Abstract
In 2015 a mine dam with Mn-Fe-rich tailings collapsed releasing million tons of sediments over an estuary, in the Southwest of Brazil. The tailings have a high concentration of metals that contaminated soil until the present day. The high contaminant concentrations possibly caused a selection for microorganisms able to strive in such harsh conditions. Here, we isolated metal(loid) and anti-biotic resistance bacteria from the contaminated estuarine soil. After 16S rDNA sequencing to identify the strains, we selected the Mucilaginibacter sp. strain for a whole-genome sequence due to the bioprospective potential of the genus and the high resistance profile. We obtained a complete genome and a genome-guided characterization. Our finding suggests that the 21p strain is possibly a new species of the genus. The species presented genes for resistance for metals (i.e., As, Zn, Co, Cd, and Mn) beyond resistance and cross-resistance for antibiotics (i.e., quinolone, aminoglycoside, β-lactamase, sulphonamide, tetracycline). The Mucilaginibacter sp. 21p description as new species should be further explored, as their extracellular polymeric substances and the potential of this strain as bioremediation and as a growth promoter in high met-al(loid) contaminated soil.
1. Introduction
Metal pollution is one of the emerging environmental contamination problems, mainly due to anthropogenic activities such as mine dam collapses [1]. One of the largest mine tailing disasters occurred in Brazil in 2015. The Fundão Dam spilled millions of tons of tailing on the Doce river basin, which later reached its estuary region enriching with exogenous metal(loid) [2]. The presence of tailing is still a problem for the population in 39 Brazilian cities [3]. The metal(loid) contamination in the estuary area is even higher due to the soil redox process, which may facilitate metal remobilization to the aquatic biota [4]. Long-term monitoring of this ecosystem revealed soils with an elevated concentration of As, Cd, Pb, Zn, and Mn sediments and water beyond the threshold, causing cumulative effects on fishes and along their other trophic levels [5].
Investigating sediment bacteria is an opportunity to understand resistance/tolerance adaptation and molecular mechanisms involved, with important applications in the field of bioremediation and biostimulation [6]. Metals exert selective pressure on microbial communities, driving processes in the evolution of metal resistance determinants [7]. Indigenous bacteria isolated from the indigenous group of bacteria from the contaminated area is usually the best cost-effective, eco-friendly bioremediation technology. One mechanism of metal resistance is the production of extracellular polymeric substances (EPS). Some bioremediation bacteria were described absorbing metalloids (e.g., Cd, Cu) and increasing plants growth due to EPS production [8].
Mucilaginibacter is a recently described bacterial genus (named in 2008), characterized by species with a high capacity for EPS production [9]. The genus was described as encompassing bacteria that play a key role in precipitating metal and growth-promoting in plants in contaminated and stressed environments. Beyond this, components of Mucilaginibacter have a versatile metabolic characteristic allowing a broad ecological niche and habits as lakes [10,11], marine sand [12], phyllosphere [13], mine soil [14], wood [15], straw [16], etc., what is favorable for it use as bioremediation.
In this study, we aimed to isolate and identify indigenous bacteria with resistance to metal(loid) in estuarine soils and explore indigenous bacteria’s resistance mechanism for metal(loid) exposition through the genome. We hypothesized that metal(loid) contamination could increase the resistance genes of antibiotics and metals by co-selection.
2. Materials and Methods
2.1. Site Description and Sampling
The Rio Doce is a major river in southeastern Brazil. The basin has been modified for several decades of human activities, e.g., waste discharge, agriculture, aquaculture, industrial and mining activities. In November 2015, a large amount of Fe-enriched mine tailings mostly composed by highly crystalline Fe oxyhydroxides (e.g., hematite and goethite) were dumped into Rio Doce Basin after the Fundão Dam collapse, reaching the Rio Doce Estuary. The presence of tailing was visible in the water and in first 10 cm of the soil.
For this study, contaminated soil was collected in December 2018 from the Rio Doce estuary on the southwest coast of Brazil (latitude 19°23′28″ S, longitude 40°04′20″ O), 24 months after the disaster, (Figure 1). The soils were sampled using polyvinyl chloride tubes attached to a sampler used flooded soils. After the tubes were hermetically sealed and transported in a vertical position (at approximately 4 °C). We collected the first 5–10 cm depth from the core, where mine tailing was visible and homogenized in Ziplock bag and kept in cold stored chamber (5 °C) for 5 days. Soil redox potentials (Eh) were measured during sample collection using a Pt electrode, with the Eh values adjusted to a calomel reference electrode by adding +244 mV. Soil pH values were recorded with a glass electrode, previously calibrated with standard solutions of pH 4.0 and 7.0. A sub-sample was collected for total metal contents analysis, determined by plasma atomic emission spectroscopy (ICP-OES) after microwave-assisted triacid digestion (HCl + HNO3 + HF; USEPA, Washington, DC, USA, 1996).
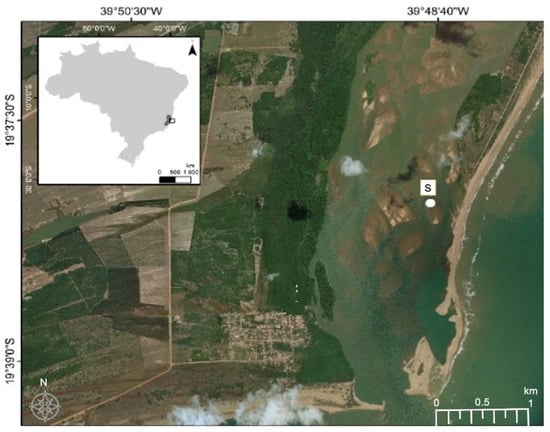
Figure 1.
Image of the Rio Doce estuary indicating the locations of sampling sites (S). Coordinate System. GCS WGS 1984 Datum: WGS 1984.
2.2. Isolation and Inhibitory Tests
In the laboratory, we add 100 mL of sterile water to 5 g of soil and incubated at 30 °C with 150 rpm agitation for 6 h. The bacterial isolation was performed by spread plate technique using soil sample dilutions in 10% Tryptic Soy Agar (Merck) solid media with 1 mg/mL of nystatin and Mn (1.6 mg/mL) [17]. The TSB agar was made up of 10% of the recommended concentration normally used to create a low nutrient media simulating the low nutrients in the water samples [18]. The plates were incubated for 72 h at 35 °C. The colonies that survived in this concentration of Mn, were reselected for further metal resistance test including Zn (500 mg/mL); Cd (150 mg/mL); Co (800 mg/mL). Due to the high frequency of cross-resistance among metals and antibiotics, we also conducted antibiotic resistance test for the 21 strains isolated using five class of antibiotic: β-lactam (ampicillin 10, 200, 300 µg/mL), aminoglycosides (kanamycin 50, 100 µg/mL, streptomycin 20, 50, 100 µg/mL, tetracycline 10, 30, 50 µg/mL, amphenicols (chloramphenicol 10, 50, 70 µg/mL) quinolones (nalidixic acid 50, 100, 200 µg/mL) [19,20]. Metals and antibiotics solutions were added to the medium after sterilization by filtering using a 0.22 m membrane (Millipore nitrocellulose GSWP 04700) [17]. All the solution were added in warm media avoiding precipitation. The antibiotics and metal were diluted in sterile water, except for chloramphenicol which was diluted in ethanol 70 %. All 21 isolates were inoculated in the plate and the ones that presented minimum grown were considered resistant. After the tests we stored in tryptic soy broth (TSB; Fisher Scientific Inc., Hampton, NH) with 20 % glycerol at −80 °C.
2.3. DNA Extraction and 16S rDNA Sequencing
A colony of each isolate was cultured in tryptic soy broth incubated at 30 °C with 150 rpm agitation for 24 h. The DNA was extracted using the Fenol-Chloroform method [18]. The 16S rDNA was amplified using the PCR method with primers 1492R (TACGGYTACCTTGTTACGACT) 27F (GAGAGTTTGATCCTGGCTCA) [20]. The PCR products were purified with Charge Switch™ PCR Clean-Up Kit (Invitrogen™, Carlsbad, CA, USA) then sequenced and ran on the ABI 3730XL capillary DNA. A contiguous sequence was constructed with forward and reverse sequencing data resulting in a fragment of approximately 900 bp, with DNA Baser Sequence Assembler v4 (2013) (Heracle BioSoft, Arges, Romania).
