Identification of Differentially Expressed miRNAs in Porcine Adipose Tissues and Evaluation of Their Effects on Feed Efficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and RNA Isolation
2.2. Small RNA Library Construction and Sequencing
2.3. Analysis of miRNA Sequencing Data
2.4. Identification of DEMs and Their qRT-PCR Validation
2.5. Prediction of miRNA Target Gene, Gene Ontology (GO) and KEGG Pathway Enrichment Analyses
3. Results
3.1. Characterization of miRNA-Sequencing Data
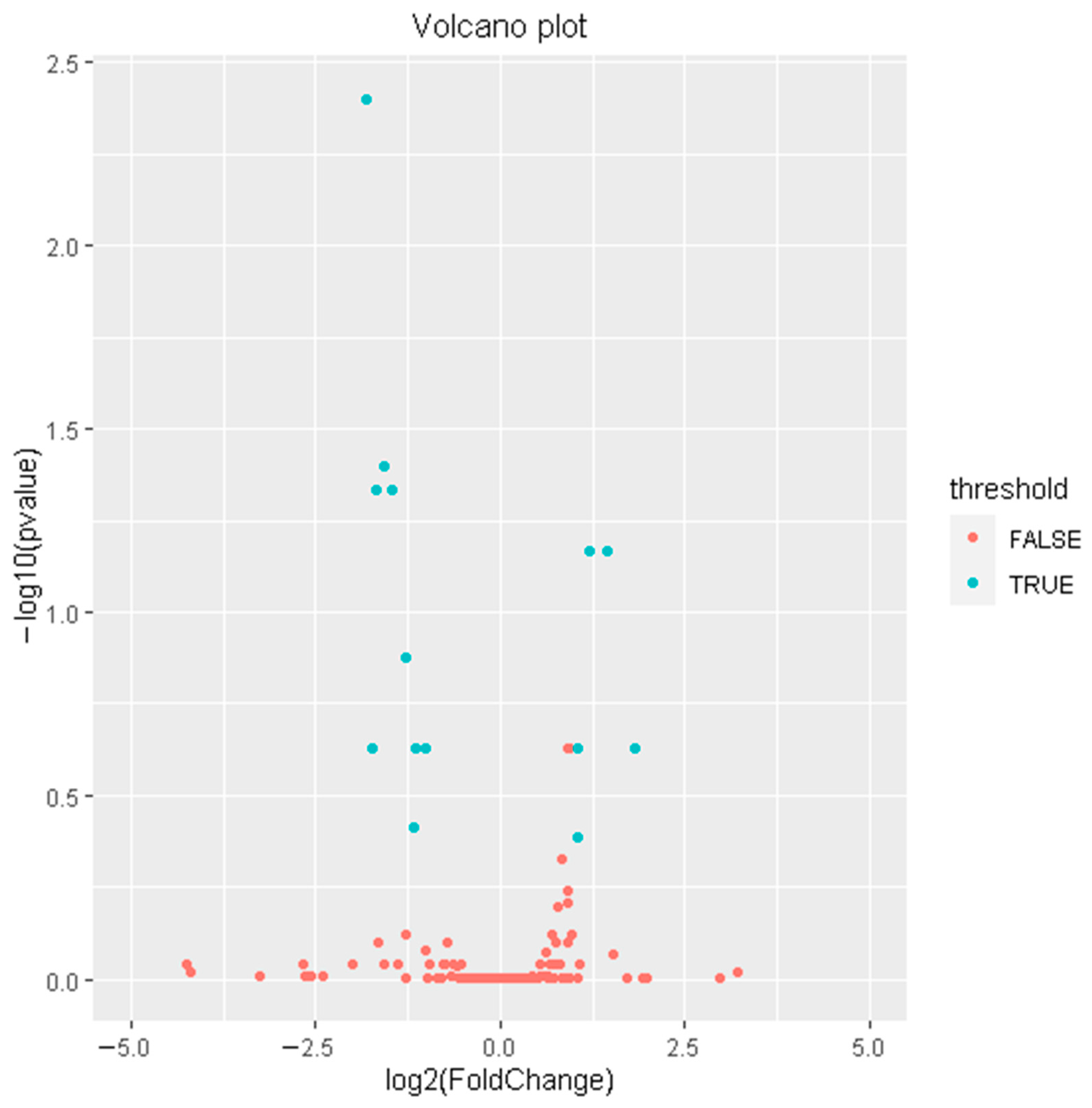
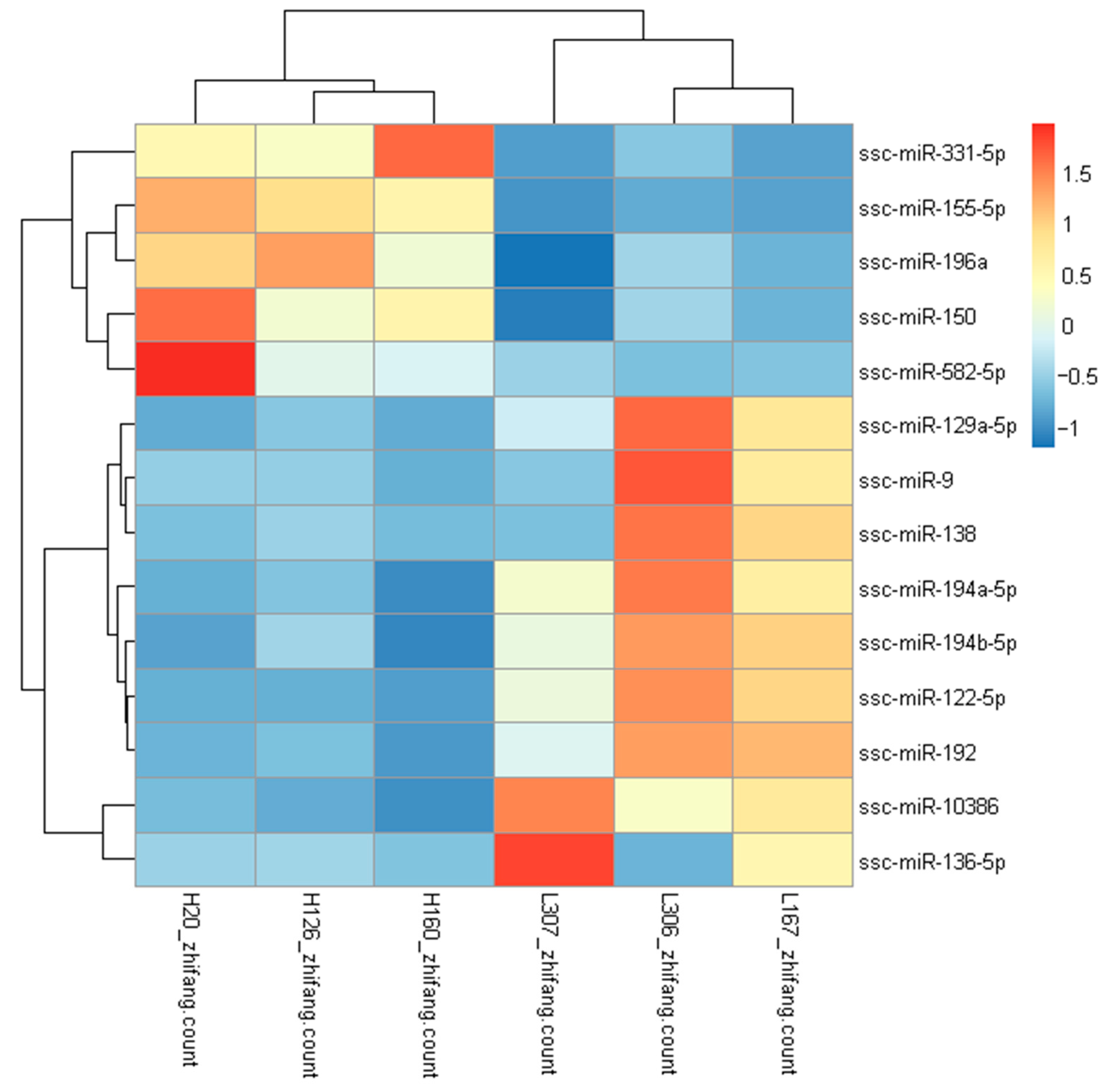
3.2. Differentially Expressed miRNAs between High-FE and Low-FE Pigs
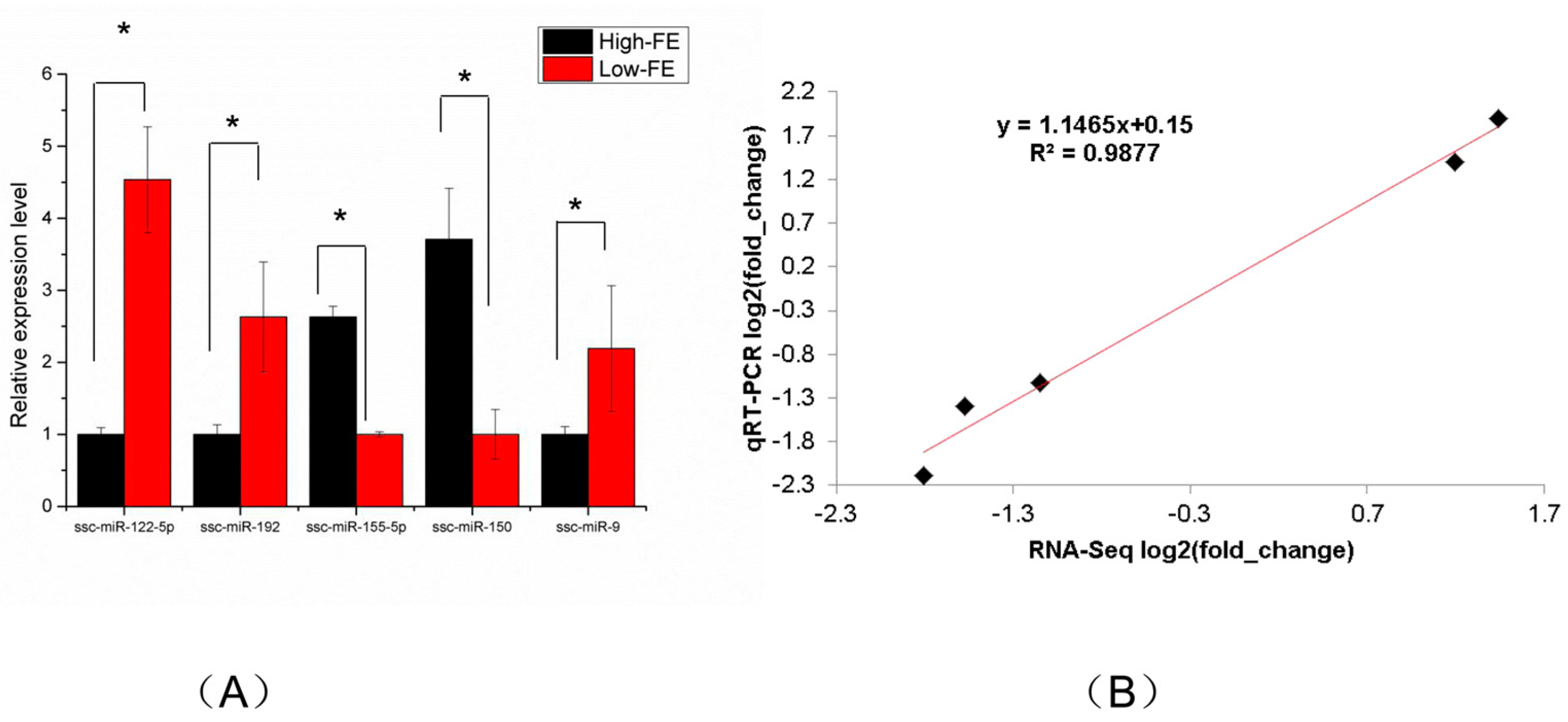
3.3. Verification of miRNA Sequencing Data by qRT-PCR
3.4. Target-Gene Prediction of DEMs
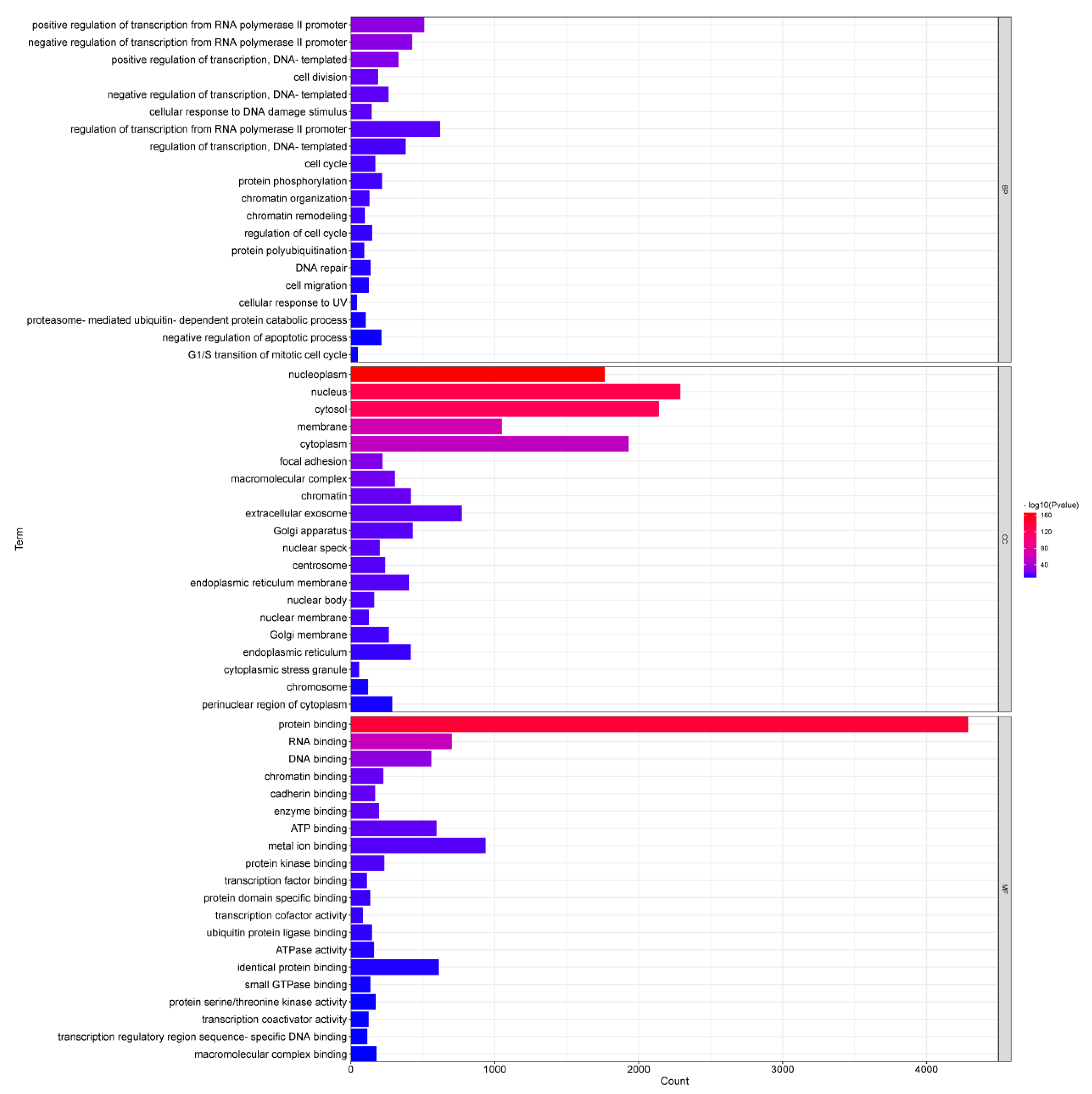
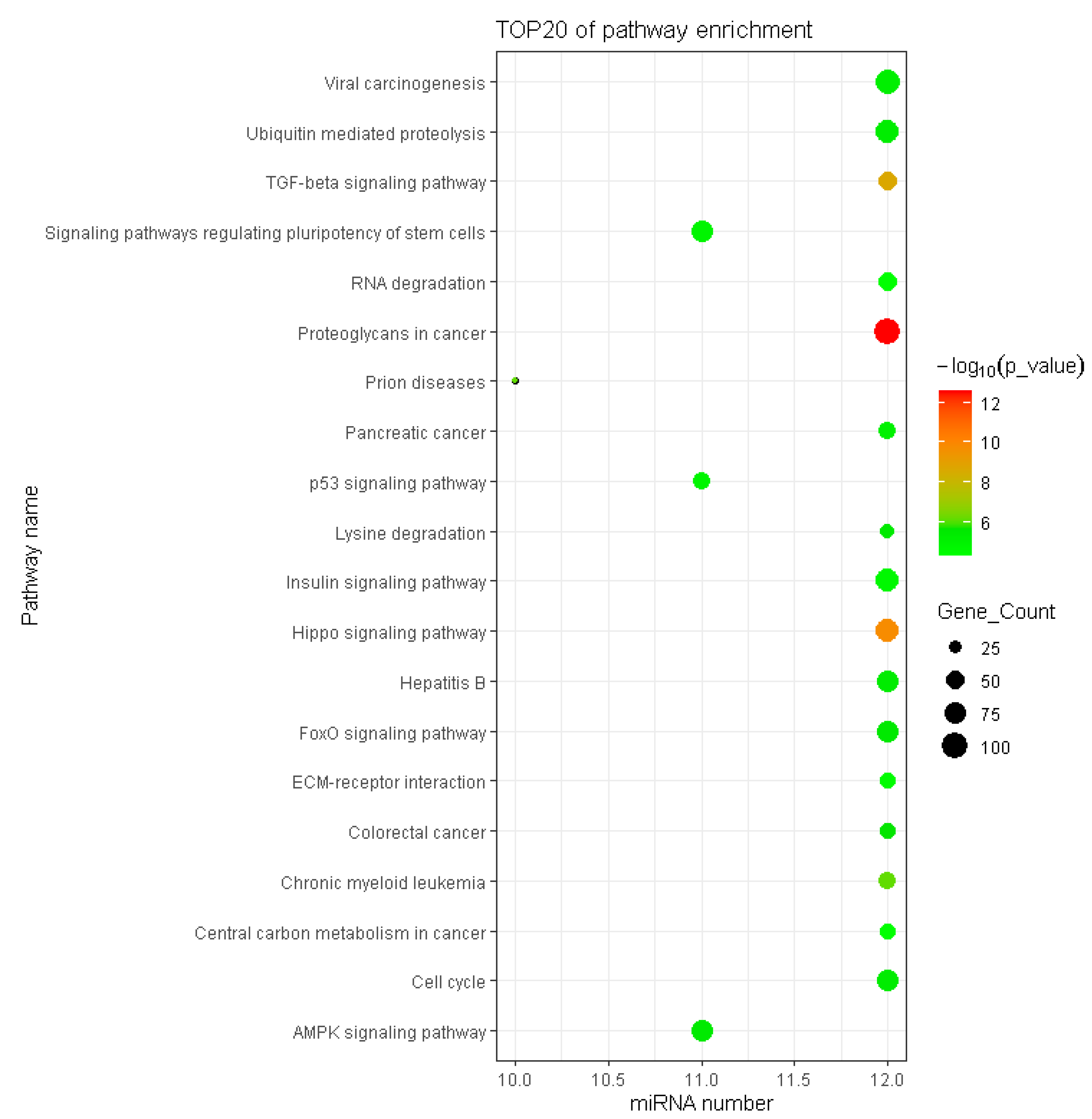
3.5. Functional Enrichment Analysis of DEMs in Adipose Tissues
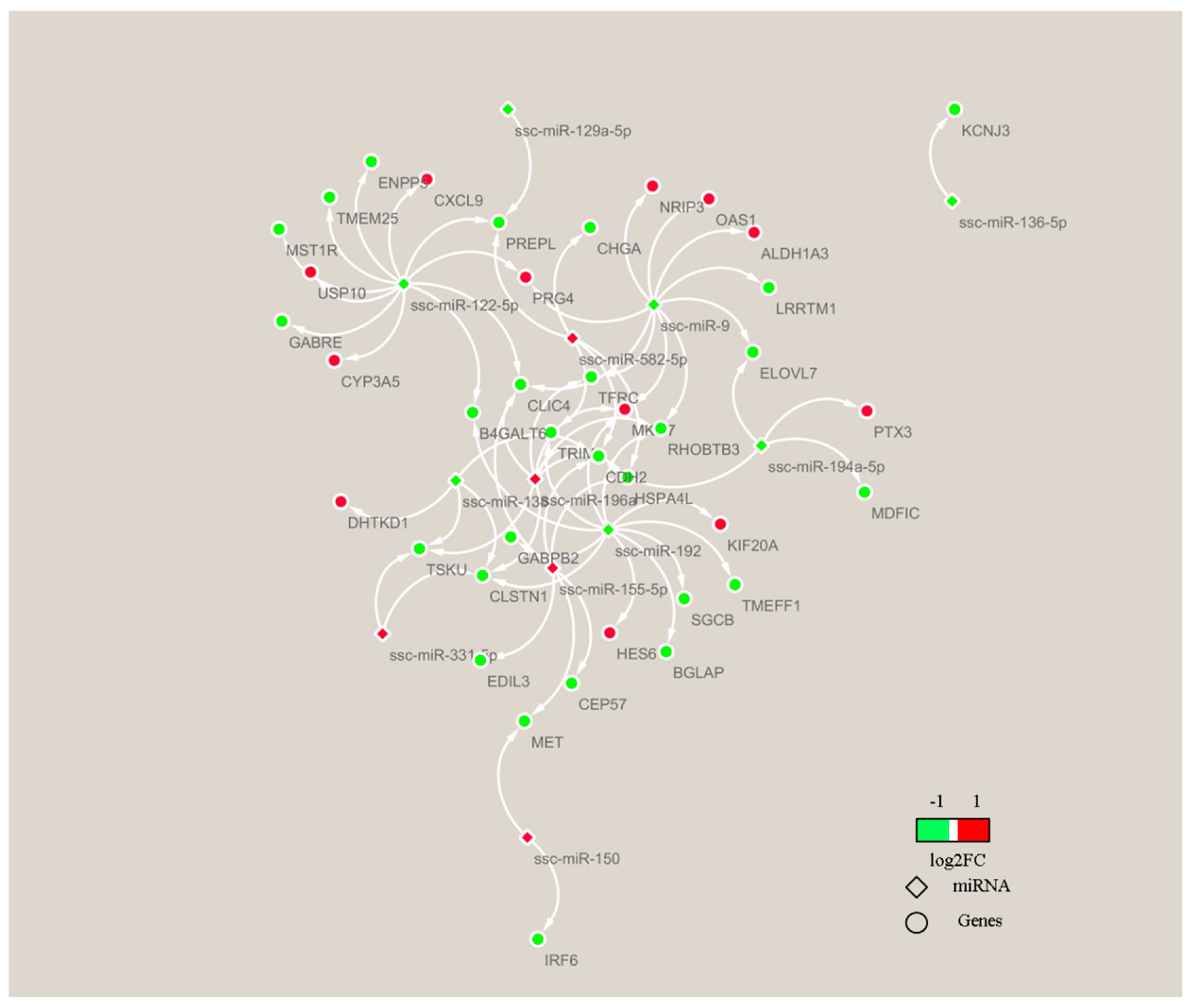
3.6. miRNA–mRNA Association Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hume, D.A.; Whitelaw, C.B.A.; Archibald, A.L. Foresight project on global food and farming futures the future of animal production: Improving productivity and sustainability. J. Anim. Sci. 2011, 149, 9–16. [Google Scholar]
- Patience, J.F.; Rossoni-Serao, M.C.; Gutierrez, N.A. A review of feed efficiency in swine: Biology and application. J. Anim. Sci. Biotechnol. 2015, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Koch, R.L.A.S.; Chambers, D.K.E.G. Efficiency of feed use in beef cattle. J. Anim. Sci. 1963, 22, 486–494. [Google Scholar] [CrossRef]
- Kennedy, B.W.; van der Werf, J.H.; Meuwissen, T.H. Genetic and statistical properties of residual feed intake. J. Anim. Sci. 1993, 71, 3239–3250. [Google Scholar] [CrossRef]
- Herd, R.M.; Arthur, P.F. Physiological basis for residual feed intake. J. Anim. Sci. 2009, 87, 64–71. [Google Scholar] [CrossRef]
