Abstract
MicroRNA (miRNA) plays essential roles in post-transcriptional regulation of protein coding genes, and the quantitative real-time polymerase chain reaction (qRT-PCR) is the powerful and broadly employed tool to conduct studies of miRNA expression. Identifying appropriate references to normalize quantitative data is a prerequisite to ensure the qRT-PCR accuracy. Until now, there has been no report about miRNA reference for qRT-PCR in Japanese flounder (Paralichthys olivaceus), one important marine cultured fish along the coast of Northern Asia. In this study, combined with miRNA-Seq analysis and literature search, 10 candidates (miR-34a-5p, miR-205-5p, miR-101a-3p, miR-22-3p, miR-23a-3p, miR-210-5p, miR-30c-5p, U6, 5S rRNA, and 18S rRNA) were chosen as potential references to test their expression stability among P. olivaceus tissues, and in livers of P. olivaceus infected with Edwardsiella tarda at different time points. The expression stability of these candidates was analyzed by qRT-PCR and evaluated with Delta CT, BestKeeper, geNorm, as well as NormFinder methods, and RefFinder was employed to estimate the comprehensive ranking according to the four methods. As the result, miR-22-3p and miR-23a-3p were proved to be the suitable combination as reference miRNAs for both P. olivaceus normal tissues and livers infected with E. tarda, and they were successfully applied to normalize miR-7a and miR-221-5p expression in P. olivaceus livers in response to E. tarda infection. All these results provide valuable information for P. olivaceus miRNA quantitative expression analysis in the future.
1. Introduction
MicroRNA (miRNA) is the small non-coding RNA (ncRNA) with a length of about 22–24 nucleotide (nt), which exists broadly in diverse organisms and plays important regulatory roles in gene expression at the post-transcription level [1]. Mature miRNA pairs completely or incompletely with mRNA 3’ un-translated region (UTR) through the 2–7 nt seed region to degrade mRNA or inhibit translation [2]. Since miRNA was first found in Caenorhabditis elegans, an increasing number of studies have reported that miRNA could regulate various biological processes, such as cell proliferation and differentiation [3,4], embryonic development [5,6], immunology [7], reproduction [8], as well as stress responses [9], in many eukaryotes.
To explore the function of miRNA, its expression pattern commonly needs to be verified. There are several methods to quantify miRNA expression, including northern blot, microarray, and quantitative real-time PCR (qRT-PCR) [10,11,12]. qRT-PCR is one of the most widely used technique to quantify miRNA expression due to several advantages, such as good accuracy, high sensitivity, quick reaction, broad application, as well as low cost [13]. However, differences in RNA quality and reverse transcription efficiency may lead to inaccuracy in qRT-PCR. Therefore, it is essential to employ an appropriate reference to normalize the expression of the target RNA molecules [14].
Endogenous reference of the same RNA class can better normalize the expression level of targeting RNA [15]. Recently, small ncRNA families, including the small nuclear RNA (snRNA) or the small nucleolar RNA (snoRNA), as well as the ribosomal RNA (rRNA), are the most frequently used references for miRNA quantification in vertebrates [16], such as small nuclear RNA U6 (~107 nt) and 5S rRNA (~121 nt). However, it is vital that the references employed for normalization should generally have a similar length, structure, as well as the same nature of the interest targets to assure accurate results, including the comparable efficiency in RNA isolation, cDNA synthesis, and quantification in qRT-PCR [17]. Moreover, recently, there were studies reported that U6 and 5S rRNA displayed less stable than some miRNAs across different tissues and experimental conditions, indicating the inadvisable to use U6 or 5S rRNA as the reference without evaluation [18].
However, at present, the application of evaluated miRNA as qRT-PCR reference has not been commonly adopted, although it is essential that the references selected need to have the similar nature as the studied molecules. In this manner, limited studies have verified the expression stability of certain miRNAs in model organisms [13,15,18,19], as well as in teleost species. For example, in common carp (Cyprinus carpio. var), when considering different tissues and developmental stages, let-7a and miR-23a were proved to be the most stable miRNAs [16]; in Atlantic salmon (Salmo salar), miR-25-3p and miR-455-5p were the most suitable combination as references among tissues and with infections [20]; miR-101a was verified to be the stable miRNA among different tissues and developmental stages of grass carp (Ctenopharyngodon idella) [21]; miR-22 and miR-23a were the most suited candidates for various tissues and developmental stages in Chinese perch (Siniperca chuatsi) [22]. Moreover, in Atlantic cod (Gadus morhua), miR-17-1-5p, miR-25-3p, and miR-210-5p were the best combinations for different tissues [23], and in blunt snout bream (Megalobrama amblycephala), miR-221-3p and miR-103-3p were the best combination for different tissues and various stresses [24].
Japanese flounder (Paralichthys olivaceus) is one of the most widely cultured marine fish along the coast of Northern China as well as in Japan and Korea. Until now, most studies about the expression of Japanese flounder protein-coding genes with qRT-PCR usually use β-actin and ubiquitin-conjugating enzyme (UBCE) as references [25,26], whereas studies about miRNA expression commonly use U6 as reference [27], with no report about the evaluation of miRNA reference. Therefore, the goal of this study was to systematically choose and evaluate suitable references for normalizing miRNA qRT-PCR among Japanese flounder normal tissues as well as in livers with bacterial infection. We first selected 10 candidate references according to miRNA-Seq data and a literature survey, and then identified the most suitable reference miRNAs across adult tissues and bacterial infectious conditions with qRT-PCR. Our findings suggest using appropriate reference miRNAs, which will be beneficial in qRT-PCR studies of miRNA expression in Japanese flounder under normal and infectious conditions.
2. Materials and Methods
2.1. Sample Collection
Nine tissue samples (heart, liver, spleen, kidney, brain, gill, intestine, ovary and testis) were collected from three 2-years old normal Japanese flounder, with ovaries and testes collected from six 2-years old Japanese flounder. Bacterial challenge experiments were described previously by Liu et al. [27]. Briefly, Japanese flounder individuals were collected and acclimated at 19 °C aerated seawater (pH 7.5, salinity 30 ppt) for one week, and were classified into three experimental groups, as the blank control (BC) group, Ringer’s solution (RS) group, and Edwardsiella tarda-challenged (EC) group. The BC group was not treated with any solution, while RS and EC groups were intraperitoneally injected with 1 mL Ringer’s solution or E. tarda suspension (2 × 107 CFU/mL), respectively. After 3 and 24 h post injection, the livers from EC and RS groups were sampled. Three individuals from each time point of the BC, RS and EC groups were sampled. A final 15 individuals were collected, and about 3 g of liver was sampled and stored at −80 °C for RNA isolation later.
2.2. Candidate miRNA References Selection
The candidate miRNA references for Japanese flounder were selected through several ways.
2.2.1. miRNA-Seq for Normal Tissues and Bacterial Infected Livers of Japanese Flounder
We previously generated miRNA-Seq data for 11 adult tissues (brain, kidney, gill, heart, liver, spleen, stomach, muscle, intestine, testis, and ovary) of Japanese flounder, as well as miRNA-Seq data for 15 liver tissues (triplicate for BC-0h, RS-3h, RS-24h, EC-3h, and EC-24h) [27]. For each tissue sample, only miRNAs with the expression level higher than five transcripts per million (TPM) were included for the expression stability analyses due to the difficulty to detect and quantify low-expressing miRNAs with qRT-PCR. Two methods were employed to analyze the miRNA expression stability: the fold change (FC) comparison [28] and the coefficient of variation (CV) evaluation [29].