2.4. Bioinformatic Analysis of 16S and Phylogeny
The sequences were trimmed (phred > 20) by Codoncode Aligner v. 2.0.4 and SeaView v.4. The sequences were aligned using MAFFT version 7 with L-INS-I toll [21]. The highest similarities (as percentage similarities) and accession numbers are given in Table S1. Sequences were compared with those in the NCBI database (GenBank) using the BLAST Sequence Analysis Tool (BLASTN). Isolates with a >99% match to the published sequences were identified to the species level, and those with a >97% match was identified to the genus level. For phylogenetic tree construction we used IQ-tree [22] and MEGA 7 [23]. Tree visualization was performed with ITOL [24].
2.5. DNA Extraction and Whole Genome Sequencing of Mucilaginibacter sp.
The selected isolated bacterium (strain 21p) was incubated in 40 mL of liquid tryptic soy broth 10% for 48 h, 30 °C and 400 rpm (SK-O330). The media was centrifuged until 1 mL of bacteria pellet was obtained. DNA was extracted with Wizard® Genomic DNA Purification System (Promega, Leiden, The Netherlands) using the manufacturer’s protocol. DNA integrity was confirmed in the electrophoreses gel. DNA quantity and quality were assessed by fluorometry using a QuantiFluor® ONE dsDNA System (Promega Corporation, Fitchburg, WI, USA) and ratios 260/280 and 260/230 absorbance by spectrophotometry.
The genome was sequenced using Illumina, for high quality short reads sequencing and MinION, for long reads to achieve a complete genomic sequence. For Illumina, the genomic DNA library was constructed using a Nextera XT library prep kit (Illumina, Inc., San Diego, CA, USA) with paired-end reads (2 × 150 bp) on a MiSeq v3 platform 1GBps (Illumina, San Diego, CA, USA). For MiniON sequencing, libraries were prepared with Rapid Barcoding Sequencing QK-RBK004 (Oxford Nanopore Technologies [ONT], Oxford, UK) platform using the MinKNOW software, Version 4.5.0 (Figure S1).
2.6. Whole Genome Hybrid Assembly
The bioinformatic workflow of the hybrid, MinION and MiSeq assemblages (Figure S1) Illumina sequencing data were converted to fastq format using the MiSeq reports program, producing 14,288,200 raw reads. To perform quality trimming (phred < 20) and adapter removal, pre-processing was carried out with the fastp tool [25]. Besides, for long ONT-reads fastp data files were obtained using Guppy base-calling v.3.6.0 (https://staff.aist.go.jp/yutaka.ueno/guppy/, accessed on 20 October 2020). The mean read quality of the raw long reads was scored using NanoPlot 1.0.0 and trimmed with NanoFilt and Porechop [26]. Trimmed ONT and Illumina reads were then de novo assembled with the hybrid assembly method in the Unicycler 0.4.8 pipeline, which functions mainly as an optimizer of SPAdes 3.13.1 [27], but includes the long ONT sequences to complete the assembly. The genome was deposit in GenBank, SUB8286515, under the BioProject accession number PRJNA667924.
2.7. Taxonomic Affiliation and Phylogenetic Analysis
Whole genome comparison was performed through Average Nucleotide Identity using BLAST algorithm (ANIb) [28]. Two comparisons were made one with the 32 Mucilaginibacter representative sequence available at NCBI—excluding partial and anomalous results and the other with 60 complete assembly sequences deposited in NCBI using OrthoANI v1.4 [29].
Furthermore, phylogenetic analysis was performed based on 120 single-copy conserved marker proteins (amino acid sequence) from bacteria, selected and aligned with GTDB-Tk v0.3.2 based on 94,759 bacterial genomes [30]. Maximum likelihood phylogenomic tree was constructed with RAxML v8.0.0 [21] with 1000 bootstraps using the PROTGAMMAIGTR model. IQ-TREE web server applied Akaike information criterion (Kalyaanamoorthy) to find the most accurate substitution model method using the following parameters: GTR substitution model, 1000 bootstrapped data sets, 4 substitution rate categories for across site rate variation, estimated γ distribution parameter, optimized variable sites and empirical nucleotide equilibrium frequencies, a heuristic search of starter tree with BioNJ algorithms, and tree topology search with NNIs. The phylogenetic tree was visualized using FAST Tree program [31].
2.8. Annotation, Pan-Genome Analysis, and Genomic Resistance Profile
The sequence was annotated through RAST (Rapid Annotation using Subsystem Technology) database (http://rast.nmpdr.org/, accessed on 10 December 2020). The 32 species RefSeq genomes used for ANI determination were downloaded from NCBI and were annotated through Prokka 1.14.0 [32]. The genome annotation files GFF3 format were used for pan-genome analysis using Pirate [33]. The genetic source of the resistance was evaluated using comparative functions embedded within the PATRIC pipeline and a Comprehensive Antibiotic Resistance Database (CARD, http://arpcard.mcmaster.ca, accessed 28 December 2020) [34]. In addition, Island Viewer was used to identifying genomic islands (GEIs) containing the resistance genes [35]. For the pangenome figures, a new analysis was performed using Anvi’o [36] using the meta-pangenomic workflow with the standard parameters. Genes in the pan-genome were annotated using NCBI’s Clusters of Orthologous Groups.
3. Results
3.1. Diversity of Culturable Resistant Bacteria from Mine Tailing Contained Soil
A total of 21 bacteria strains were isolated from soil samples belonging to two genera: 18 Bacillus (from Firmicutes phylum) and three Mucilaginibacter (from Bacteroidetes phylum). All the strains showed resistance to Mn and Co and other multiple resistance to metals and antibiotics (Table 1 and Table S2). Proteobacteria, Firmicutes, and Bacteroidetes are the most common phyla in the contaminated environment [37]. Despite Proteobacteria being a frequent prevailing phylum, encompassing more than 40% of all prokaryotic genera [38], no strain of this phyla was isolated in the present work. This could be related to the selection pressure exerted by Mn and other metals in the community [39]. In fact, a study in Cu-mine in Brazil reported that Firmicutes was the main isolated phyla [40], a similar response that occurred in the present study.

Table 1.
Resistance profile of environmental isolates from contaminated soil and colonies characteristics.
Based on 16S rDNA analyses, we identified six species of Bacillus: B. aerophilous, B. pumilus, B. licheniformis, B. subtilis, B. aryabhattai, B. megaterium. The most common ones were B. subtilis, B. megaterium and B. pumilus. Bacillus is a genus characterized as aerobic endospore-forming bacteria, known to resist stressing environments like heat, radiation. The genus also participates in biogeochemical cycling of metals, facilitating oxidation/reduction processes [41]. Observing the phylogenetic tree (Figure S2), some clusters of Bacillus (6p, 11p, 12p, 17p, 18p) are less clustered in related to others. One possible cause of this isolation could be related to specialization due to the high metal concentration present in this environment.
All Bacillus species isolated in this study have been described with multiple-metal and antibiotic resistance, mainly isolated from the wastewater treatment, or associated with plant growth-promoting. Among them, B. subtilis and B. licheniformis are the most investigated, with high resistance of oxidative stress and with bioprospection application [42]. Specifically for Mn resistance, strains of B. subtilis were described to be involved in manganese oxidation, having an important role in precipitating Mn [43]. Other species also have strains already described with such potential, including as growth-promoters. B. aerophilous (strain TR15c), with multiple-metal tolerance including Cu (1750 mg kg−1), antibiotic resistance (ampicillin, kanamycin, chloramphenicol, penicillin, tetracycline, and streptomycin), and plant growth-promoting attributes (phosphate solubilization and indole-3-acetic acid production) [44]. B. aryabhattai, strain AB211, was described utilizing the root exudates and other organic materials as an energy source, has genes for heat and cold shock antibiotic/metal resistance that enable bacteria to survive biotic/abiotic stress [45] B. megaterium (MNSH1-9K-1) showed an elevated resistance to potentially toxic elements and possesses the ability to remove metals like Ni, Hg, and V from liquid media [46].
