Variation in Genotype and DNA Methylation Patterns Based on Alcohol Use and CVD in the Korean Genome and Epidemiology Study (KoGES)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subjects and Data Source
2.2. Study Design
2.3. Definition and Measurement of Lifestyle Factors
2.4. SNP Genotypes
2.5. DNA Methylation Analysis
2.6. Statistical Analysis of Genotypes and Methylation
3. Results
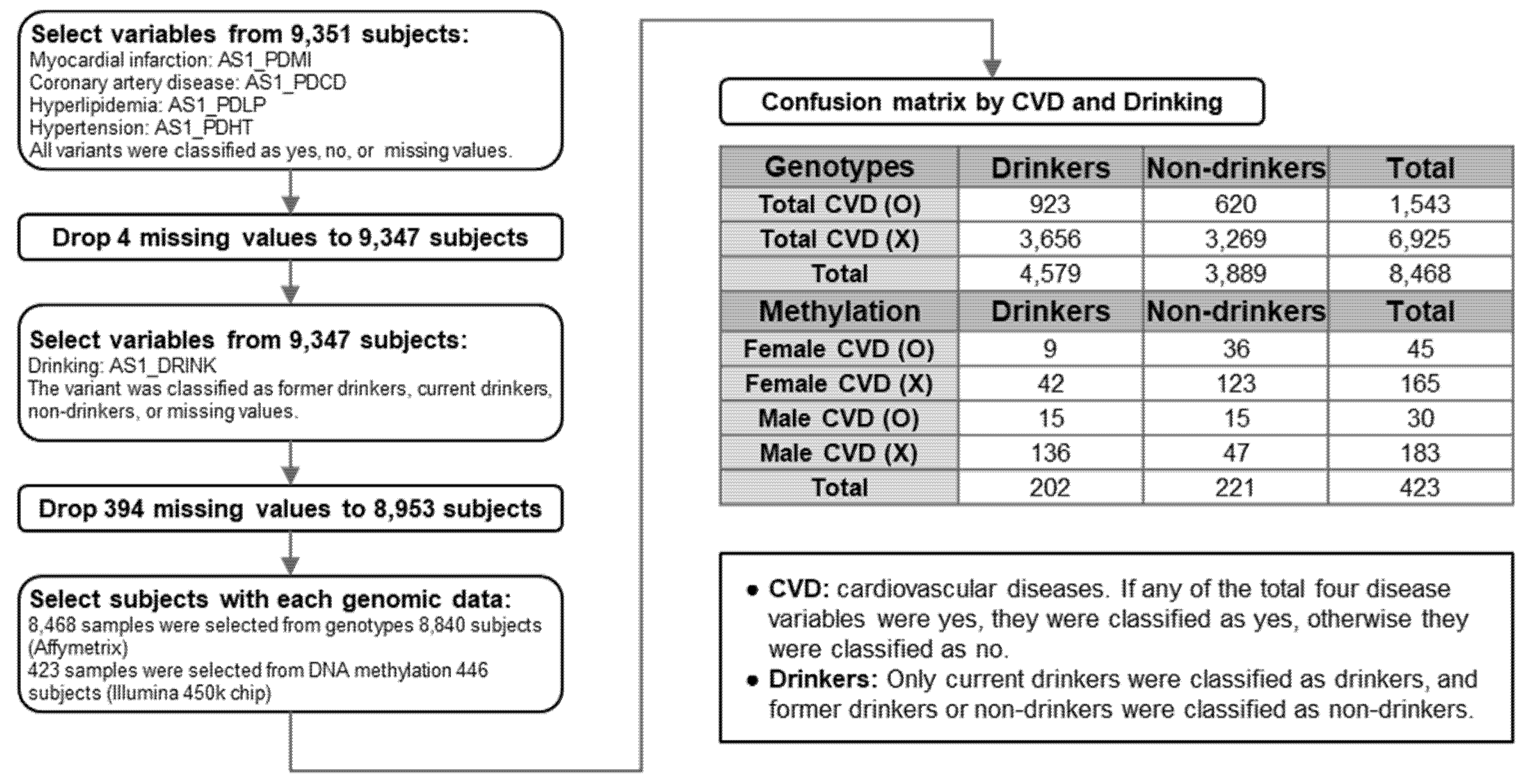
3.1. Study Processes
3.2. Identification of Genotype Patterns between the Four Selected Conditions
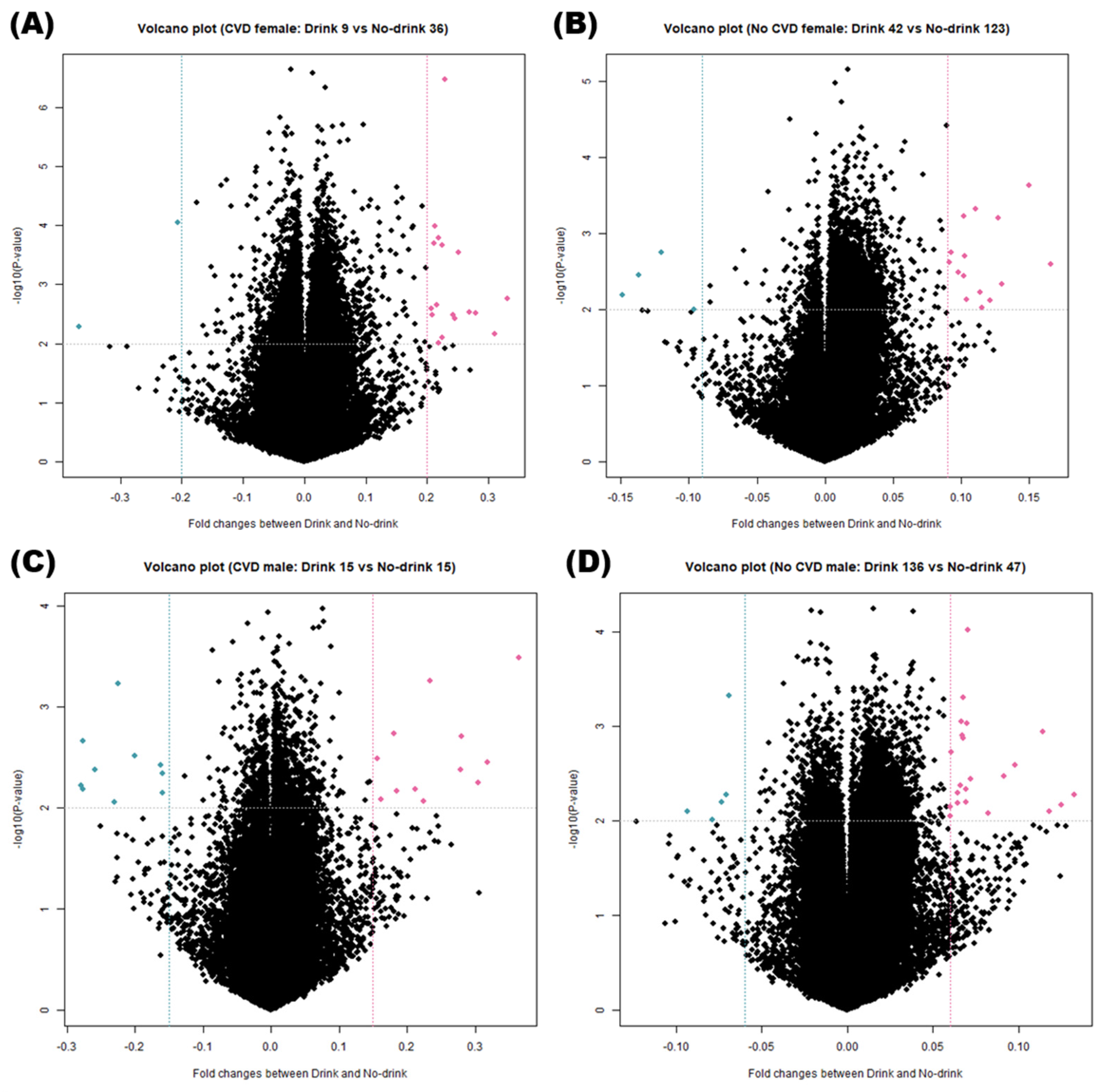
3.3. DNA Methylation Analysis of Various Groups
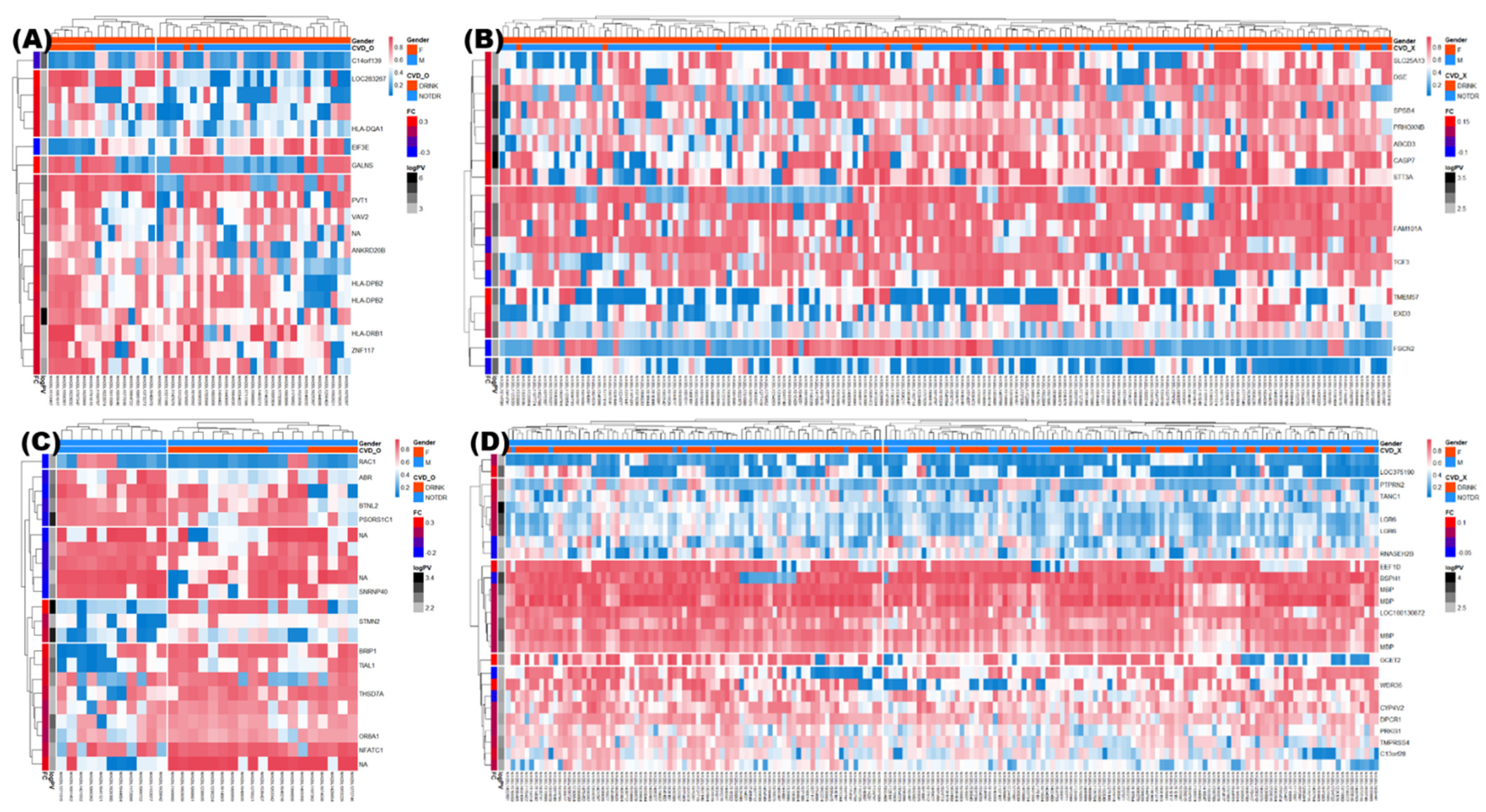
3.4. Identification of DMRs
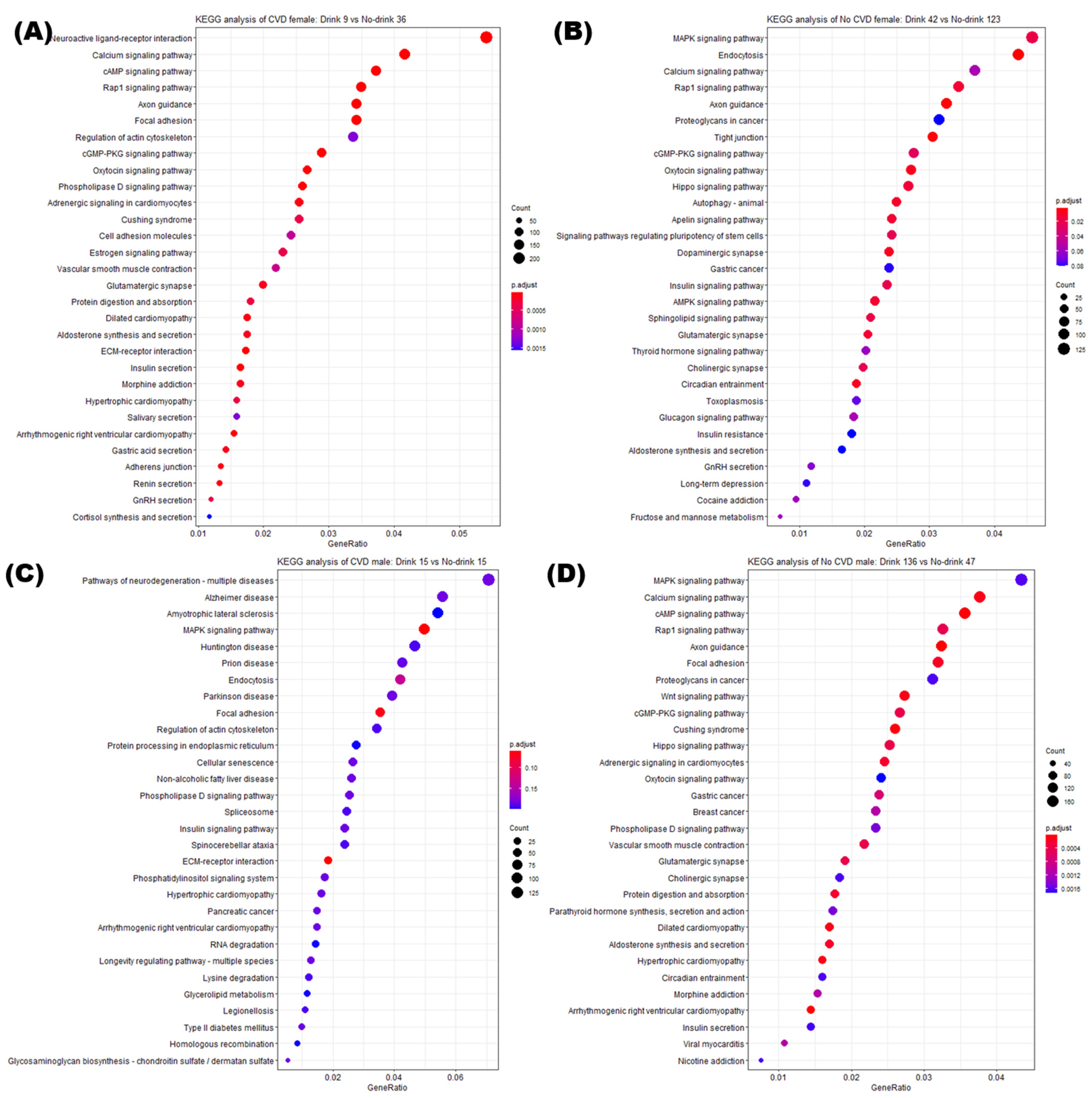
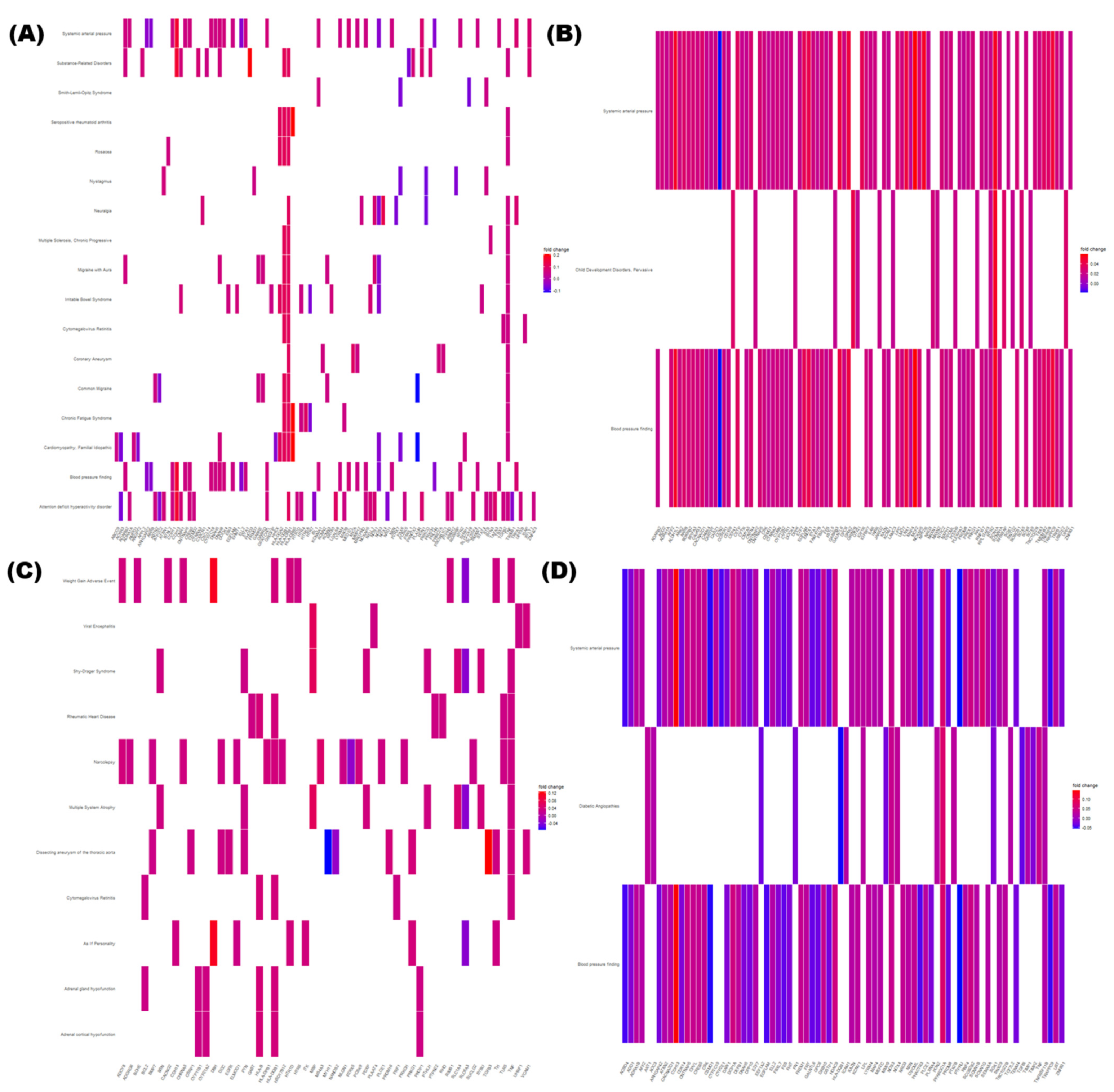
3.5. Enrichment Analysis of DMRs
3.6. Network Analysis of DMRs
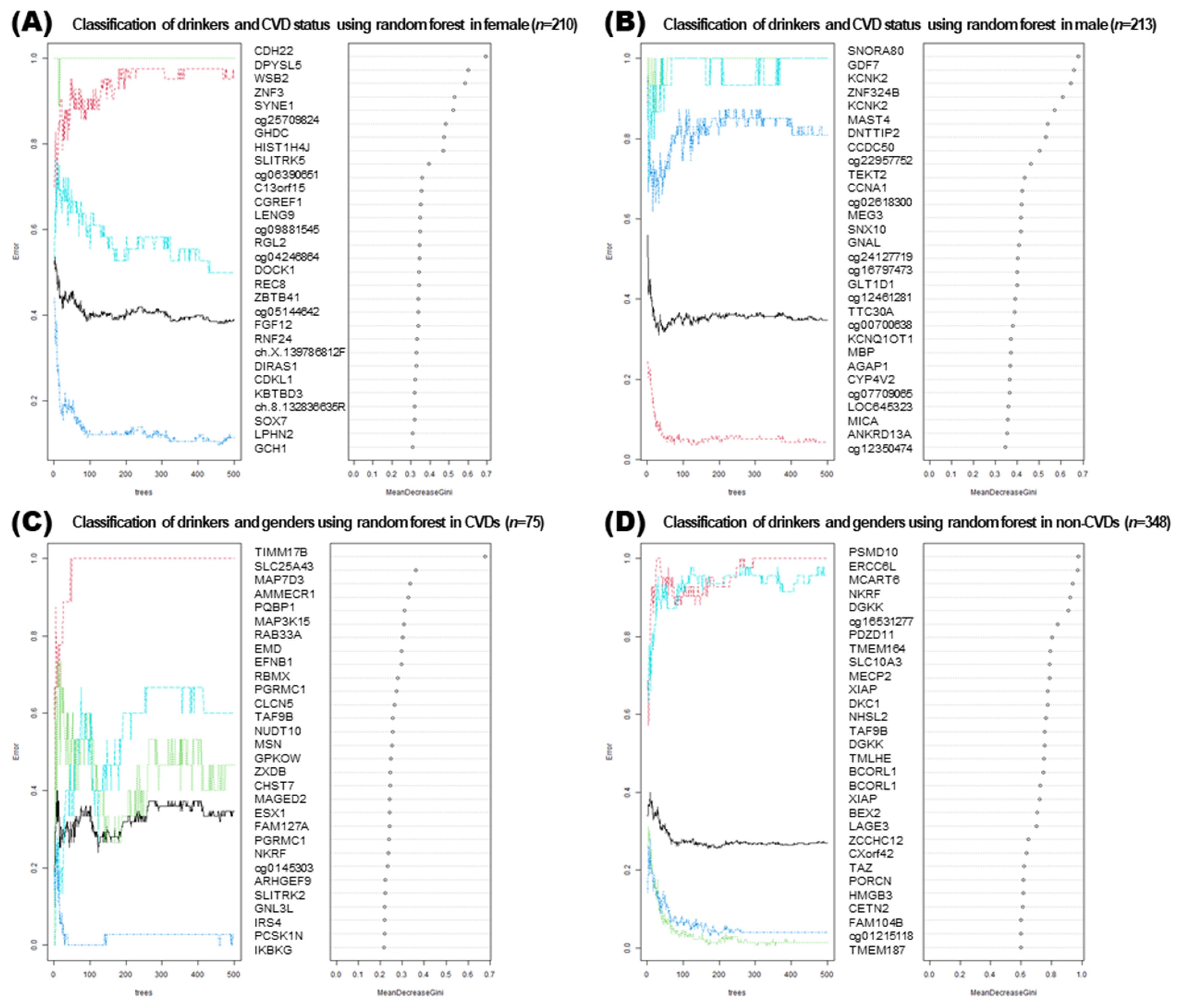
3.7. Machine Learning Approaches for Analyzing DMRs
4. Discussion
4.1. DNA Methylation in Male vs. Female
4.2. Alcohol Consumption and DNA Methylation
4.3. SNP and DNA Methylation
4.4. Limitations of This Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GBD 2015 Risk Factors Collaborators. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1659–1724. [Google Scholar] [CrossRef] [Green Version]
- Alkerwi, A.; Boutsen, M.; Vaillant, M.; Barre, J.; Lair, M.L.; Albert, A.; Guillaume, M.; Dramaix, M. Alcohol Consumption and the Prevalence of Metabolic Syndrome: A Meta-Analysis of Observational Studies. Atherosclerosis 2009, 204, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Lopez, A.D. The Global Burden of Disease: A Comprehensive Assessment of Mortality and Disability from Disease Injuries and Risk Factors in 1990 and Projected to 2002; Harvard School of Public Health: Boston, MA, USA, 1996. [Google Scholar]