- Zhen-Qiang, X.U.; Chen, J.; Zhang, Y.; Cong-Liang, J.I.; Zhang, D.X.; Zhang, X.Q. Determination of residual feed intake and its associations with single nucleotide polymorphism in chickens. J. Integr. Agric. 2014, 13, 148–157. [Google Scholar]
- Montagne, L.; Gilbert, H.; Muller, N.; Le Floc’h, N. Physiological response to the weaning in two pig lines divergently selected for residual feed intake. J. Anim. Physiol. Anim. Nutr. 2021, 106, 802–812. [Google Scholar] [CrossRef]
- Delpuech, E.; Aliakbari, A.; Labrune, Y.; Feve, K.; Billon, Y.; Gilbert, H.; Riquet, J. Identification of genomic regions affecting production traits in pigs divergently selected for feed efficiency. Genet. Sel. Evol. 2021, 53, 49. [Google Scholar] [CrossRef]
- Faure, J.; Lefaucheur, L.; Bonhomme, N.; Ecolan, P.; Meteau, K.; Coustard, S.M.; Kouba, M.; Gilbert, H.; Lebret, B. Consequences of divergent selection for residual feed intake in pigs on muscle energy metabolism and meat quality. Meat Sci. 2013, 93, 37–45. [Google Scholar] [CrossRef]
- Gilbert, H.; Alaïn, S.; Sellier, P.; Lagant, H.; Brossard, L. Relations génétiques entre efficacité alimentaire et cinétiques de croissance et d’ingestion chez le porc Large White. Journées Rech. Porc. 2009, 41, 1–6. [Google Scholar]
- Le Naou, T.; Le Floc’h, N.; Louveau, I.; Gilbert, H.; Gondret, F. Metabolic changes and tissue responses to selection on residual feed intake in growing pigs. J. Anim. Sci. 2012, 90, 4771–4780. [Google Scholar] [CrossRef] [PubMed]
- Barea, R.; Dubois, S.; Gilbert, H.; Sellier, P.; van Milgen, J.; Noblet, J. Energy utilization in pigs selected for high and low residual feed intake. J. Anim. Sci. 2010, 88, 2062–2072. [Google Scholar] [CrossRef] [PubMed]
- Renaudeau, D.; Gourdine, J.L.; Fleury, J.; Ferchaud, S.; Billon, Y.; Noblet, J.; Gilbert, H. Selection for residual feed intake in growing pigs: Effects on sow performance in a tropical climate. J. Anim. Sci. 2014, 92, 3568–3579. [Google Scholar] [CrossRef] [PubMed]