The content of soil metals where the strain was isolated was as follows: 30 g kg−1 for Fe and 460, 3.1, 9, 30, 10.7, 10.1, and 48 mg·kg−1 for Mn, Cd, Co, Cu, Ni, and Zn respectively. Despite the results below the threshold value according to Brazilian legislation [47] soil metals content is 3–50 times higher compared to values from the soil before the disaster [48] (Table S2). Studies in the same contained area reported a high concentration of As, Cd, Pb, Zn, and Mn in sediments and water, causing cumulative effects in the muscle’s fishes after the disaster [5,49]. Queiroz et al. (2021) highlighted an increase of 880% in the concentration of dissolved Mn [50]. This could explain the presence of a wide bride resistance profile and the numerous resistance gene, mainly to Mn.
We found two isolates affiliated to the Mucilaginibacter genus, with the high resistance profile, where we highlighted the isolate 21p, that presented resistance to almost all the tested metal and antibiotics. Mucilaginibacter is a recent genus, was described for the first time in 2007, as a member of the Bacteroidetes phylum [9]. Bacteroidetes is known for multiply resistance due to natural resistance to aminoglycosides as well as resistance that is acquired during horizontal gene transference (HGT) [51]. The genus is specialized in the degradation of complex organic matter and is known due to the production of large amounts of extracellular polymeric substances (EPS) [9]. EPS was already described as: sorption of metals, growth promotor, and osmoprotector in plants roots [52]. Some strains of this genus are known for their resistance to metals, including Mn since strains of Mucilaginibacter were already isolates from Mn-mines presented high levels of metal(loids) resistance [52,53]. Recent studies have highlighted the genera as a root endophytic bacterium in wide broad varieties of plants, promoting growth in stress environments (e.g., salty, water restriction, metal-contaminated) [52,54,55].
In general, both genera isolated in our survey, Bacillus and Mucilaginibacter, have bioremediation potential. However, Bacillus has been more studied with multiple uses, including the production of polymeric compounds and biosolvants, a well-known for sorption and complex pollutants [56]. Mucilaginibacter, on the other hand, is a new genus with a low number of identified species with a potentiality that is still not well explored, which supports our decision to investigate more about bacteria from this genus in the present work (Table 1). All of them were resistance to Mn and Co to the maximum concentration and just a few for Cd and Zn. Resistance of NA was rarely found. Isolate 21p were the colony that presented higher resistance among metals and antibiotic and were selected for the futures steps.
3.2. A potential Novel Metal(loid) Resistant Species from Mucilaginibacter Genus
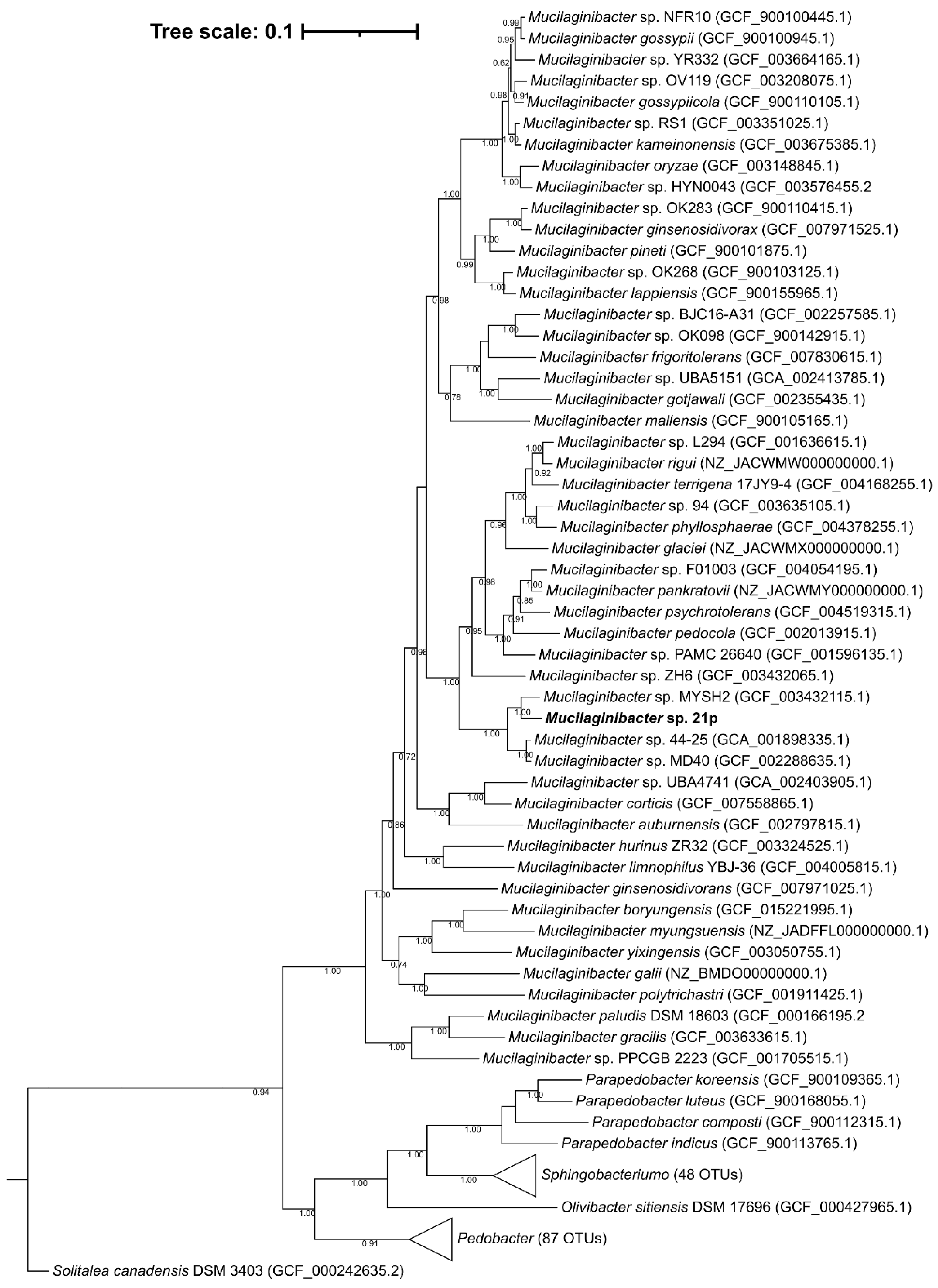
Mucilaginibacter sp. 21p was addressed by the sequencing of the complete circular genome with one contig with 4,739,655 bp. The genomic GC content of strain 21p, directly calculated from its genome sequence, was determined to be 43.2% which is in the range of the previously described species of the genus Mucilaginibacter, from 39.1 to 47.8%. It was annotated 4334 coding sequencing in RAST and 45.47% CDS in PATRIC (RAST tool kit) [9]. Average nucleotide identity (ANI) presents low similarity with all 60 Mucilaginiobacter genomes from GenBank, inferior to 95%—proposed threshold for species identity [57]. The highest identity was obtained with Mucilaginibacter sp. MYSH2 (accession GCA_003432115) with 80.08%, isolated from beach sand in Yanfyand, South Korea (Table S3). The lowest identity was with M. ginsenosidivorans (3017T) with only 70.22%, isolated from soil of ginseng field, Pocheon province, South Korea [58]. Based on the species Reference Genomes (Table S3), the strain with up to 74.1% identity was obtained with M. rigui, a strain isolated from a freshwater from a wetland, also in South Korea [59] and lowest, we have the same results, being M. ginsenosidivorans the most distance species from the genus. These results suggest that our strain 21p belongs to genus Mucilaginibacter, and that might be representative of a new species.
A phylogenetic tree of 16S rDNA including representative sequences supports the affiliation of this strain at Mucilaginibacter genus (Figure S2), as can be seen in a well-defined genus clade with maximum support of the branch. The similarity between strain 21p and other members of the genus Mucilaginibacter was also supported by the phylogenetic tree’s topology of the complete genome (Figure 2). The 21p showed higher stronger lineage (100% bootstrap value) with seven described species besides M. rigui as showed by ANI evidence; those include M. pedocola, M. pychrotolerans, M. pankratovii, M. glaciei, M. phyllosphaerae, M. rigui, M. terrigena. Despite the closer distance of 21p with those species, our assembly seems to be arranged in a separate independent clade, suggesting that it might have gone through recent speciation events. And among the non-described genomes, the stronger lineage with 21p ones is M. sp. MYSH2, described above, M. sp. 44–25, and M. sp. MD40. For instance, altogether, this genomic evidence strongly suggests that the 21p strain could be a new member of the Mucilaginibacter genus.