- Yusuf, S.; Reddy, S.; Ounpuu, S.; Anand, S. Global burden of cardiovascular diseases: Part I: General considerations, the epidemiologic transition, risk factors, and impact of urbanization. Circulation 2001, 104, 2746–2753. [Google Scholar] [CrossRef] [Green Version]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; de Ferranti, S.; Despres, J.P.; Fullerton, H.J.; Howard, W.J.; et al. Heart disease and stroke statistics—2015 update: A report from the American Heart Association. Circulation 2015, 131, e29–e322. [Google Scholar] [CrossRef] [Green Version]
- Sipos, B.; Jirak, P.; Paar, V.; Rezar, R.; Mirna, M.; Kopp, K.; Hoppe, U.C.; Berezin, A.E.; Lichtenauer, M. Promising Novel Biomarkers in Cardiovascular Diseases. Appl. Sci. 2021, 11, 3654. [Google Scholar] [CrossRef]
- Vrablik, M.; Dlouha, D.; Todorovova, V.; Stefler, D.; Hubacek, J.A. Genetics of Cardiovascular Disease: How Far Are We from Personalized CVD Risk Prediction and Management? Int. J. Mol. Sci. 2021, 22, 4182. [Google Scholar] [CrossRef] [PubMed]
- Gheorghe, A.; Griffiths, U.; Murphy, A.; Legido-Quigley, H.; Lamptey, P.; Perel, P. The economic burden of cardiovascular disease and hypertension in low- and middle-income countries: A systematic review. BMC Public Health 2018, 18, 975. [Google Scholar] [CrossRef] [Green Version]
- Hurt, R.D.; Offord, K.P.; Croghan, I.T.; Gomez-Dahl, L.; Kottke, T.E.; Morse, R.M.; Melton, L.J., III. Mortality following inpatient addictions treatment. JAMA 1996, 275, 1097–1103. [Google Scholar] [CrossRef]
- Jarvis, C.M.; Hayman, L.L.; Braun, L.T.; Schwertz, D.W.; Ferrans, C.E.; Piano, M.R. Cardiovascular Risk Factors and Metabolic Syndrome in Alcohol- and Nicotine-Dependent Men and Women. J. Cardiovasc. Nurs. 2007, 22, 429–435. [Google Scholar] [CrossRef]