For the FC method, log2FC was calculated in pairwise samples to compare the differential expression [30]. MiRNAs with a high log2FC (≥1 among 11 tissue types and ≥1.2 in differentially treated livers) were excluded from the list of stably expressed miRNAs (Supplementary Table S1). For the CV method, the mean expression (mean), standard deviation (SD), as well as the CV of each miRNA were calculated. Mean and SD were evaluated among the 11 normal tissues or differentially treated livers with three biological replicates, respectively. CV was then represented as SD/mean.
2.2.2. Published miRNA References for Teleost Species
A few conserved miRNAs have been verified as qRT-PCR references in the studies of teleost species (Table S2). For example, miR-101a for different tissues and developmental stages in grass carp (C. idella) [21]; miR-22 and miR-23a for different tissues and developmental stages in Chinese perch (S. chuatsi) [22]; miR-210-5p for different tissues in Atlantic cod (G. morhua) [23]; miR-23a-5p for different tissues and various stress in Atlantic salmon (S. salar) [31]; and miR-23a for different tissues and developmental stages in common carp (C. carpio. var) [16]. Some miRNAs from the studies above (Supplementary Table S2) were differentially expressed among Japanese flounder tissues, including let-7a, miR-25-3p, miR-99-5p, and miR-17, and they were excluded from further analysis.
2.2.3. Commonly Used miRNA References for qRT-PCR: U6, 5S rRNA, and 18S rRNA
Furthermore, 5S rRNA, 18S rRNA, as well as U6 were also included to be evaluated as references due to their common usage in miRNA qRT-PCR. In summary, miR-34a-5p, miR-205-5p, miR-101a-3p, miR-22-3p, miR-23a-3p, miR-210-5p, miR30c-5p, U6, 5S rRNA, and 18S rRNA were finally chosen to evaluate their expression stability in the following analysis.
2.3. RNA Extraction and miRNA cDNA Synthesis
Total RNA was first isolated with TRIzolTM reagents (Invitrogen, Carlsbad, CA, USA). After RNA isolation and purification, NanoPhotometer Pearl (Implen, Munich, Germany) and 1.5% agarose gel electrophoresis were employed to measure the concentration and integrity of the RNA. MiRNA cDNA was synthesized by the Mir-XTM miRNA First-Strand Synthesis Kit (clontech, Mountain View, CA, USA) with the reaction volume of 10 μL. After the reaction, 90 μL DEPC treated H2O was added to make the final concentration of miRNA cDNA 10 ng/μL, and then stored in −20 °C.
2.4. qRT-PCR Analysis
The expression of the selected candidate references was quantified by qRT-PCR. The forward primers of miR-34a-5p, miR-205-5p, miR-101a-3p, miR-22-3p, miR-23a-3p, miR-210-5p, and miR30c-5p were the same as their mature miRNA sequences (replace U with T, Table 1). The reverse miRNA primer was the mRQ 3′ primer from the Mir-XTM miRNA First-Strand Synthesis Kit (clontech, Mountain View, CA, USA). Primers for 5S rRNA, 18S rRNA, and U6 were retrieved from studies related to miRNA expression [24,32,33]. qRT-PCR was carried out in 384-well plates with 2 × SYBR Green qPCR Master Mix (US Everbright® Inc., Suzhou, Jiangsu, China) using the LightCycler® 480 real time PCR system (Roche Molecular Biochemical, Mannheim, Germany). Each sample was conducted with three biological replicates and each biological replicate with three technical repetitions. Standard curves were established from 10-fold serial dilutions with the pool of miRNA cDNA of all samples. The total volume of each reaction system was 20 μL, including 2 μL miRNA cDNA (5 ng/μL), 10 μL LightCycler® 480 SYBR Green I Master, 0.4 μL of each primer (10 μM), and 7.2 μL DEPC treated H2O. The reaction program was 95 °C 5 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 20 s.

Table 1.
Primer sequences and amplification efficiency of the candidate references.
2.5. Statistical Analysis
The melting curves and cycle threshold (Ct) values (Supplementary Table S3) of qRT-PCR were exported from the LightCycler® 480 real time PCR system software, and the performance of each candidate reference was evaluated according to the Delta CT [34], BestKeeper [35], NormFinder [36], and geNorm [37] algorithm. RefFinder (https://www.heartcure.com.au/reffinder/, accessed on 18 March 2021) was also employed to rank the comprehensive stability of the candidate references [38].
2.6. Reference miRNA Validation
To confirm the reliability of the potential reference genes, miR-7a and miR-221-5p were selected from our previous miRNA-Seq study [27], because they exhibited upregulated expression after E. tarda infection in Japanese flounder liver. The expression of miRNAs was normalized with both the most and the least stable references in E. tarda-infected Japanese flounder livers. The conditions of qRT-PCR were the same as those described above. The relative expression of both miRNAs was computed based on the 2−ΔCt method.