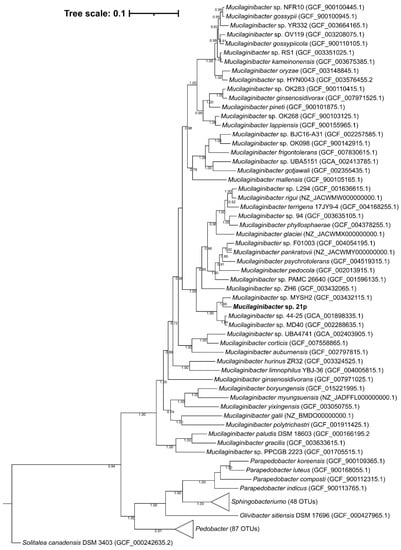
Figure 2.
Phylogenetic positioning of the isolate Mucilaginibacter sp. 21p (highlighted in bold) based on the maximum likelihood phylogenomic tree based on 120 single-copy conserved amino acid sequences. GenBank Assembly accession code in parentheses. Percentages of bootstraps values above 50% are presented. Bar: 0.1 substitutions per position.
Species of this genus has been isolated from a wide range of terrestrial and aquatic habitats. The genus Mucilaginibacter has a strictly aerobic or facultatively anaerobic metabolism, variable for catalase and oxidase, and high activities of extracellular polymeric substances [10]. This versatile metabolic characteristic allows the genus to be distributed in a wide ecological niche, having a key role in degrading various biopolymers as M. gynuensis isolated from wood [60] and oxidating metal as M. rubeus and M. kameinonensis isolated from contaminated soil [61].
Among the seven closest described species at the phylogenetic tree, M. pedocola was the only isolated in metal contaminated areas. M. psychrotolerant, M. glaciei, M. pankratovii, M. rigui were isolated in flesh water or soils with high moisture, as peatland, similar areas than 21p were isolated (Figure 3). For this reason, the elicitation of the mechanism of adaption of this strain could be important for the understanding of the impact of metal contamination in the sampled environment.
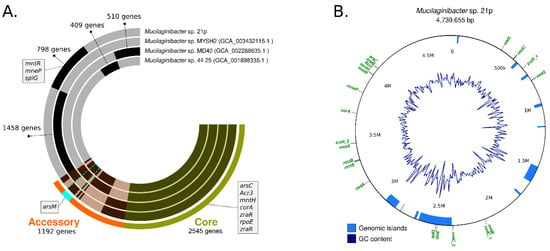
Figure 3.
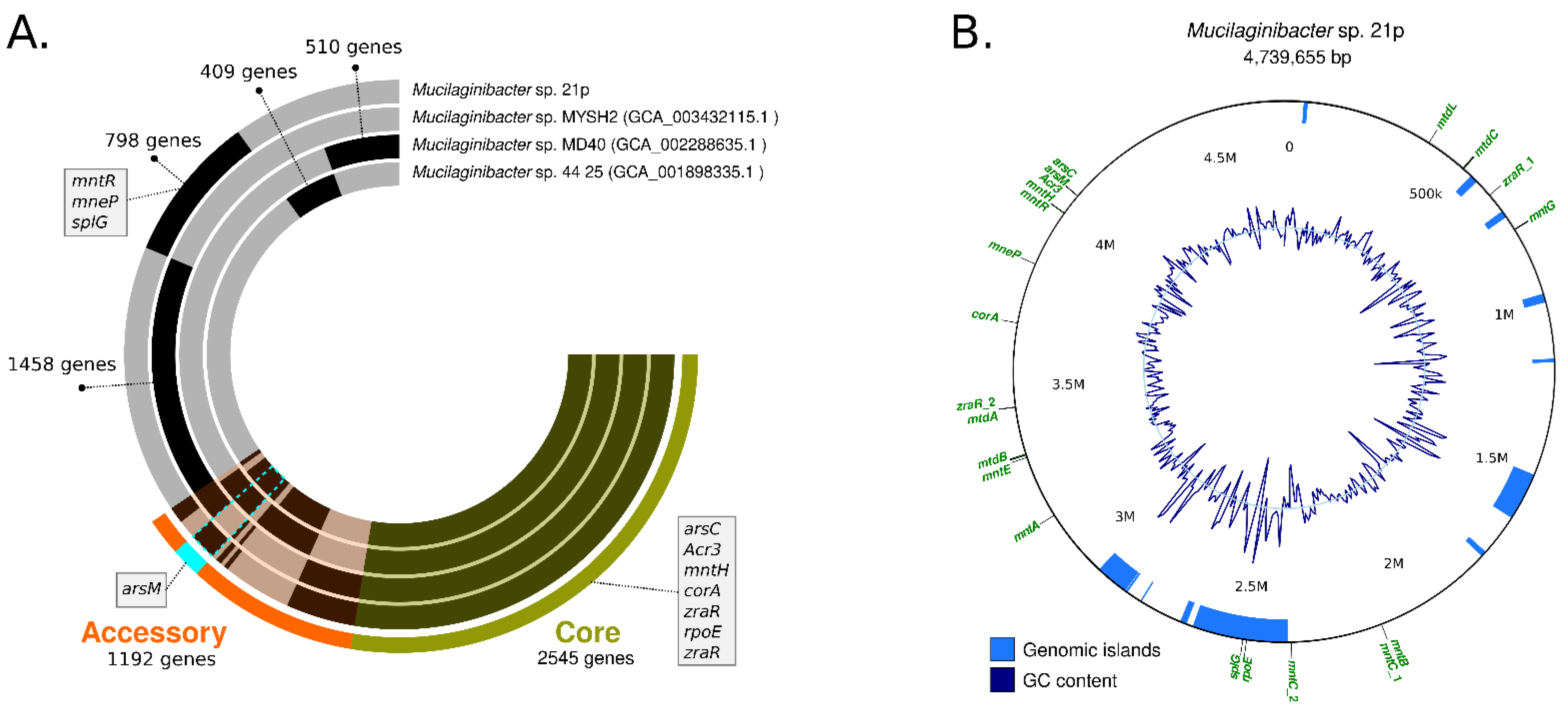
(A) Circular visualization of the genomes from Mucilaginibacter sp. 21p and the three closest strains according to the phylogenomic tree. Genomes are represented in each radial layer with the black bars representing clusters of genes. Environmental resistance genes present among clustered gene calls are indicated; (B) Circular visualization of the genomic island of Mucilaginibacter sp. 21p, GC content, and some resistance genes distributed in the genome.
In order to characterize the functional genomic variation of the 21p strain we conducted a pangenome analysis for the determination of the orthologous gene family’s presence/absence across 32 representative Mucilaginibacter species. Pangenome results revealed the presence of gene coding resistance in our assembly that is shared with just one or two other species as yojl and cusC, related with efflux plumps, important for the resistance of bacteria (Table 2) and the absence of others genes (Table S4).

Table 2.
List of rare or unique genes of M. 21p according to Pirate.
Our assembly 21p also has unique alleles, as dltC, rpoE_4, menE, mmgB, rfbD and glmE and some of them are located in GI, suggesting that were horizontally transferred (Table 2). Bastiat (2012) described rpoE_4 as an extra-cytoplasmic function sigma factor, activated during the generation of sulfite compounds (thiosulfate and taurine) that controls a response required for efficient growth in the presence of sulfite [62] (Table 2). The same authors, also suggesting that it may be advantageous for bacteria in the stationary phase by providing either a sulfite detoxification function or an energy input through sulfite respiration [62]. The other unique resistance gene is glmE, related to cobalamin production (vitamin B12)—a cobalt-containing tetrapyrrole cofactor involved in intramolecular rearrangement reactions [63]. Recently some authors have been suggested cobalamin contribute to maintaining the redox balance under highly oxidizing conditions, providing a specific advantage in extremely acidic and highly metal-loaded environments by increasing the tolerance and fitness of these microorganisms [63]. Both unique genes seem to be compatible with the Fe and Mn contaminated environment where 21p were isolated.