- Harada, S.; Agarwal, D.P.; Goedde, H.W.; Tagaki, S.; Ishikawa, B. Possible Protective Role Against Alcoholism for Aldehyde Dehydrogenase Isozyme Deficiency in Japan. Lancet 1982, 2, 827. [Google Scholar] [CrossRef]
- Cook, W.K.; Tam, C.C.; Luczak, S.E.; Kerr, W.C.; Mulia, N.; Lui, C.; Li, L. Alcohol Consumption, Cardiovascular-Related Conditions, and ALDH2*2 Ethnic Group Prevalence in Asian Americans. Alcohol. Clin. Exp. Res. 2021, 45, 418–428. [Google Scholar] [CrossRef]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Agha, G.; Baccarelli, A.A. The Role of DNA Methylation in Cardiovascular Risk and Disease: Methodological Aspects, Study Design, and Data Analysis for Epidemiological Studies. Circ. Res. 2016, 118, 119–131. [Google Scholar] [CrossRef] [Green Version]
- Prasher, D.; Greenway, S.C.; Singh, R.B. The impact of epigenetics on cardiovascular disease. Biochem. Cell. Biol. 2020, 98, 12–22. [Google Scholar] [CrossRef]
- Kim, M.; Long, T.I.; Arakawa, K.; Wang, R.; Yu, M.C.; Laird, P.W. DNA methylation as a biomarker for cardiovascular disease risk. PLoS ONE 2010, 5, e9692. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Kumar, J.; Garg, G.; Kumar, A.; Patowary, A.; Karthikeyan, G.; Ramakrishnan, L.; Brahmachari, V.; Sengupta, S. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 2008, 27, 357–365. [Google Scholar] [CrossRef]
- Kaminsky, Z.A.; Tang, T.; Wang, S.C.; Ptak, C.; Oh, G.H.; Wong, A.H.; Feldcamp, L.A.; Virtanen, C.; Halfvarson, J.; Tysk, C.; et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat. Genet. 2009, 41, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Marioni, R.E.; Hedman, Å.K.; Pfeiffer, L.; Tsai, P.