3. Results
3.1. Candidate Reference Selection for Japanese Flounder miRNA Quantification
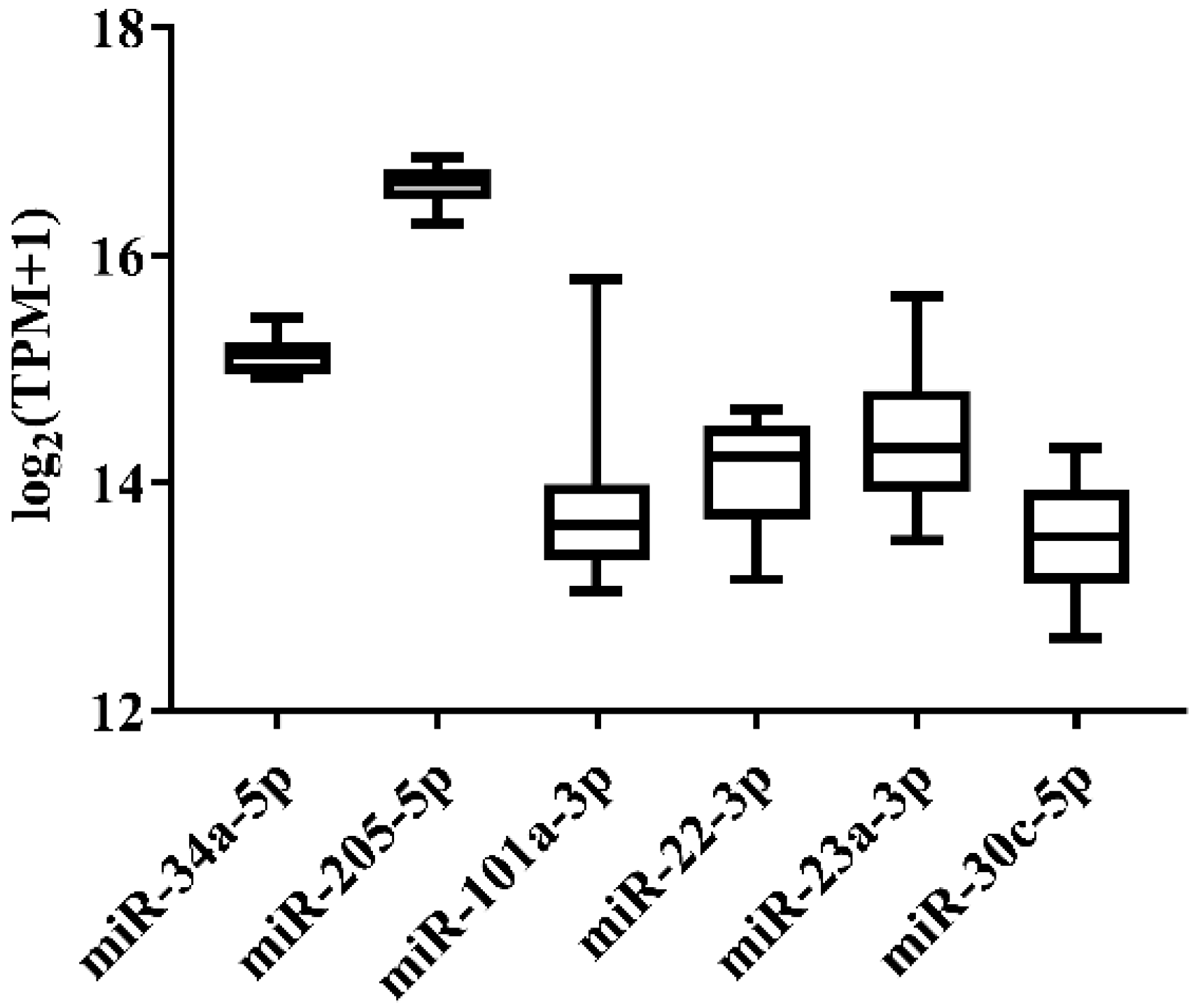
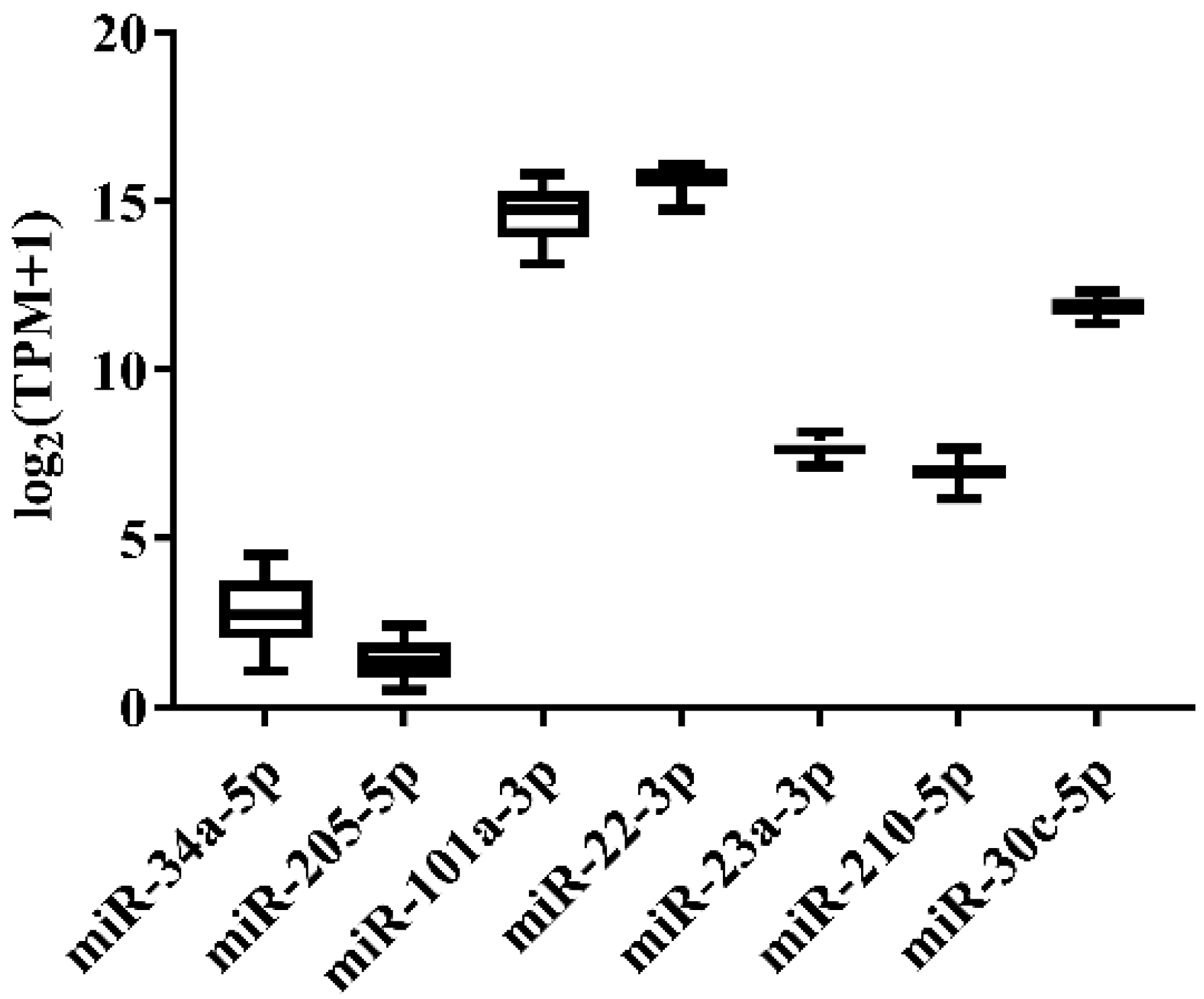
In order to select the candidate qRT-PCR reference miRNA, two methods (log2FC and CV) were first performed to evaluate miRNA expression variation across miRNA-Seq data of 11 Japanese flounder normal tissues and livers infected with Ringer’s solution or E. tarda. Two criteria were employed: (a) exclusion of differentially expressed miRNAs through the log2FC pairwise comparison of tissues or experimental groups (Supplementary Table S1); (b) inclusion of miRNA with the lowest overall CV across all tissues or experimental groups (Supplementary Table S1). For Japanese flounder normal tissues, subject to log2FC < 1, the CV values were ranked, and miR-34a-5p (CV = 0.1230) and miR-205-5p (CV = 0.1278) with the lowest CV were selected as the candidate references (Figure 1 and Supplementary Table S1). For Japanese flounder livers infected with E. tarda or Ringer’s solution at 3 h and 24 h, subject to log2FC < 1.2, miR-202-3p (CV = 0.1093), miR-30c-5p (CV = 0.1659), miR-203a-5p (CV = 0.1702), and miR-23a-3p (CV = 0.1817) were the most stably expressed among the experimental groups (Figure 2 and Supplementary Table S1). However, the expression levels of miR-202-3p and miR-203a-5p among the infection experimental groups were lower than one TPM (Supplementary Table S1), and thus, they were not suitable as references and were excluded from the further analysis. Therefore, miR-30c-5p and miR-23a-3p were included as the candidates.