Other unique genes integrated important operons that seem to be incomplete in 21p genome as: srf responsible for surfactin biosynthesis srfAA, srfAB, missing srfAC; menE, for melaquinone synthesis, missing menB, menC; and rfbC, dTDP-Rhamnose synthesis, missing rfbA. The loss of this genes along generations should be investigated, mainly surfactin biosynthesis operon since it has an important role in bioremediation. Some of the unique genes, as rpoE_4 and splG, are located among 2.55–2.36 M, area associated with one of eleven Genomic Island (GI) harbored in the genome (Figure 3B). Typically, GEIs have been shown to be associated with host-beneficial adaptive traits such as bioremediation, virulence, antibiotic resistance, and metabolism [35,64] (Figure 3B).
Among the unique genes of the three closest strains, there is splG gene that encodes a thermophilic spore photoproduct lyase. This lyase belongs to a family of radical S-adenosylmethionine (AdoMet) enzymes [65]. This enzyme is normally present in species, from Bacillus and Clostridium genus, that are extremely resistant to harsh (physical, chemical, and biological) conditions allowing them to survive. Since 21p was resistant to a high level of metals and antibiotics, the presence of splG collaborated with the resistance profile [65]. Other genes related to metal resistant as arsC, acr3, mntH, corA, zraR, rpoE, are present in the other strains, integrating the core genome.
3.3. Antibiotic Resistance Genetic Profile of Mucilaginibacter 21p
To gain insight into the genetic basis of how the strain was able to deal with high antibiotic concentrations, we study the annotated genes. Draft genomes were automatically annotated through the RAST database by PATRIC. Putative resistance genes of six antibiotic resistance family were found in our assembly: quinolone (gyrA), aminoglycoside (katG, gidB, str), β-lactamase (bLc, ampG, ampH, pse2, ybxl, CS), sulfonamide (folP, folA), tetracycline (s10p, tet), daptomycin (gdpD), and other (pxyR, kasA, ef-tu, Iso-tRNA, ddl, dxr, fabl) (Table 2). Fluoroquinolone and tetracycline resistance was a strict match two classes antibiotic for resistance gene database (CARD).
Some mechanisms of resistance could not be restricted just to antibiotics. When a single mechanism confers resistance to both an antibiotic and metal is called cross-resistance. The mdtABC, a multiple drug transporter, is a plasmid-encoded efflux pump that confers resistance for a high concentration of metals (zinc and copper) and β-lactam antibiotic [66,67]. In czcABC—operon that confers resistance to zinc, cadmium, and cobalt. This pump efflux, when co-regulatory with gene czcR, also present in 21p can confer resistance carbapenems, a class of last resort antibiotic, by repressing the expression of the oprD porin the route of entry for these antibiotics to the bacterial cell [68].
The mechanism as cross-resistance is responsible for bacteria co-selection, which could maintain and promote antibiotic resistance in indigenous bacterial populations even in the absence of antibiotics [69,70]. This topic is a public health concern since soil bacteria could transfer ARG to pathogenic bacteria of clinic relevance [71,72]. Is already known that the exposition of multiple metal increase antibiotic resistance horizontal transference [73]. Exposure to multiple metals, as happens in this contaminated area, has been shown to be more effective for co-selection for antibiotic resistance than exposure to a single metal [74,75]. Recent studies also described those mechanisms of cross-resistance are related to the sub-lethal concentration of metals. So that, this led us understand that antibiotic resistance in 21p could be a consequence of the rich metal(loid) environment where the strain was isolated (Table S2).
Most of the metal(loid) resistance is supported by biosorption, without energy cost process, so that, to defend themselves from metal toxicity, microbes have evolved a lot of mechanisms of adaption and resistance. Essentially, the diverse determinants maintaining metal(loid) homeostasis can be divided into four categories: pump efflux (ATPase, RND, CDF family), enzymatic detoxification (redox and (de)methylation), intracellular sequestration, and reduction of uptake. Other unspecific mechanisms enhancing bacterial resistance to potential toxic elements are extracellular polymers (EPS) or siderophores secreted by bacteria to trap metal(loid)s, reducing its bioavailability and further alleviating stress [76,77].
The strain 21p isolated presented two major mechanisms of resistance: efflux pump, a mechanism that helps the microorganism to extrude antibiotics and disinfectants and EPS in an efficient manner. On its genome, it is observed genes related with five families of efflux pumps: (i) major facilitator superfamily (MFS); (ii) the ATP-binding cassette superfamily (ABC transport); (iii) the small multidrug resistance (SMR) family; (iv) the resistance-nodulation-division (RND) superfamily; and (v) the multidrug and toxic compound extrusion (MATE) family [78]. Our assembly has at least one representative mechanism from each family, which can explain the wide metal resistance (Table 2).
Studies in the same estuarine area reported a high concentration of As, Cd, Pb, Zn, Mn in sediments and water, causing cumulative effects in the muscle’s fishes after the disaster [5,50,79,80]. Queiroz et al. (2021) highlighted an increase of 880% in the concentration of dissolved Mn. 21p has resistance genes to all these five metals cited in the studies: As (arsC, arsM, Acr3), Zn (zra, zraR, Xref regulator Zn sigma dependent, putative zinc protease, Zn resistance associates’ protein), Mn (mneP and manganese efflux: mntR, mnH, mnmACEG); Co (corA, cobalt/magnesium transport, and the operon czc (czcA, czcB, czcD, czcR), cobalt, zinc, and cadmium). We can observe that Mn resistance is the one that presented more diversity of genes related to efflux, (operon mnm + mntR, mnH), what can be correlating to Mn be the metal(loid) with the highest concentration.
Other species from the genus isolated in soils with high concentrations of metal(loids) M. pedocola, M. rubeus, M. kameinonensis, do not present such a wide resistance profile. The resistance is multicopper oxidases, ars operons, RND transport systems (czc, cus, ncc). Bacteria exposed at metal(loid) can develop resistance through mutation, but mainly to horizontal transference since requires specific multigene resistances. HT is promoted by multiple metal(loid) contamination and biofilm formation, conditions that occurred here related to the recent evolution of the genus.
The other way to resist metals is promoted by using extracellular polymeric substances (EPS). The genes related to EPS are formation, as mannose, flippase, lipopolysaccharide formation and export system, and glycosyltransferase (Excell EPS- Vide Notion EPS) are abundant in the 21p genome. EPS was one most important characteristics of the genus, being even inference in the name. Bioprospection application of EPS in the genus is already described for (i) bioremediation, as metal(loid) removal, and (ii) growth promoter in extreme environments [81,82].
Microbial EPS binds metal(loids) in sites present in the cellular polymeric structure, complexing metals and micro-precipitated them, without the involvement of energy [83]. Strain TBZ30T from the genus, M. Pedocola, adsorb nearly 60% of Zn2+ and 55% of Cd2+, the medium (added with 0.3 mM ZnSO4 and 0.25 mM CdCl2, respectively), which is possibly intermediated by the production of EPS [84].
M. gossypii sp. and M. gossypiicola sp. were described as growth promoter in cotton plants [85]. Some field studies the genus is highlighted as growth promoting in different plants [86], alleviating salt stress [84], water deficiency soil [87] and metal(loid) contaminated environments [53]. The EPS effect is not related to auxin hormone but osmoprotectants changing ion balance in the soil environment and nutrient solubilizing ability [88,89]. Biofilm formation in the genus should be better understand, probably the genus count with an alternative to chemical signaling, as electrical, since no quorum sensing genes have been described in the genus [90,91,92].