-C.; Reynolds, L.M.; Just, A.C.; Duan, Q.; Boer, C.G.; Tanaka, T.; et al. A DNA methylation biomarker of alcohol consumption. Mol. Psychiatry 2018, 23, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Bönsch, D.; Lenz, B.; Fiszer, R.; Frieling, H.; Kornhuber, J.; Bleich, S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J. Neural Transm. 2006, 113, 1299–1304. [Google Scholar] [CrossRef] [PubMed]
- Bönsch, D.; Lenz, B.; Reulbach, U.; Kornhuber, J.; Bleich, S. Homocysteine associated genomic DNA hypermethylation in patients with chronic alcoholism. J. Neural Transm. 2004, 111, 1611–1616. [Google Scholar] [CrossRef]
- Huan, T.; Joehanes, R.; Song, C.; Peng, F.; Guo, Y.; Mendelson, M.; Yao, C.; Liu, C.; Ma, J.; Richard, M.; et al. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat. Commun. 2019, 10, 4267. [Google Scholar] [CrossRef] [Green Version]
- Monti, M.C.; Lonsdale, J.T.; Montomoli, C.; Montross, R.; Schlag, E.; Greenberg, D.A. Familial risk factors for microvascular complications and differential male-female risk in a large cohort of American families with type 1 diabetes. J. Clin. Endocrinol. Metab. 2007, 92, 4650–4655. [Google Scholar] [CrossRef]
- Cotton, A.M.; Price, E.M.; Jones, M.J.; Balaton, B.P.; Kobor, M.S.; Brown, C.J. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 2015, 24, 1528–1539. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Han, B.G.; KoGES Group. Cohort profile: The Korean genome and epidemiology study (KoGES) consortium. Int. J. Epidemiol. 2017, 46, e20. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, J.; Lim, J.E.; Oh, B.; Won, S.; Kim, M.K. Genome Wide Interaction Study of Single-Nucleotide Polymorphisms and Alcohol Consumption on Blood Pressure the Ansan and Ansung Study of the Korean Genome and Epidemiology Study (KoGES). Genet. Epidemiol. 2020, 44, 300–310. [Google Scholar] [CrossRef]