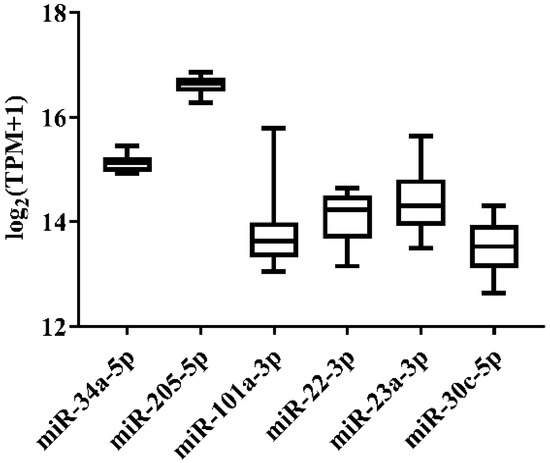
Figure 1.
Expression variation of miRNA reference candidates among 11 normal tissues of Japanese flounder from miRNA-Seq data.
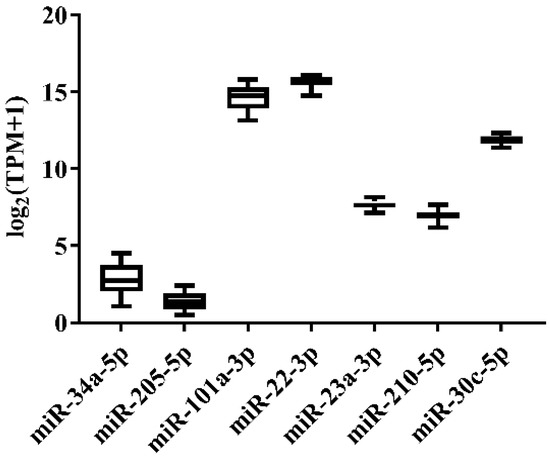
Figure 2.
Expression variation of miRNA reference candidates in the livers of Japanese flounder injected with E. tarda or Ringer’s solution from miRNA-Seq data.
According to the survey of published miRNA references in teleost species (Supplementary Table S2), such as in Atlantic salmon (S. salar) [20], grass carp (C. idella) [21], Chinese perch (S. chuatsi) [22], common carp (C. carpio. var) [16], and Atlantic cod (G. morhua) [23], miR-101a-3p, miR-22-3p, miR-23a-3p, miR-210-5p, and 18S rRNA represented relatively high expression stability among Japanese flounder tissues, and they showed conserved sequence identity among teleost species as well. 5S rRNA and U6 were also included, as they were commonly used as references in studies about miRNA expression [27,33]. In summary, a total of 10 reference candidates were selected, including miR-34a-5p, miR-205-5p, miR-101a-3p, miR-22-3p, miR-23a-3p, miR-210-5p, miR-30c-5p, U6, 5S rRNA, and 18S rRNA, in Japanese flounder for the following qRT-PCR analysis. The primer sequences of these candidate references are shown in Table 1.
The reverse primer employed for miRNA quantification was universal primer (mRQ 3′ Primer, Takara).
3.2. Amplification Efficiency of the Candidate References by qRT-PCR
Amplification efficiency of the 10 selected candidates was analyzed by the standard curve method, with 10-fold serial dilutions of a pool for all samples (101 ng/μL, 100 ng/μL, 10−1 ng/μL, 10−2 ng/μL, and 10−3 ng/μL) as templates to perform qRT-PCR, respectively. As a result, the amplification efficiency of the candidate references in Japanese flounder was from 83% to 204% (Table 1), and miR-205-5p, miR-101a-3p, miR-22-3p, and miR-23a-3p generally complied with the normal requirements of about 90–110%.
3.3. Expression Stability of the Candidate References among Normal Tissues of Japanese Flounder by qRT-PCR
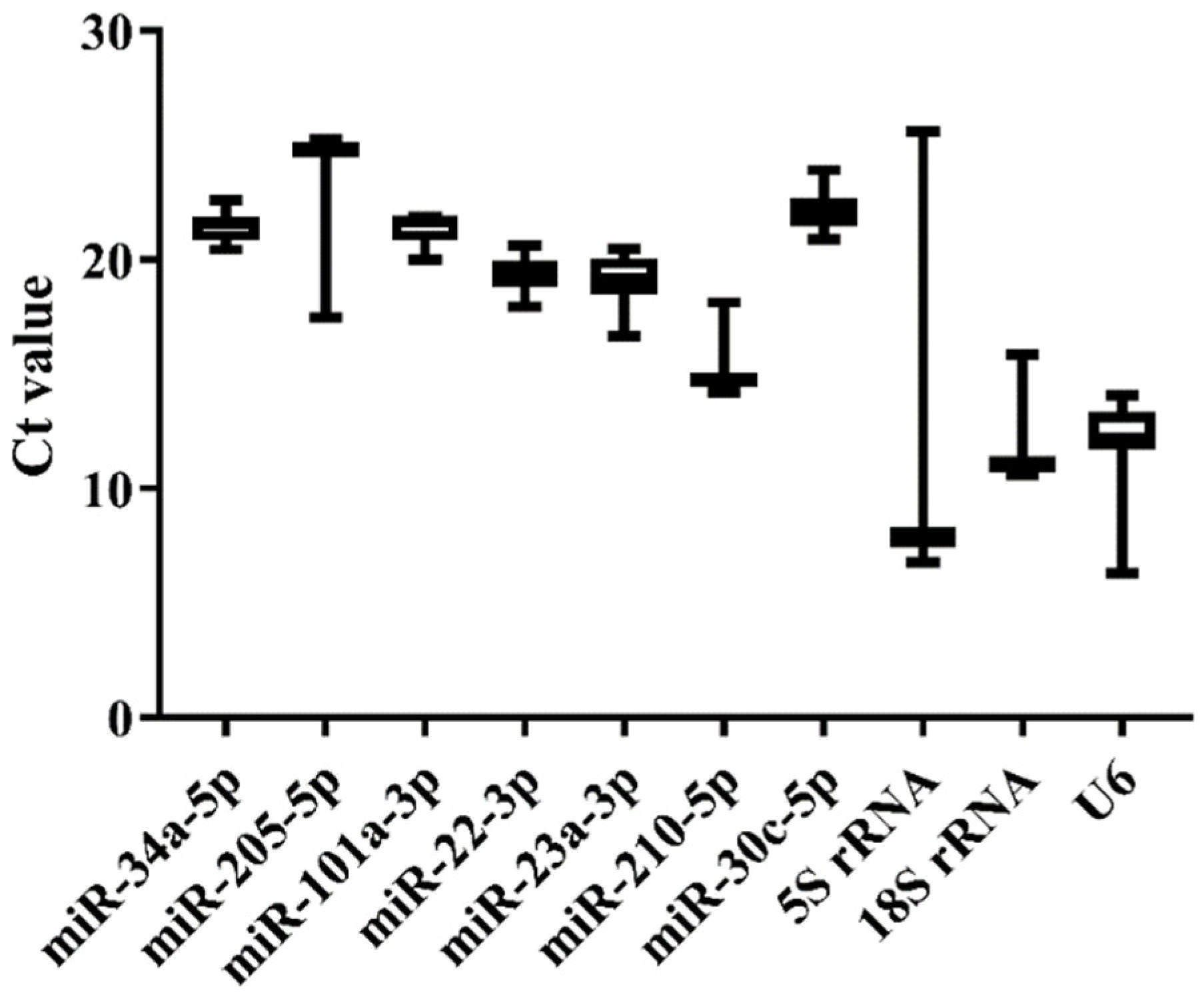
We selected nine tissues (heart, liver, spleen, brain, kidney, gill, intestine, testis, and ovary) of normal Japanese flounder to estimate the expression stability of the 10 candidate references by qRT-PCR. The melting curves of the candidates from qRT-PCR indicated that miR-34a-5p, miR-205-5p, miR-22-3p, miR-23a-3p, and 5S rRNA generally represented a single peak with specific product among tissues (Supplementary Figure S1). The Ct values from qRT-PCR also varied among the 10 candidate references (Figure 3 and Supplementary Table S3). The commonly used genes such as 5S rRNA, 18S rRNA, and U6 showed higher expression level (low Ct value) and lower stability (high Ct value variation) than other miRNA candidates, among which 5S rRNA displayed the highest variation of Ct values (Figure 3). Moreover, miR-34a-5p, miR-101a-3p, miR-22-3p, miR-23a-3p, and miR-30c-5p presented lower variation of Ct values, indicating their higher expression stability (Figure 3). Among these candidates with high stability in Ct values, miR-22-3p and miR-23a-3p had lower Ct values, indicating their higher expression level.
