org/10.3390/genes13020224
Chicago/Turabian StyleTian, Tian, Lijuan Chen, Yufang Ai, and Huaqin He. 2022. "Selection of Candidate Genes Conferring Blast Resistance and Heat Tolerance in Rice through Integration of Meta-QTLs and RNA-Seq" Genes 13, no. 2: 224. https://doi.org/10.3390/genes13020224
APA StyleTian, T., Chen, L., Ai, Y., & He, H. (2022). Selection of Candidate Genes Conferring Blast Resistance and Heat Tolerance in Rice through Integration of Meta-QTLs and RNA-Seq. Genes, 13(2), 224. https://doi.org/10.3390/genes13020224