- Song, Y.; Choi, J.E.; Kwon, Y.J.; Chang, H.J.; Kim, J.O.; Park, D.H.; Park, J.M.; Kim, S.J.; Lee, J.W.; Hong, K.W. Identification of Susceptibility Loci for Cardiovascular Disease in Adults with Hypertension, Diabetes, and Dyslipidemia. J. Transl. Med. 2021, 19, 85. [Google Scholar] [CrossRef]
- Azman, N.F.; Abdullah, W.Z.; Hanafi, S.; Diana, R.; Bahar, R.; Johan, M.F.; Zilfalil, B.A.; Hassan, R. Genetic polymorphisms of HbE/beta thalassemia related to clinical presentation: Implications for clinical diversity. Ann. Hematol. 2020, 99, 729–735. [Google Scholar] [CrossRef]
- Rakyan, V.K.; Down, T.A.; Balding, D.J.; Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011, 12, 529–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, A.J.; Stathaki, E.; Migliavacca, E.; Brahmachary, M.; Montgomery, S.B.; Dupre, Y.; Antonarakis, S.E. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011, 21, 1592–1600. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, Y.; Zhang, J.; Tang, X.; Qian, Y.; Gao, P.; Zhu, D. Association of a functional single- nucleotide polymorphism in the ALDH2 gene with essential hypertension depends on drinking behavior in a Chinese Han population. J. Hum. Hypertens. 2013, 27, 181–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, C.G.; Grimm, S.A.; Morgan, D.L.; Bushel, P.R.; Bennett, B.D.; NISC Comparative Sequencing Program; Roberts, J.D.; Tyson, F.L.; Merrick, B.A.; Wade, P.A. Dosage compensation and DNA methylation landscape of the X chromosome in mouse liver. Sci. Rep. 2018, 8, 10138. [Google Scholar] [CrossRef]