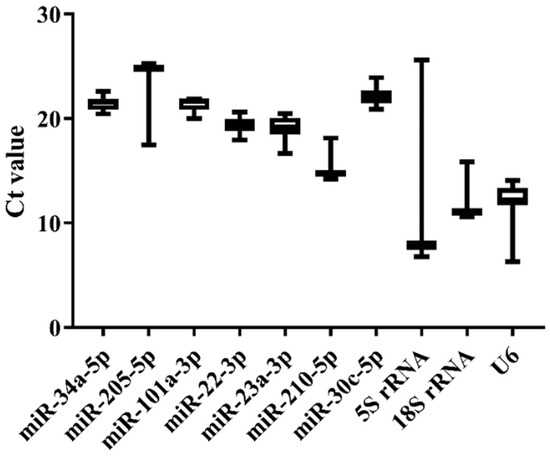
Figure 3.
Ct value variability for the candidate references in normal tissues of Japanese flounder by qRT-PCR.
Furthermore, Delta CT, BestKeeper, NormFinder, and geNorm were employed to calculate and sort the expression stability of candidate references (Table 2). As a result, miR-22-3p, miR-23a-3p, and miR-101a-3p were the most stable miRNAs by Delta CT, NormFinder, and geNorm methods, while miR-34a-5p was the best from BestKeeper. 5S rRNA and U6 were the least stable candidates among Japanese flounder tissues with the lowest ranking values. For the geNorm stability value (M), all candidate references were less than 1.5 except 5S rRNA and U6 (Table 3), as the most stable reference should have the lowest M value. According to the comprehensive ranking of RefFinder and M value of geNorm, miR-22-3p and miR-23a-3p were recommended as the most suitable combined references for miRNA quantification among normal Japanese flounder tissues (Table 2 and Table 3).

Table 2.
Expression stability ranking of the candidate references for normal tissues of Japanese flounder. The best performers, miR-22-3p and miR-23a-3p, are shown in bold.

Table 3.
Expression stability values (M) calculated by geNorm for normal tissues of Japanese flounder.
3.4. Expression Stability of Candidate References in Japanese Flounder Livers Injected with E. tarda or Ringer’s Solution by qRT-PCR
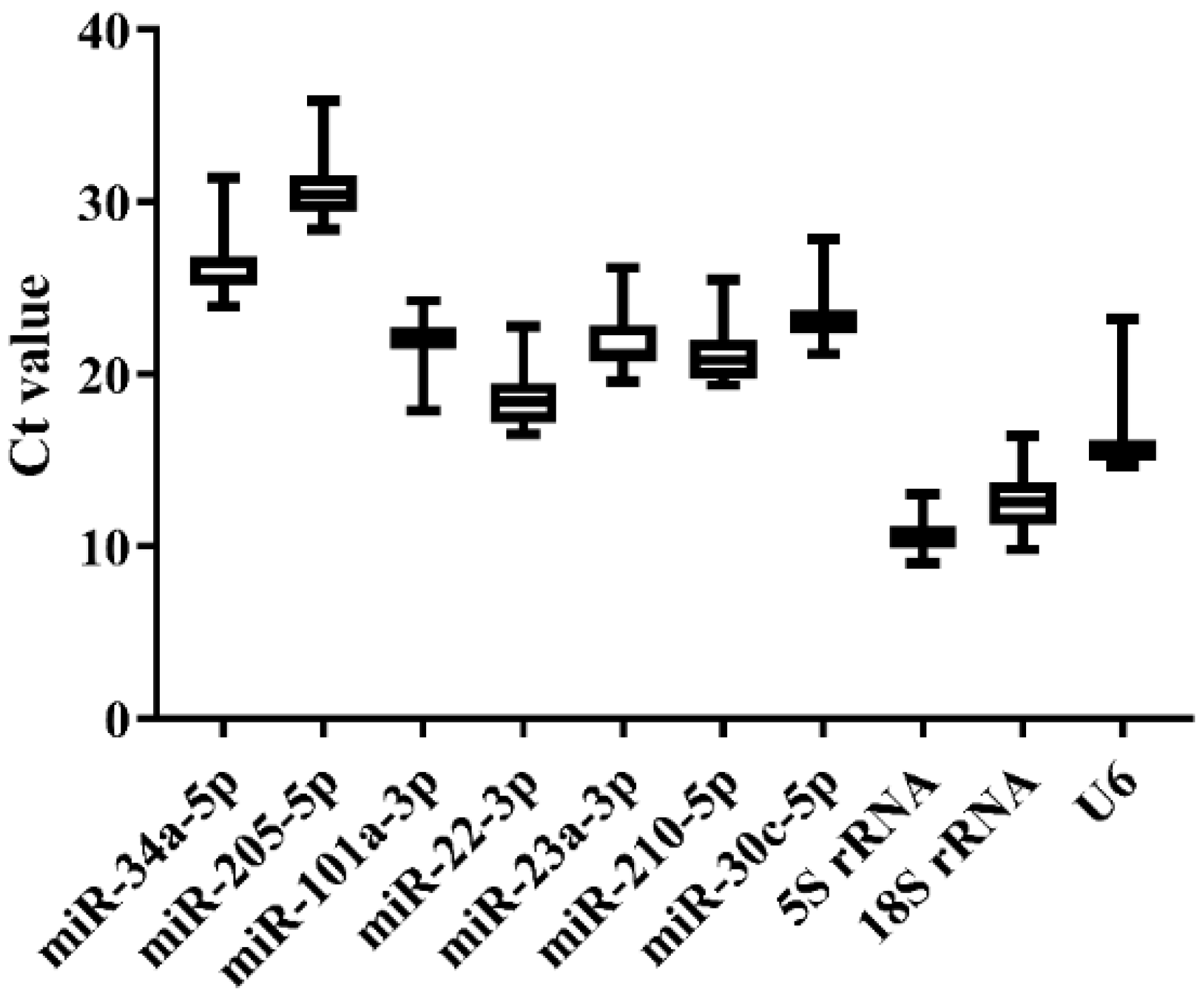
Three groups of liver samples in the challenge experiment, including the blank control group (BC-0h), Ringer’s solution group (RS-3h and -24h), and E. tarda-challenged group (EC-3h and -24h), were used for qRT-PCR analysis. As a result, melting curves displayed that miR-101a-3p, miR-22-3p, miR-23a-3p, 5S rRNA, and 18S rRNA generally presented a single peak (Supplementary Figure S2). The Ct value variation among experimental groups was generally similar across the 10 candidate references (Figure 4 and Supplementary Table S3). Consistent with normal tissues, the expression level of 5S rRNA, 18S rRNA, and U6 was higher (low Ct values) than other miRNA references, while miR-34a-5p and miR-205-5p were lowly expressed (high Ct values) across all groups (Figure 4). The stability of the 10 candidates was calculated and ranked by the Delta CT, BestKeeper, NormFinder, and geNorm methods, and the comprehensive ranking was carried out by the RefFinder method. As a result, miR-22-3p, miR-23a-3p, and miR-210-5p were the steadiest miRNAs by the Delta CT, NormFinder, and geNorm methods, while 5S rRNA was the most stable one by BestKeeper (Table 4). The M value of 5S rRNA was greater than 1.5, which was not appropriate as a reference (Table 5). According to the stability order and melting curves, miR-22-3p and miR-23a-3p were still the best reference combination in Japanese flounder livers injected with E. tarda and Ringer’s solution (Table 4 and Table 5).