4. Conclusions
We successfully isolated multiple resistant bacteria from mine tailing contaminated soils in the Rio Doce estuary. The bacterial genera obtained were Bacillus and Mucilaginibacter. We depicted the genome of the most resistant bacteria obtained. Our results suggest that this strain is possible a new species of the genus. A high number of genes related efflux plump with metal(loid) and antibiotic resistance and EPS production was found. This study describes multiple resistance genes of the 21p of and provides the better understanding of the genes related efflux plump with metal(loid) and antibiotic resistance and EPS production. Our assembly presented some genomic island with rare and unique genes among the genus, what could suggest a recent speciation process due to the high concentration of metal in the soil. A further experimental investigation is required to described Mucilaginibacter sp. 21p as a new species and explore the functionality of these strain in bioremediation and/or growth promoter in high metal(loid) contaminated soil. New studies should be considered to better understand the potential of this specie on bioremediation process and the capacity of horizontal transference of these resistances.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13020174/s1, Figure S1. Bioinformatic workflow of the hybrid, MinION and MiSeq assembly, Figure S2. Neighbor-joining phylogenetic tree constructed based on the 16S ribosomal RNA (rRNA) gene sequences from bacteria isolated showing the phylogenetic relationships between all isolated strains including Mucilaginibacter sp. ALS21, Table S1. Direct submission of sequence data to GenBank, Table S2. Chemical Analysis of the soil where the strains were selected, Table S3: Average Nucleotide Identification (Blast algorithm), Table S4. List of genes that is not present in assembly M. 21p e according to Pirate.
Author Contributions
Conceptualization, F.D.A.; methodology F.D.A. and A.C.F.D.; validation, K.N.-M. and E.D.; formal analysis, A.L.S.V., E.D. and K.N.-M.; investigation, A.L.S.V. and T.D.; data curation, A.L.S.V. and F.A.G.; writing—original draft preparation, research structure, F.D.A. and A.L.S.V.; writing—review and editing, F.D.A., L.B., A.F.B. and K.N.-M.; visualization, E.D. and A.L.S.V.; supervision, F.D.A. and K.N.-M.; project administration, A.F.B.; funding acquisition, A.F.B., F.D.A. and L.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundação de Amparo do Espirito Santo grant FAPES/CNPq/CAPES Rio Doce 77683544/2017 to A.F.B.; Coordenação de Aperfeiçoamento dePessoal de Nível Superior grant CAPES/PRINT 88887.370137/2019-00 to A.L.S.V.; andInstituto Antártico Chileno (INACH) grant INACH DG_01-19, Universidad de la Fronteragrant DI20-2018, Network for Extreme Environments Research (NEXER) grant NXR17-0003, andCONICYT grant CONICYT-PFCHA/Doctorado Nacional/2017–21170263 to K.N.-M. The fundershad no role in study design, data collection and interpretation, or the decision to submit thework for publication.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
The authors like to thank Andrios Consultoria for supplies donations and Julia Franke, Sonia Pires, Denise Mescolotti and Fernado Baldesin for the technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Naidu, R.; Biswas, B.; Willett, I.R.; Cribb, J.; Singh, B.K.; Nathanail, C.P.; Coulon, F.; Semple, K.T.; Jones, K.C.; Barclay, A.; et al. Chemical pollution: A growing peril and potential catastrophic risk to humanity. Environ. Int. 2021, 156, 106616. [Google Scholar] [CrossRef]
- Escobar, H. In Brazil, researchers struggle to fend off deepening budget cuts. Science 2017, 90, 1–2. [Google Scholar] [CrossRef]
- Segura, F.R.; Nunes, E.A.; Paniz, F.P.; Paulelli, A.C.C.; Rodrigues, G.B.; Braga, G.Ú.L.; dos Reis Pedreira Filho, W.; Barbosa, F.; Cerchiaro, G.; Silva, F.F.; et al. Potential risks of the residue from samarco’s mine dam burst (Bento Rodrigues, Brazil). Environ. Pollut. 2016, 218, 813–825. [Google Scholar] [CrossRef]
- Queiroz, H.M.; Nóbrega, G.N.; Ferreira, T.O.; Almeida, L.S.; Romero, T.B.; Santaella, S.T.; Bernardino, A.F.; Otero, X.L. The samarco mine tailing disaster: A possible time-bomb for heavy metals contamination? Sci. Total Environ. 2018, 637, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, F.; Hauser-Davis, R.A.; Soares, L.; Mazzuco, A.C.A.; Chavez Rocha, R.C.; Saint Pierre, T.D.; Saggioro, E.; Correia, F.V.; Ferreira, T.O.; Bernardino, A.F. Contamination and oxidative stress biomarkers in estuarine fish following a mine tailing disaster. PeerJ 2020, 8, e10266. [Google Scholar] [CrossRef] [PubMed]
- Carpio, I.E.M.; Franco, D.C.; Sato, M.I.Z.; Sakata, S.; Pellizari, V.H.; Ferreira Filho, S.S.; Rodrigues, D.F. Biostimulation of metal-resistant microbial consortium to remove zinc from contaminated environments. Sci. Total Environ. 2016, 550, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Bruins, M.R.; Kapil, S.; Oehme, F.W. Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 2000, 45, 198–207. [Google Scholar] [CrossRef]
- Fang, L.; Wei, X.; Cai, P.; Huang, Q.; Chen, H.; Liang, W.; Rong, X. Role of extracellular polymeric substances in Cu(II) adsorption on Bacillus Subtilis and Pseudomonas Putida. Bioresour. Technol. 2011, 102, 1137–1141. [Google Scholar] [CrossRef] [PubMed]
- Pankratov, T.A.; Tindall, B.J.; Liesack, W.; Dedysh, S.N. Mucilaginibacter paludis gen. nov., sp. nov. and Mucilaginibacter Gracilis sp. nov., pectin-, xylan and laminarin-degrading members of the family Sphingobacteriaceae from acidic sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 2007, 57, 2349–2354. [Google Scholar] [CrossRef]
- Joung, Y.; Kim, H.; Kang, H.; Lee, B.-I.; Ahn, T.S.; Joh, K. Mucilaginibacter soyangensis sp. nov., isolated from a lake. Int. J. Syst. Evol. Microbiol. 2014, 64, 413–416. [Google Scholar] [CrossRef]
- Sheu, S.Y.; Xie, Y.R.; Chen, W.M. Mucilaginibacter Limnophilus sp. Nov., Isolated from a Lake. J. Microbiol. 2019, 57, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kang, S.J.; Jung, Y.T.; Oh, T.K.; Yoon, J.H. Mucilaginibacter lutimaris sp. nov., isolated from a tidal flat sediment. Int. J. Syst. Evol. Microbiol. 2012, 62, 515–519. [Google Scholar] [CrossRef]
- Aydogan, E.L.; Busse, H.J.; Moser, G.; Müller, C.; Kämpfer, P.; Glaeser, S.P. Proposal of mucilaginibacter phyllosphaerae sp. nov. isolated from the phyllosphere of galium album. Int. J. Syst. Evol. Microbiol. 2016, 66, 4138–4147. [Google Scholar] [CrossRef]
- Zhou, Z.; Dong, Y.; Xia, X.; Wu, S.; Huang, Y.; Liao, S.; Wang, G. Mucilaginibacter terrenus sp. nov., isolated from manganese mine soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 3074–3079. [Google Scholar] [CrossRef]
- Khan, S.R.; Abbasi, M.K.; Hussan, A.U. Effect of induced soil compaction on changes in soil properties and wheat productivity under sandy loam and sandy clay loam soils: A greenhouse experiment. Commun. Soil Sci. Plant Anal. 2012, 43, 2550–2563. [Google Scholar] [CrossRef]
- An, D.S.; Yin, C.R.; Lee, S.T.; Cho, C.H. Mucilaginibacter daejeonensis sp. nov., isolated from dried rice straw. Int. J. Syst. Evol. Microbiol. 2009, 59, 1122–1125. [Google Scholar] [CrossRef]