- Labussière, E.; Dubois, S.; Gilbert, H.; Thibault, J.N.; Le Floc’h, N.; Noblet, J.; Van Milgen, J. Effect of inflammation stimulation on energy and nutrient utilization in piglets selected for low and high residual feed intake. Animal 2015, 9, 1653–1661. [Google Scholar] [CrossRef]
- Vincent, A.; Louveau, I.; Gondret, F.; Trefeu, C.; Gilbert, H.; Lefaucheur, L. Divergent selection for residual feed intake affects the transcriptomic and proteomic profiles of pig skeletal muscle. J. Anim. Sci. 2015, 93, 2745–2758. [Google Scholar] [CrossRef]
- Grubbs, J.K.; Fritchen, A.N.; Huff-Lonergan, E.; Dekkers, J.C.; Gabler, N.K.; Lonergan, S.M. Divergent genetic selection for residual feed intake impacts mitochondria reactive oxygen species production in pigs. J. Anim. Sci. 2013, 91, 2133–2140. [Google Scholar] [CrossRef]
- Carrington, J.C.; Ambros, V. Role of microRNAs in plant and animal development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Shen, L.; Gan, M.; Chen, L.; Zhao, Y.; Niu, L.; Tang, G.; Jiang, Y.; Zhang, T.; Zhang, S.; Zhu, L. miR-152 targets pyruvate kinase to regulate the glycolytic activity of pig skeletal muscles and affects pork quality. Meat Sci. 2022, 185, 108707. [Google Scholar] [CrossRef]
- Hu, M.; Kuang, R.; Guo, Y.; Ma, R.; Hou, Y.; Xu, Y.; Qi, X.; Wang, D.; Zhou, H.; Xiong, Y.; et al. Epigenomics analysis of miRNA cis-regulatory elements in pig muscle and fat tissues. Genomics 2022, 114, 110276. [Google Scholar] [CrossRef]
- Herrera-Uribe, J.; Zaldívar-López, S.; Aguilar, C.; Entrenas-García, C.; Bautista, R.; Claros, M.G.; Garrido, J.J. Study of microRNA expression in Salmonella Typhimurium-infected porcine ileum reveals miR-194a-5p as an important regulator of the TLR4-mediated inflammatory response. Vet. Res. 2022, 53, 35. [Google Scholar] [CrossRef]
- De Oliveira, P.S.; Coutinho, L.L.; Tizioto, P.C.; Cesar, A.S.; de Oliveira, G.B.; Diniz, W.J.D.S.; De Lima, A.O.; Reecy, J.M.; Mourão, G.B.; Zerlotini, A.; et al. An integrative transcriptome analysis indicates regulatory mRNA-miRNA networks for residual feed intake in Nelore cattle. Sci. Rep. 2018, 8, 17072. [Google Scholar] [CrossRef] [PubMed]