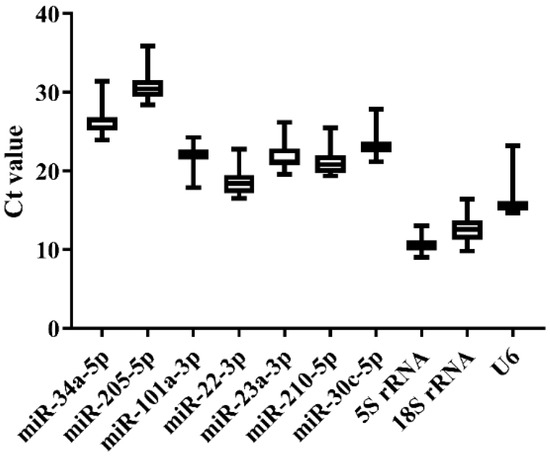
Figure 4.
Ct value variability for the candidate references in Japanese flounder livers with Ringer’s solution or E. tarda injection at different time points by qRT-PCR.

Table 4.
Expression stability ranking of the candidate references in Japanese flounder livers with Ringer’s solution or E. tarda injection at different time points. The best performers, miR-22-3p and miR-23a-3p, are shown in bold.

Table 5.
Expression stability values (M) calculated by geNorm in Japanese flounder livers with Ringer’s solution or E. tarda injection at different time points.
3.5. Reference miRNA Validation in Japanese Flounder Livers Infected with E. tarda by qRT-PCR
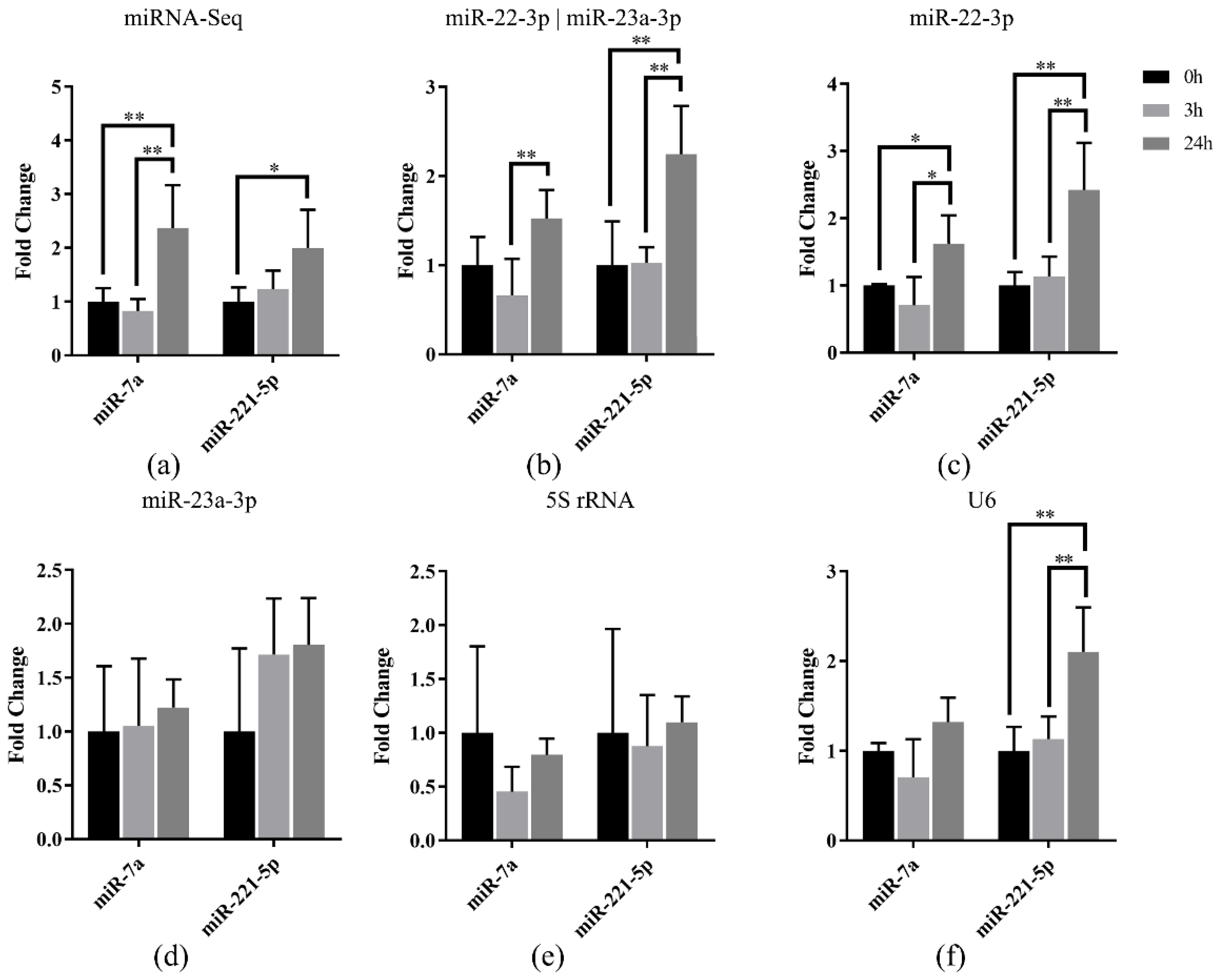
To validate the selected miRNA references in Japanese flounder, the relative expression of miR-7a and miR-221-5p were normalized using the selected references in Japanese flounder livers with E. tarda infection. Our previous miRNA-Seq result [27] showed that miR-7a and miR-221-5p were upregulated in livers after 3 h or 24 h infection of E. tarda (Figure 5a). Data normalization using the most stably expressed references, miR-22-3p and the combination of miR-22-3p and miR-23a-3p, resulted in generally consistent miRNA expression patterns along the infection time points (Figure 5b,c), whereas the result referenced with only miR-23a-3p was not as stable as the combined references (Figure 5d).