- Matyar, F.; Kaya, A.; Dinçer, S. Antibacterial agents and heavy metal resistance in gram-negative bacteria isolated from seawater, shrimp and sediment in Iskenderun Bay, Turkey. Sci. Total Environ. 2008, 407, 279–285. [Google Scholar] [CrossRef]
- Jardine, J.L.; Abia, A.L.K.; Mavumengwana, V.; Ubomba-Jaswa, E. Phylogenetic analysis and antimicrobial profiles of cultured emerging opportunistic pathogens (phyla actinobacteria and proteobacteria) identified in hot springs. Int. J. Environ. Res. Public Health 2017, 14, 1070. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor: New York, NY, USA, 1989. [Google Scholar]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid Determination of 16S Ribosomal RNA Sequences for Phylogenetic Analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (ITOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Price Morgan, N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Bayliss, S.C.; Thorpe, H.A.; Coyle, N.M.; Sheppard, S.K.; Feil, E.J. PIRATE: A fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. GigaScience 2019, 8, 10. [Google Scholar] [CrossRef]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Eren, A.M.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L. A filtering method to generate high quality short reads using illumina paired-end technology. PLoS ONE 2013, 8, 6. [Google Scholar] [CrossRef]
- Jiang, B.; Adebayo, A.; Jia, J.; Xing, Y.; Deng, S.; Guo, L.; Liang, Y.; Zhang, D. Impacts of heavy metals and soil properties at a nigerian e-waste site on soil microbial community. J. Hazard. Mater. 2019, 362, 187–195. [Google Scholar] [CrossRef]
- Kersters, K.; de Vos, P.; Gillis, M.; Swings, J.; Vandamme, P.; Stackebrandt, E. Introduction to the proteobacteria. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 3–37. ISBN 978-0-387-30745-9. [Google Scholar]
- Bernardino, A.F.; Pais, F.S.; Oliveira, L.S.; Gabriel, F.A.; Ferreira, T.O.; Queiroz, H.M.; Mazzuco, A.C.A. Chronic trace metals effects of mine tailings on estuarine assemblages revealed by environmental DNA. PeerJ 2019, 7, e8042. [Google Scholar] [CrossRef] [PubMed]
- Domingues, V.S.; de Souza Monteiro, A.; Júlio, A.D.L.; Queiroz, A.L.L.; dos Santos, V.L. Diversity of metal-resistant and tensoactive-producing culturable heterotrophic bacteria isolated from a copper mine in Brazilian Amazonia. Sci. Rep. 2020, 10, 6171. [Google Scholar] [CrossRef]
- Dick, G.J.; Torpey, J.W.; Beveridge, T.J.; Tebo, B.M. Direct identification of a bacterial manganese(II) oxidase, the multicopper oxidase MnxG, from spores of several different marine bacillus species. Appl. Environ. Microbiol. 2008, 74, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Handtke, S.; Schroeter, R.; Jürgen, B.; Methling, K.; Schlüter, R.; Albrecht, D.; van Hijum, S.A.F.T.; Bongaerts, J.; Maurer, K.H.; Lalk, M.; et al. Bacillus Pumilus reveals a remarkably high resistance to hydrogen peroxide provoked oxidative stress. PLoS ONE 2014, 9, e85625. [Google Scholar] [CrossRef]
- Sathiyanarayanan, G.; Filippidou, S.; Junier, T.; Muñoz Rufatt, P.; Jeanneret, N.; Wunderlin, T.; Sieber, N.; Dorador, C.; Junier, P. Manganese-II oxidation and copper-II resistance in endospore forming firmicutes isolated from uncontaminated environmental sites. AIMS Environ. Sci. 2016, 3, 220–238. [Google Scholar] [CrossRef]
- Kumar, A.; Tripti; Maleva, M.; Bruno, L.B.; Rajkumar, M. Synergistic effect of ACC deaminase producing Pseudomonas sp. TR15a and siderophore producing bacillus aerophilus TR15c for enhanced growth and copper accumulation in Helianthus annuus L. Chemosphere 2021, 276, 130038. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, C.; Bakshi, U.; Mallick, I.; Mukherji, S.; Bera, B.; Ghosh, A. Genome-guided insights into the plant growth promotion capabilities of the physiologically versatile bacillus aryabhattai strain AB211. Front. Microbiol. 2017, 8, 411. [Google Scholar] [CrossRef]
- Rivas-Castillo, A.; Mejía-Escobedo, Y.; Rojas-Avelizapa, N. Study of bacillus megaterium potential application for high metal content residues biotreatment. Open J. Bacteriol. 2018, 2, 4–8. [Google Scholar] [CrossRef]
- CONAMA Resolução No 420, de 28 de Dezembro de 2009; Brazilian National Environment Council: Brasília, Brazil, 2009.
- Gomes, L.E.D.O.; Correa, L.B.; Sá, F.; Neto, R.R.; Bernardino, A.F.; Rodrigues-Neto, R.; Bernardino, A.F. The impacts of the samarco mine tailing spill on the rio doce estuary, Eastern Brazil. Mar. Pollut. Bull. 2017, 120, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.F.; de Freitas, M.B.D.; Szinwelski, N.; Vicente, N.; Medeiros, L.C.C.; Schaefer, C.E.G.R.; Dergam, J.A.; Sperber, C.F. Impacts of the samarco tailing dam collapse on metals and arsenic concentration in freshwater fish muscle from Doce river, Southeastern Brazil. Integr. Environ. Assess. Manag. 2020, 16, 622–630. [Google Scholar] [CrossRef]
- Queiroz, H.M.; Ying, S.C.; Abernathy, M.; Barcellos, D.; Gabriel, F.A.; Otero, X.L.; Nóbrega, G.N.; Bernardino, A.F.; Ferreira, T.O. Manganese: The overlooked contaminant in the world largest mine tailings dam collapse. Environ. Int. 2021, 146, 106284. [Google Scholar] [CrossRef] [PubMed]
- Husain, F.; Tang, K.; Veeranagouda, Y.; Boente, R.; Patrick, S.; Blakely, G.; Wexler, H.M. Novel large-scale chromosomal transfer in bacteroides fragilis contributes to its pan-genome and rapid environmental adaptation. Microb. Genom. 2017, 3, e000136. [Google Scholar] [CrossRef]
- Banach, A.M.; Kuźniar, A.; Grządziel, J.; Wolińska, A. Azolla Filiculoides L. as a source of metal-tolerant microorganisms. PLoS ONE 2020, 15, e0232699. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, J.S.; Singh, D.P. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 2011, 21, 214–222. [Google Scholar] [CrossRef]
- Luo, L.Y.; Xie, L.L.; Jin, D.C.; Mi, B.B.; Wang, D.H.; Li, X.F.; Dai, X.Z.; Zou, X.X.; Zhang, Z.; Ma, Y.Q.; et al. Bacterial community response to cadmium contamination of agricultural paddy soil. Appl. Soil Ecol. 2019, 139, 100–106. [Google Scholar] [CrossRef]
- Soares, R.; Trejo, J.; Lorite, M.J.; Figueira, E.; Sanjuán, J.; Castro, I.V.E. Diversity, phylogeny and plant growth promotion traits of nodule associated bacteria isolated from Lotus Parviflorus. Microorganisms 2020, 8, 499. [Google Scholar] [CrossRef]
- Singh, A.K.; Cameotra, S.S. Efficiency of lipopeptide biosurfactants in removal of petroleum hydrocarbons and heavy metals from contaminated soil. Environ. Sci. Pollut. Res. 2013, 20, 7367–7376. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Kim, M.M.; Siddiqi, M.Z.; Im, W.-T. Mucilaginibacter ginsenosidivorans sp. nov., isolated from soil of ginseng field. Curr. Microbiol. 2017, 74, 1382–1388. [Google Scholar] [CrossRef] [PubMed]
- Baik, K.S.; Park, S.C.; Kim, E.M.; Lim, C.H.; Seong, C.N. Mucilaginibacter rigui sp. nov., isolated from Wetland freshwater, and emended description of the genus Mucilaginibacter. Int. J. Syst. Evol. Microbiol. 2010, 60, 134–139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Floyd, M.M.; Tang, J.; Kane, M.; Emerson, D. Captured diversity in a culture collection: Case study of the geographic and habitat distributions of environmental isolates held at the american type culture collection. Appl. Environ. Microbiol. 2005, 71, 2813–2823. [Google Scholar] [CrossRef]