- Al-Husseini, W.; Chen, Y.; Gondro, C.; Herd, R.M.; Gibson, J.P.; Arthur, P.F. Characterization and profiling of liver microRNAs by RNA-sequencing in cattle divergently selected for residual feed intake. Asian-Australas. J. Anim. Sci. 2016, 29, 1371–1382. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Hou, Y.; Wu, H.; Miao, Y.; Li, X.; Cao, J.; Brameld, J.; Parr, T.; Zhao, S. Transcriptome analysis of mRNA and miRNA in skeletal muscle indicates an important network for differential Residual Feed Intake in pigs. Sci. Rep. 2015, 5, 11953. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Fu, C.; Liao, M.; Fang, F. Differences in liver microRNA profiling in pigs with low and high feed efficiency. J. Anim. Sci. Technol. 2022, 64, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Romacho, T.; Elsen, M.; Rohrborn, D.; Eckel, J. Adipose tissue and its role in organ crosstalk. Acta Physiol. 2014, 210, 733–753. [Google Scholar] [CrossRef]
- Louveau, I.; Perruchot, M.H.; Bonnet, M.; Gondret, F. Invited review: Pre- and postnatal adipose tissue development in farm animals: From stem cells to adipocyte physiology. Animal 2016, 10, 1839–1847. [Google Scholar] [CrossRef]
- Lkhagvadorj, S.; Qu, L.; Cai, W.; Couture, O.P.; Barb, C.R.; Hausman, G.J.; Nettleton, D.; Anderson, L.L.; Dekkers, J.C.M.; Tuggle, C.K. Gene expression profiling of the short-term adaptive response to acute caloric restriction in liver and adipose tissues of pigs differing in feed efficiency. Am. J. Physiol. Integr. Comp. Physiol. 2010, 298, R494–R507. [Google Scholar] [CrossRef]
- Louveau, I.; Vincent, A.; Tacher, S.; Gilbert, H.; Gondret, F. Increased expressions of genes and proteins involved in mitochondrial oxidation and antioxidant pathway in adipose tissue of pigs selected for a low residual feed intake. J. Anim. Sci. 2016, 94, 5042–5054. [Google Scholar] [CrossRef]