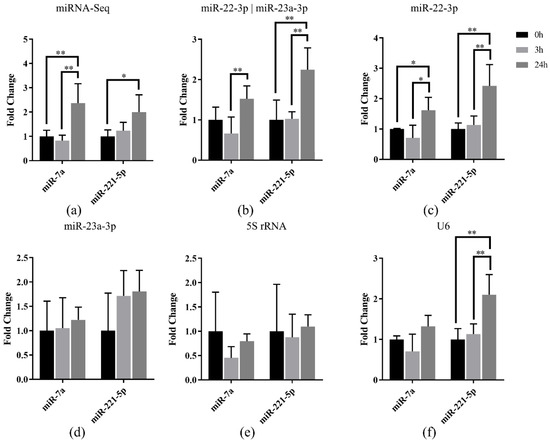
Figure 5.
Relative quantification of miR-7a and miR-221-5p expression in livers of Japanese flounder infected with E. tarda at different time points by (a) miRNA-Seq and qRT-PCR referenced with (b) miR-22-3p and miR-23a-3p, (c) miR-22-3p, (d) miR-23a-3p, (e) 5S rRNA, and (f) U6, respectively. Data are shown as mean ± SD (n = 3). Asterisks indicate statistical significance between groups (* p < 0.1 and ** p < 0.05).
Moreover, the results normalized using the unstable reference genes 5S rRNA and U6 showed that there was no induced expression of miR-7a and miR-221-5p after infection referenced with 5S rRNA (Figure 5e), while there was induced expression only for miR-221-5p when U6 was the reference (Figure 5f). The above results indicated that the most stable reference (miR-22-3p) and the most suitable combination (miR-22-3p and miR-23a-3p) showed similar miRNA expression patterns with miRNA-Seq data, so that they could be used for normalization of miRNA in Japanese flounder.
4. Discussion
MiRNA plays important regulatory roles in almost all aspects of development and physiology. It is inevitable to invest the miRNA expression pattern across different samples if we need to study its function. Although there are several approaches, qRT-PCR is one of the most widely applied techniques to evaluate miRNA expression. The accuracy of the qRT-PCR result could be easily affected due to the sensitivity of the experimental process; therefore, a suitable reference is required to normalize qRT-PCR data to ensure the validity of miRNA expression level [13,14]. Commonly used in miRNA qRT-PCR, the references are relatively short genes, such as 5S rRNA, 18S rRNA, and U6 [39,40]. There has been a lot of controversies about the stability of these commonly used references, and many experiments have reported that their expression was not constant across different species, tissues, and experimental treatments, which made them unsuitable as default miRNA references [23,24,26,31]. Hence, the expression level and stability were examined for 10 potential references in Japanese flounder normal tissues as well as livers under experimental conditions to reduce avoidable errors in miRNA qRT-PCR analysis. Owing to discrepant statistical algorithms, the ranking of candidate references obtained by the three methods (Delta CT, NormFinder, and geNorm) was generally similar, but different from the result from BestKeeper. Consistent with the argument above, U6, 5S rRNA, and 18S rRNA all performed less stability with way much higher expression levels than miR-22-3p and miR-23a-3p in both normal tissues and infected livers, only with 5S rRNA performing better with the BestKeeper method (Table 2 and Table 4). In addition, the amplification efficiency of the three commonly used reference genes (U6, 5S rRNA, and 18S rRNA) were all lower than 90%, while the ideal value should be around 90–110%. Another reason that rRNA and U6 are unsuitable as a reference is that their expression levels are much higher than most miRNAs, which could lead to the deviated qRT-PCR result of targeting miRNA with low expression [22,41]. In our analysis, U6 and rRNA genes represented the highest expression among all candidate references (Figure 3 and Figure 4). To sum up, U6, 5S rRNA, and 18S rRNA are not suitable as references for miRNA quantification in Japanese flounder.
In general, no single reference gene can be stable across all samples [42], which emphasizes the significance of identifying adequate references to normalize miRNA expression under various experiment conditions. For example, in common carp (C. carpio), 5S rRNA and 18S rRNA were deemed as the most suitable references for normalizing miRNA expression during early developmental stages, whereas let-7a and miR-23a were the best combination in different developmental gonads [16]. In Chinese perch (S. chuatsi), there were various suitable references under different conditions. For instance, among different tissues, the best combination was miR-22-3p and miR-23a-3p, and for developmental stages of muscle, the most appropriate references were let-7a and miR-26a, whereas for different embryonic developmental stages, the suited combination was miR-22-3p and miR-146a, and for fasting-refeeding treated livers, the best combination was miR-26a and miR-23a [22]. In Japanese flounder, the expression stability of candidate references in normal tissues and E. tarda-infected livers were analyzed, and the three best performers in normal tissues were identified as miR-22-3p, miR-23a-3p, and miR-101a-3p, whereas in E. tarda-infected livers, the best candidates were miR-22-3p, miR-23a-3p, and miR-210-5p. However, miR-101a and miR-210-5p were discarded due to their non-specific amplification and high amplification efficiency (Table 1 and Supplementary Figure S1). Therefore, miR-22-3p and miR-23a were concluded to be the best combination of reference miRNAs for Japanese flounder in both normal tissues and E. tarda-infected livers, which were also successfully validated in Japanese flounder livers for miR-7a and miR-221-5p expression in response to E. tarda infection (Figure 5).
5. Conclusions
In this study, 10 candidate references were identified for Japanese flounder miRNA quantification through both miRNA-Seq data and literature search. With qRT-PCR and statistical evaluation, commonly used U6 and 5S rRNA were not suitable as references, whereas miR-22-3p and miR-23a-3p were the most stably expressed miRNAs in both Japanese flounder normal tissues and livers challenged by E. tarda, which could be the most suitable combined references for Japanese flounder miRNA quantification in future studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes13020175/s1, Table S1. MiRNA reference selection from miRNA-Seq data with CV and log2FC methods. Table S2: Literature survey of candidate miRNA for qRT-PCR in other teleost species. Table S3: Ct values generated from miRNA qRT-PCR of Japanese flounder normal tissues and livers injected with E. tarda and Ringer’s solution. Figure S1: Melting curves of the candidate references in normal tissues of Japanese flounder by qRT-PCR. Figure S2: Melting curves of the candidate references in Japanese flounder livers with Ringer’s solution and E. tarda injection at different times by qRT-PCR.
Author Contributions
J.C. and Q.Z. designed the study; S.L., H.S., Z.L., and W.L. conducted the reference identification and qRT-PCR analysis; J.C. and S.L. conducted the transcriptome analysis and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31702331) and the National Infrastructure of Fishery Germplasm Resources.