- Liu, J.; Morgan, M.; Hutchison, K.; Calhoun, V.D. A study of the influence of sex on genome wide methylation. PLoS ONE 2010, 5, e10028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousefi, P.; Huen, K.; Davé, V.; Barcellos, L.; Eskenazi, B.; Holland, N. Sex differences in DNA methylation assessed by 450K BeadChip in newborns. BMC Genom. 2015, 16, 911. [Google Scholar] [CrossRef] [Green Version]
- Ward-Caviness, C.K.; Agha, G.; Chen, B.H.; Pfeiffer, L.; Wilcon, R.; Wolf, P.; Gieger, C.; Schwartz, J.; Vokonas, P.S.; Hou, L.; et al. Analysis of repeated leukocyte DNA methylation assessments reveals persistent epigenetic alterations after an incident myocardial infarction. Clin. Epigenet. 2018, 10, 161. [Google Scholar] [CrossRef]
- Stephenson, M.; Bollepalli, S.; Cazaly, E.; Salvatore, J.E.; Barr, P.; Rose, R.J.; Dick, D.; Kaprio, J.; Ollikainen, M. Associations of Alcohol Consumption with Epigenome-Wide DNA Methylation and Epigenetic Age Acceleration: Individual-Level and Co-twin Comparison Analyses. Alcohol Clin. Exp. Res. 2021, 45, 318–328. [Google Scholar] [CrossRef]
- Rosen, A.D.; Robertson, K.D.; Hlady, R.A.; Muench, C.; Lee, J.; Philibert, R.; Horvath, S.; Kaminsky, Z.A.; Lohoff, F.W. DNA methylation age is accelerated in alcohol dependence. Transl. Psychiatry 2018, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Hu, Y.; Zang, T.; Wang, Y. Integrate GWAS, eQTL, and mQTL data to identify Alzheimer’s disease-related genes. Front. Genet. 2019, 10, 1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciuculete, D.M.; Boström, A.E.; Voisin, S.; Philipps, H.; Titova, O.E.; Bandstein, M.; Nikontovic, L.; Williams, M.J.; Mwinyi, J.; Schiöth, H.B. A methylome-wide mQTL analysis reveals associations of methylation sites with GAD1 and HDAC3 SNPs and a general psychiatric risk score. Transl. Psychiatry 2017, 7, e1002. [Google Scholar] [CrossRef] [PubMed]