- Gondret, F.; Vincent, A.; Houée-Bigot, M.; Siegel, A.; Lagarrigue, S.; Causeur, D.; Gilbert, H.; Louveau, I. A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs. BMC Genom. 2017, 18, 244. [Google Scholar] [CrossRef]
- Fu, L.; Jiang, Y.; Wang, C.; Mei, M.; Zhou, Z.; Jiang, Y.; Song, H.; Ding, X. A genome-wide association study on feed efficiency related traits in landrace pigs. Front. Genet. 2020, 11, 692. [Google Scholar] [CrossRef] [PubMed]
- Hermesch, S.; Kanis, E.; Eissen, J.J. Economic weights for feed intake in the growing pig derived from a growth model and an economic model. J. Anim. Sci. 2003, 81, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Maltecca, C.; Gray, K.A.; Cassady, J.P. Feed intake, average daily gain, feed efficiency, and real-time ultrasound traits in Duroc pigs: I. Genetic parameter estimation and accuracy of genomic prediction. J. Anim. Sci. 2014, 92, 2377–2386. [Google Scholar] [CrossRef] [PubMed]
- Young, J.M.; Cai, W.; Dekkers, J.C. Effect of selection for residual feed intake on feeding behavior and daily feed intake patterns in Yorkshire swine. J. Anim. Sci. 2011, 89, 639–647. [Google Scholar] [CrossRef]
- Gilbert, H.; Billon, Y.; Brossard, L.; Faure, J.; Gatellier, P.; Gondret, F.; Labussière, E.; Lebret, B.; Lefaucheur, L.; Le Floch, N.; et al. Review: Divergent selection for residual feed intake in the growing pig. Animal 2017, 11, 1427–1439. [Google Scholar] [CrossRef]
- Mackowiak, S.D. Identification of novel and known miRNAs in deep-sequencing data with miRDeep2. Curr. Protoc. Bioinform. 2011, 36, 12.10.1–12.10.15. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Xu, Y.; Qi, X.; Hu, M.; Lin, R.; Hou, Y.; Wang, Z.; Zhou, H.; Zhao, Y.; Luan, Y.; Zhao, S.; et al. Transcriptome analysis of adipose tissue indicates that the cAMP signaling pathway affects the feed efficiency of pigs. Genes 2018, 9, 336. [Google Scholar] [CrossRef]