Institutional Review Board Statement
This study was conducted in accordance with the Institutional Animal Care and Use Committee of Ocean University of China (OUC-IACUC), and it does not contain any studies with human participants.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw sequencing data was obtained at NCBI with accession number SRP135934. The TPM values and expression results of miRNAs were in Supplementary Table S1.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Flynt, A.S.; Lai, E.C. Biological principles of microRNA-mediated regulation: Shared themes amid diversity. Nat. Rev. Genet. 2008, 9, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Tilahun, B.T.; Igor, B. MicroRNA in Teleost Fish. Genome Biol. Evol. 2014, 6, 1911–1937. [Google Scholar] [CrossRef]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Leaman, D.; Chen, P.Y.; Fak, J.; Yalcin, A.; Pearce, M.; Unnerstall, U.; Marks, D.S.; Sander, C.; Tuschl, T.; Gaul, U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 2005, 121, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Wienholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H.A. MicroRNA expression in zebrafish embryonic development. Science 2005, 309, 310–311. [Google Scholar] [CrossRef]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Reza, A.M.M.T.; Choi, Y.J.; Han, S.G.; Song, H.; Park, C.; Hong, K.; Kim, J.H. Roles of microRNAs in mammalian reproduction: From the commitment of germ cells to per-implantation embryos. Biol. Rev. 2019, 94, 415–438. [Google Scholar] [CrossRef]
- Olejniczak, M.; Kotowska-Zimmer, A.; Krzyzosiak, W. Stress-induced changes in miRNA biogenesis and functioning. Cell Mol. Life Sci. 2018, 75, 177–191. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, 853–858. [Google Scholar] [CrossRef]
- Sun, Y.; Koo, S.; White, N.; Peralta, E.; Perera, R.J. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004, 32, e188. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Davoren, P.A.; Mcneill, R.E.; Lowery, A.J.; Kerin, M.J.; Miller, N. Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol. Biol. 2008, 9, 76–86. [Google Scholar] [CrossRef]
- Mackay, I.M.; Arden, K.E.; Andreas, N. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef] [PubMed]
- Oriol, T.; Ingrid, B.; Sarai, C.; Anna, C.; Armand, S.; Atsushi, A. Determination of Reference microRNAs for Relative Quantification in Porcine Tissues. PLoS ONE 2012, 7, e44413. [Google Scholar] [CrossRef]
- Wang, F.; Yang, Q.W.; Zhao, W.J.; Du, Q.Y.; Chang, Z.J. Selection of suitable candidate genes for miRNA expression normalization in Yellow River Carp (Cyprinus carpio. var). Sci. Rep. 2019, 9, 8691. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef]
- Latham, P.G.J. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA 2008, 14, 844–852. [Google Scholar] [CrossRef]
- Lardizábal, M.; Nocito, A.L.; Daniele, S.M.; Ornella, L.A.; Veggi, L.M. Reference Genes for Real-Time PCR Quantification of MicroRNAs and Messenger RNAs in Rat Models of Hepatotoxicity. PLoS ONE 2012, 7, e36323. [Google Scholar] [CrossRef] [PubMed]
- Johansen, I.; Andreassen, R. Validation of miRNA genes suitable as reference genes in qPCR analyses of miRNA gene expression in Atlantic salmon (Salmo salar). BMC Res. Notes 2014, 7, 281–297. [Google Scholar] [CrossRef]
- Xu, X.Y.; Shen, Y.B.; Fu, J.J.; Lu, L.Q.; Li, J.L. Determination of reference microRNAs for relative quantification in grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2014, 36, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, Y.L.; Chen, D.X.; Wu, P.; Yi, T. Selection of Reference Genes for MicroRNA Quantitative Expression Analysis in Chinese Perch, Siniperca chuatsi. Int. J. Mol. Sci. 2015, 16, 8310–8323. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, R.; Rangnes, F.; Sivertsen, M.; Chiang, M.; Tran, M.; Molton, W.M.; Prunet, P. Discovery of miRNAs and Their Corresponding miRNA Genes in Atlantic Cod (Gadus morhua): Use of Stable miRNAs as Reference Genes Reveals Subgroups of miRNAs That Are Highly Expressed in Particular Organs. PLoS ONE 2016, 11, e0153324. [Google Scholar] [CrossRef]
- Liu, J.; Jia, E.; Shi, H.; Li, X.; Jiang, G.; Chi, C.; Liu, W.; Zhang, D. Selection of reference genes for miRNA quantitative PCR and its application in miR-34a/Sirtuin-1 mediated energy metabolism in Megalobrama amblycephala. Fish Physiol. Biochem. 2019, 45, 1663–1681. [Google Scholar] [CrossRef]
- Ni, F.F.; Yu, H.Y.; Liu, Y.Z.; Meng, L.H.; Yan, W.J.; Zhang, Q.Q.; Yu, H.Y.; Wang, X.B. Roles of piwil1 gene in gonad development and gametogenesis in Japanese flounder, Paralichthys olivaceus. Gene 2019, 701, 104–112. [Google Scholar] [CrossRef]
- Song, H.F.; Xing, C.J.; Lu, W.; Liu, Z.Y.; Wang, X.B.; Cheng, J.; Zhang, Q.Q. Rapid evolution of piRNA pathway and its transposon targets in Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100609. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Z.; Liu, Y.X.; Han, M.; Du, X.X.; Liu, X.M.; Zhang, Q.Q.; Liu, J.X. Edwardsiella tarda-induced miR-7a functions as a suppressor in PI3K/AKT/GSK3β signaling pathway by targeting insulin receptor substrate-2 (IRS2a and IRS2b) in Paralichthys olivaceus. Fish Shellfish Immunol. 2019, 89, 477–485. [Google Scholar] [CrossRef]
- Yang, H.L.; Liu, J.; Huang, S.M.; Guo, T.T.; Deng, L.B.; Hua, W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 2014, 538, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R.D. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Zavala, E.; Reyes, D.; Deerenberg, R.; Vidal, R. Selection of reference genes for microRNA analysis associated to early stress response to handling and confinement in Salmo salar. Sci. Rep. 2017, 7, 1756. [Google Scholar] [CrossRef]
- Sun, Y.Y.; Zhang, Q.Q.; Qi, J.; Wang, Z.G.; Chen, Y.J.; Li, C.M.; Zhong, Q.W. Cloning and Expression Analysis of DMRT 1 Gene in Cynoglossus semilaevis. Nat. Sci. Ed. 2008, 54, 221–226. [Google Scholar] [CrossRef]
- Fu, Y.S.; Shi, Z.Y.; Wang, G.Y.; Li, W.J.; Zhang, J.L.; Jia, L. Expression and regulation of miR-1, -133a, -206a, and MRFs by thyroid hormone during larval development in Paralichthys olivaceus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2012, 161, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-Based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; Preter, K.D.; Pattyn, F.; Poppe, B.; Roy, N.V.; Paepe, A.D.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 0034.1–0034.11. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.L.; Xiao, P.; Chen, D.L.; Xu, L.; Zhang, B.H. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Sun, Y.H.; Shi, R.; Clark, C.; Chiang, V.L. Novel and Mechanical Stress-Responsive MicroRNAs in Populus trichocarpa That Are Absent from Arabidopsis. Plant Cell 2005, 17, 2186–2203. [Google Scholar] [CrossRef] [PubMed]
- Zanca, A.S.; Vicentini, R.; Ortiz-Morea, F.A.; Bem, L.E.D.; Silva, M.J.D.; Vincentz, M.; Nogueira, F.T. Identification and expression analysis of microRNAs and targets in the biofuel crop sugarcane. BMC Plant Biol. 2010, 10, 260. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Perou, C.M.; Karaca, M.; Perreard, L. Statistical modeling for selecting housekeeper genes. Genome Biol. 2008, 5, 156–167. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).