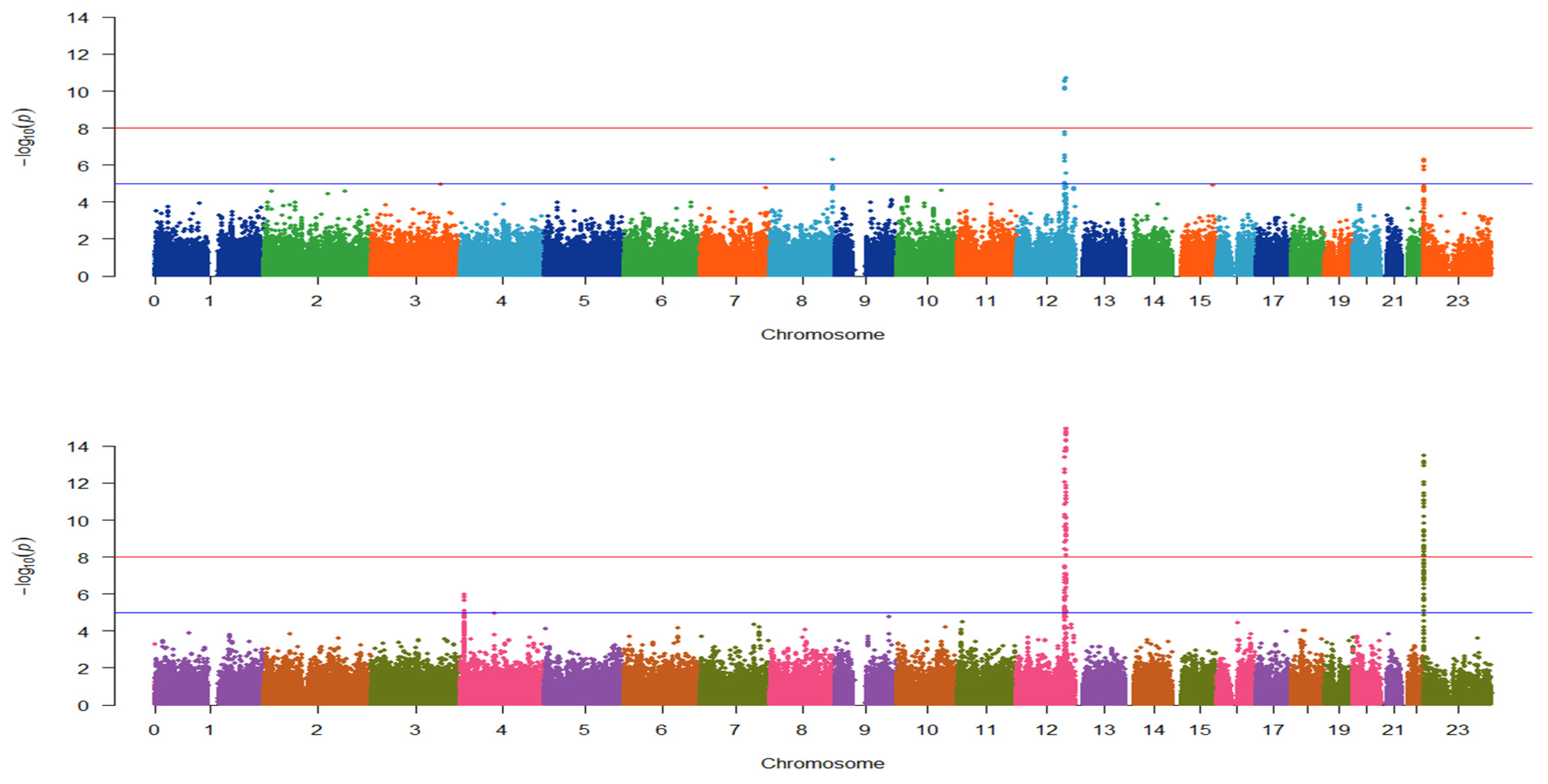
| Class | Location (hg38) | dbSNP ID | Gene | Odds Ratio | p-Value |
|---|---|---|---|---|---|
| With CVD | chr12:112207597 | rs2074356 | HECTD4: Intron Variant | 0.2402 | 1.49 × 10−26 |
| With CVD | chr12:112379979 | rs11066280 | HECTD4: Intron Variant | 0.2919 | 1.15 × 10−24 |
| With CVD | chr12:110976657 | rs12229654 | None | 0.308 | 2.01 × 10−20 |
| Without CVD | chr12:112207597 | rs2074356 | HECTD4: Intron Variant | 0.3013 | 1.41 × 10−107 |
| Without CVD | chr12:112379979 | rs11066280 | HECTD4: Intron Variant | 0.3326 | 1.95 × 10−106 |
| Without CVD | chr12:110976657 | rs12229654 | None | 0.3689 | 1.43 × 10−75 |
| Variable | CVDs (n = 75) | Non-CVDs (n = 348) | ||||
|---|---|---|---|---|---|---|
| Never Drinkers | Drinkers | p-Value | Never Drinkers | Drinkers | p-Value | |
| Sex | 15/36/29.41 | 15/9/62.5 | 0.011 | 47/123/27.65 | 136/42/76.4 | <0.001 |
| Age | 56.75 ± 7.63 | 53.62 ± 6.97 | <0.001 | 53.12 ± 8.37 | 49.57 ± 7.95 | <0.001 |
| BMI | 26.58 ± 3.05 | 25.5 ± 3.27 | <0.001 | 23.91 ± 3.21 | 24.53 ± 3.39 | <0.001 |
| Area | 34/17/66.67 | 14/10/58.33 | 0.607 | 102/68/60.00 | 72/106/40.45 | 0.001 |
| Education | 37/13/74.00 | 11/13/45.83 | 0.022 | 112/58/65.88 | 68/108/38.64 | <0.001 |
| Income | 33/16/67.35 | 8/16/33.33 | 0.011 | 97/69/58.43 | 70/108/39.33 | 0.001 |
| Exercise1 | 33/17/66.00 | 13/11/54.17 | 0.443 | 114/55/67.46 | 100/77/56.50 | 0.046 |
| Exercise2 | 12/37/24.49 | 7/17/29.17 | 0.778 | 43/125/25.60 | 35/141/19.89 | 0.246 |
| Exercise3 | 20/29/40.82 | 9/15/37.50 | 1.000 | 55/113/32.74 | 46/129/26.29 | 0.195 |