- Lu, J.; Clark, A.G. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012, 22, 1243–1254. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Varkalaite, G.; Vaitkeviciute, E.; Inciuraite, R.; Salteniene, V.; Juzenas, S.; Petkevicius, V.; Gudaityte, R.; Mickevicius, A.; Link, A.; Kupcinskas, L.; et al. Atrophic gastritis and gastric cancer tissue miRNome analysis reveals hsa-miR-129-1 and hsa-miR-196a as potential early diagnostic biomarkers. World J. Gastroenterol. 2022, 28, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Özel, H.P.; Dinç, T.; Tiryaki, R.S.; Keşküş, A.G.; Konu, Ö.; Kayilioğlu, S.I.; Coşkun, F. Targeted microRNA profiling in gastric cancer with clinical assessement. Balkan J. Med. Genet. 2022, 24, 55–64. [Google Scholar] [CrossRef]
- Zhao, Y.; Hou, Y.; Liu, F.; Liu, A.; Jing, L.; Zhao, C.; Luan, Y.; Miao, Y.; Zhao, S.; Li, X. Transcriptome analysis reveals that vitamin A metabolism in the liver affects feed efficiency in pigs. G3 Genes Genomes Genet. 2016, 6, 3615–3624. [Google Scholar] [CrossRef] [PubMed]
- Guerre-Millo, M. Adipose tissue hormones. J. Endocrinol. Investig. 2002, 25, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Horodyska, J.; Reyer, H.; Wimmers, K.; Trakooljul, N.; Lawlor, P.G.; Hamill, R.M. Transcriptome analysis of adipose tissue from pigs divergent in feed efficiency reveals alteration in gene networks related to adipose growth, lipid metabolism, extracellular matrix, and immune response. Mol. Genet. Genom. MGG 2019, 294, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Casey, D.S.; Dekkers, J.C. Selection response and genetic parameters for residual feed intake in Yorkshire swine. J. Anim. Sci. 2008, 86, 287–298. [Google Scholar] [CrossRef]
- Clare, M.; Richard, P.; Kate, K.; Sinead, W.; Mark, M.; David, K. Residual feed intake phenotype and gender affect the expression of key genes of the lipogenesis pathway in subcutaneous adipose tissue of beef cattle. J. Anim. Sci. Biotechnol. 2018, 9, 68. [Google Scholar]
- Greathead, H.M.; Dawson, J.M.; Scollan, N.D.; Buttery, P.J. In vivo measurement of lipogenesis in ruminants using [1-(14)C]acetate. Br. J. Nutr. 2001, 86, 37–44. [Google Scholar] [CrossRef]