- Li, Y.P.; Carraro, N.; Yang, N.; Liu, B.; Xia, X.; Feng, R.; Saquib, Q.; Al-Wathnani, H.A.; van der Meer, J.R.; Rensing, C. Genomic islands confer heavy metal resistance in Mucilaginibacter Kameinonensis and Mucilaginibacter Rubeus isolated from a gold/copper mine. Genes 2018, 9, 573. [Google Scholar] [CrossRef] [PubMed]
- Bastiat, B.; Sauviac, L.; Picheraux, C.; Rossignol, M.; Bruand, C. Sinorhizobium meliloti sigma factors RpoE1 and RpoE4 are activated in stationary phase in response to sulfite. PLoS ONE 2012, 7, e50768. [Google Scholar] [CrossRef]
- Osman, D.; Cooke, A.; Young, T.R.; Deery, E.; Robinson, N.J.; Warren, M.J. The requirement for cobalt in vitamin B12: A paradigm for protein metalation. Biochim. Biophys. Acta 2021, 1868, 118896. [Google Scholar] [CrossRef]
- Ferrer, A.; Rivera, J.; Zapata, C.; Norambuena, J.; Sandoval, Á.; Chávez, R.; Orellana, O.; Levicán, G. Cobalamin protection against oxidative stress in the acidophilic iron-oxidizing Bacterium Leptospirillum group II CF-1. Front. Microbiol. 2016, 7, 748. [Google Scholar] [CrossRef]
- Pieck, J.C.; Hennecke, U.; Pierik, A.J.; Friedel, M.G.; Carell, T. Characterization of a new thermophilic spore photoproduct lyase from Geobacillus Stearothermophilus (SplG) with defined lesion containing DNA substrates. J. Biol. Chem. 2006, 281, 36317–36326. [Google Scholar] [CrossRef]
- Perreten, V.; Schwarz, F.V.; Teuber, M.; Levy, S.B. Mdt(A), a new efflux protein conferring multiple antibiotic resistance in Lactococcus Lactis and Escherichia Coli. Antimicrob. Agents Chemother. 2001, 45, 1109–1114. [Google Scholar] [CrossRef]
- Nishino, K.; Nikaido, E.; Yamaguchi, A. Regulation of multidrug efflux systems involved in multidrug and metal resistance of Salmonella Enterica Serovar Typhimurium. J. Bacteriol. 2007, 189, 9066–9075. [Google Scholar] [CrossRef]
- Perron, K.; Caille, O.; Rossier, C.; van Delden, C.; Dumas, J.L.; Köhler, T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 8761–8768. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Núñez-Montero, K.; Quezada-Solís, D.; Khalil, Z.G.; Capon, R.J.; Andreote, F.D.; Barrientos, L. Genomic and metabolomic analysis of antarctic bacteria revealed culture and elicitation conditions for the production of antimicrobial compounds. Biomolecules 2020, 10, 673. [Google Scholar] [CrossRef]
- Ashbolt, N.J.; Amézquita, A.; Backhaus, T.; Borriello, P.; Brandt, K.K.; Collignon, P.; Coors, A.; Finley, R.; Gaze, W.H.; Heberer, T.; et al. Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013, 121, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Patel, S.; Gibson, M.K.; Lauber, C.L.; Fierer, N.; Dantas, G.; Biology, E. HHS public access. Ann. Glob. Health 2014, 509, 612–616. [Google Scholar] [CrossRef]
- Graham, D.W.; Olivares-Rieumont, S.; Knapp, C.W.; Lima, L.; Werner, D.; Bowen, E. Antibiotic resistance gene abundances associated with waste discharges to the Almendares river near Havana, Cuba. Environ. Sci. Technol. 2011, 45, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Elena, S.F.; Lenski, R.E. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat. Rev. Genet. 2003, 4, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Fan, Z.; Lu, S.; Ma, Y.; Nie, X.; Tong, F.; Peng, X. Changes in rhizosphere bacterial communities during remediation of heavy metal-accumulating plants around the Xikuangshan mine in Southern China. Sci. Rep. 2019, 9, 1947. [Google Scholar] [CrossRef]
- Chaturvedi, K.S.; Hung, C.S.; Crowley, J.R.; Stapleton, A.E.; Henderson, J.P. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 2012, 8, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.J.; Ceri, H.; Turner, R.J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 2007, 5, 928–938. [Google Scholar] [CrossRef]
- Pal, C.; Asiani, K.; Arya, S.; Rensing, C.; Stekel, D.J.; Larsson, D.G.J.; Hobman, J.L. Metal resistance and its association with antibiotic resistance. Adv. Microb. Physiol. 2017, 70, 261–313. [Google Scholar] [CrossRef]
- Queiroz, H.M.; Ferreira, T.O.; Barcellos, D.; Nóbrega, G.N.; Antelo, J.; Otero, X.L.; Bernardino, A.F. From sinks to sources: The role of fe oxyhydroxide transformations on phosphorus dynamics in estuarine soils. J. Environ. Manag. 2021, 278, 111575. [Google Scholar] [CrossRef] [PubMed]
- Barcellos, D.; Queiroz, H.M.; Nóbrega, G.N.; de Oliveira Filho, R.L.; Santaella, S.T.; Otero, X.L.; Ferreira, T.O. Phosphorus enriched effluents increase eutrophication risks for mangrove systems in Northeastern Brazil. Mar. Pollut. Bull. 2019, 142, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Subramanian, S.; Smith, D.L. Plant endophytes promote growth and alleviate salt stress in Arabidopsis Thaliana. Sci. Rep. 2020, 10, 12740. [Google Scholar] [CrossRef]
- Dixit, R.; Malaviya, D.; Pandiyan, K.; Singh, U.B.; Sahu, A.; Shukla, R.; Singh, B.P.; Rai, J.P.; Sharma, P.K.; Lade, H.; et al. Bioremediation of heavy metals from soil and aquatic environment: An overview of principles and criteria of fundamental processes. Sustainability 2015, 7, 2189–2212. [Google Scholar] [CrossRef]
- Fan, X.; Tang, J.; Nie, L.; Huang, J.; Wang, G. High-quality-draft genome sequence of the heavy metal resistant and exopolysaccharides producing bacterium Mucilaginibacter pedocola TBZ30T 06 biological sciences 0604 genetics. Stand. Genom. Sci. 2018, 13, 34. [Google Scholar] [CrossRef]
- Tang, J.; Huang, J.; Qiao, Z.; Wang, R.; Wang, G. Mucilaginibacter pedocola sp. nov isolated from a heavy-metal-contaminated paddy field. Int. J. Syst. Evol. Microbiol. 2016, 66, 4033–4038. [Google Scholar] [CrossRef] [PubMed]
- Madhaiyan, M.; Poonguzhali, S.; Lee, J.S.; Senthilkumar, M.; Lee, K.C.; Sundaram, S. Mucilaginibacter gossypii sp. nov. and Mucilaginibacter gossypiicola sp. nov., plant-growth-promoting bacteria isolated from cotton rhizosphere soils. Int. J. Syst. Evol. Microbiol. 2010, 60, 2451–2457. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wang, F.; Huang, Y.; Zhou, M.; Gao, J.; Yan, T.; Sheng, H.; An, L. Sphingomonas sp. cra20 increases plant growth rate and alters rhizosphere microbial community structure of arabidopsis thaliana under drought stress. Front. Microbiol. 2019, 10, 1221. [Google Scholar] [CrossRef] [PubMed]
- Son, H.J.; Park, G.T.; Cha, M.S.; Heo, M.S. Solubilization of insoluble inorganic phosphates by a novel salt- and PH-tolerant Pantoea Agglomerans R-42 isolated from soybean rhizosphere. Bioresour. Technol. 2006, 97, 204–210. [Google Scholar] [CrossRef]
- Ten, L.N.; Jeon, N.Y.; Li, W.; Cho, Y.J.; Kim, M.K.; Lee, S.Y.; Rooney, A.P.; Jung, H.Y. Mucilaginibacter terrigena sp. nov. sp., a novel member of the family Sphingobacteriaceae. Curr. Microbiol. 2019, 76, 1152–1160. [Google Scholar] [CrossRef]
- Kato, S.; Hashimoto, K.; Watanabe, K. Microbial interspecies electron transfer via electric currents through conductive minerals. Proc. Natl. Acad. Sci. USA 2012, 109, 10042–10046. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Muras, A.; Romero, M.; López, M.; Tomás, M.; Otero, A. Multiple quorum quenching enzymes are active in the nosocomial pathogen acinetobacter baumannii ATCC17978. Front. Cell. Infect. Microbiol. 2018, 8, 310. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Prindle, A.; Liu, J.; Asally, M.; Ly, S.; Garcia-Ojalvo, J.; Süel, G.M. Ion channels enable electrical communication in bacterial communities. Nature 2015, 527, 59–63. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).