| Exercise4 | 37/10/78.72 | 19/5/79.17 | 1.000 | 120/47/71.86 | 127/48/72.57 | 0.904 |
| Exercise5 | 32/16/66.67 | 18/6/75 | 0.591 | 111/58/65.68 | 125/52/70.62 | 0.356 |
| Myocardial infarction | 1/50/1.96 | 0/24/0 | <0.001 | 0/170/0 | 0/178/0 | 1.000 |
| Coronary artery disease | 3/48/5.88 | 3/21/12.50 | 0.377 | 0/170/0 | 0/178/0 | 1.000 |
| Hyperlipidemia | 3/48/5.88 | 4/20/16.67 | 0.201 | 0/170/0 | 0/178/0 | 1.000 |
| High blood pressure | 45/6/88.24 | 20/4/83.33 | 0.717 | 0/170/0 | 0/178/0 | 1.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, M.; Ahn, Y.-S.; Chang, S.-J.; Kim, C.-B.; Jeong, K.S.; Koh, S.-B.; Gim, J.-A. Variation in Genotype and DNA Methylation Patterns Based on Alcohol Use and CVD in the Korean Genome and Epidemiology Study (KoGES). Genes 2022, 13, 172. https://doi.org/10.3390/genes13020172
Jung M, Ahn Y-S, Chang S-J, Kim C-B, Jeong KS, Koh S-B, Gim J-A. Variation in Genotype and DNA Methylation Patterns Based on Alcohol Use and CVD in the Korean Genome and Epidemiology Study (KoGES). Genes. 2022; 13(2):172. https://doi.org/10.3390/genes13020172
Chicago/Turabian StyleJung, Myoungjee, Yeon-Soon Ahn, Sei-Jin Chang, Chun-Bae Kim, Kyoung Sook Jeong, Sang-Baek Koh, and Jeong-An Gim. 2022. "Variation in Genotype and DNA Methylation Patterns Based on Alcohol Use and CVD in the Korean Genome and Epidemiology Study (KoGES)" Genes 13, no. 2: 172. https://doi.org/10.3390/genes13020172
APA StyleJung, M., Ahn, Y.-S., Chang, S.-J., Kim, C.-B., Jeong, K. S., Koh, S.-B., & Gim, J.-A. (2022). Variation in Genotype and DNA Methylation Patterns Based on Alcohol Use and CVD in the Korean Genome and Epidemiology Study (KoGES). Genes, 13(2), 172. https://doi.org/10.3390/genes13020172