- Faust, I.M.; Johnson, P.R.; Hirsch, J. Surgical removal of adipose tissue alters feeding behavior and the development of obesity in rats. Science 1977, 197, 393–396. [Google Scholar] [CrossRef]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Pawlina-Tyszko, K.; Ukowski, K.; Tyra, M.; Piórkowska, K. Identification of molecular mechanisms related to pig fatness at the transcriptome and miRNAome levels. Genes 2020, 11, 600. [Google Scholar] [CrossRef]
- Rajewsky, N. microRNA target predictions in animals. Nat. Genet. 2006, 38, S8–S13. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Hu, M.; Zhou, H.; Li, C.; Li, X.; Liu, X.; Zhao, Y.; Zhao, S. Neuronal signal transduction-involved genes in pig hypothalamus affect feed efficiency as revealed by transcriptome analysis. Biomed. Res. Int. 2018, 2018, 5862571. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Luan, S.; Lu, X.; Luo, K.; Kong, J. Comparative transcriptome analysis of the Pacific White Shrimp (Litopenaeus vannamei) muscle reveals the molecular basis of residual feed intake. Sci. Rep. 2017, 7, 10483. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.M.; Yang, Z.; Wu, F.; Han, Z.Y.; Wang, G.L. Gene expression profiling of hormonal regulation related to the residual feed intake of Holstein cattle. Biochem. Biophys. Res. Commun. 2015, 465, 19–25. [Google Scholar] [CrossRef]
- Do, D.N.; Ostersen, T.; Strathe, A.B.; Mark, T.; Jensen, J.; Kadarmideen, H.N. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC Genet. 2014, 15, 27. [Google Scholar] [CrossRef]
- Banerjee, P.; Carmelo, V.A.O.; Kadarmideen, H.N. Genome-wide epistatic interaction networks affecting feed efficiency in duroc and landrace pigs. Front. Genet. 2020, 11, 121. [Google Scholar] [CrossRef]
- Fatima, A.; Lynn, D.J.; O’Boyle, P.; Seoighe, C.; Morris, D. The miRNAome of the postpartum dairy cow liver in negative energy balance. BMC Genom. 2014, 15, 279. [Google Scholar] [CrossRef]
- Castano, C.; Kalko, S.; Novials, A.; Parrizas, M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef]
- Pirola, C.J.; Gianotti, T.F.; Castaño, G.O.; Mallardi, P.; San Martino, J.; Ledesma, M.M.G.L.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015, 64, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.; Singh, A.K.; Rotllan, N.; Price, N.; Fernandez-Hernando, C. MicroRNAs and lipid metabolism. Curr. Opin. Lipidol. 2017, 28, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Heo, M.J.; Kim, T.H.; You, J.S.; Blaya, D.; Sancho-Bru, P.; Kim, S.G. Alcohol dysregulates miR-148a in hepatocytes through FoxO1, facilitating pyroptosis via TXNIP overexpression. Gut 2019, 68, 708–720. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef]
- Cirera, S.; Birck, M.; Busk, P.K.; Fredholm, M. Expression profiles of miRNA-122 and its target CAT1 in minipigs (Sus scrofa) fed a high-cholesterol diet. Comp. Med. 2010, 60, 136–141. [Google Scholar]
- Tokuda, N.; Numata, S.; Li, X.; Nomura, T.; Takizawa, M.; Kondo, Y.; Yamashita, Y.; Hashimoto, N.; Kiyono, T.; Urano, T.; et al. β4GalT6 is involved in the synthesis of lactosylceramide with less intensity than β4GalT5. Glycobiology 2013, 23, 1175–1183. [Google Scholar] [CrossRef]
- Tang, Z.; Xu, J.; Yin, L.; Yin, D.; Zhu, M.; Yu, M.; Li, X.; Zhao, S.; Liu, X. Genome-wide association study reveals candidate genes for growth relevant traits in pigs. Front. Genet. 2019, 10, 302. [Google Scholar] [CrossRef]
- Wang, S.; Pan, C.; Ma, X.; Yang, C.; Tang, L.; Huang, J.; Wei, X.; Li, H.; Ma, Y. Identification and functional verification reveals that miR-195 inhibiting THRSP to affect fat deposition in Xinyang buffalo. Front. Genet. 2021, 12, 2614. [Google Scholar] [CrossRef]
- Dai, J.; Huang, X.; Zhang, C.; Luo, X.; Cao, S.; Wang, J.; Liu, B.; Gao, J. Berberine regulates lipid metabolism via miR-192 in porcine oocytes matured in vitro. Vet. Med. Sci. 2021, 7, 950–959. [Google Scholar] [CrossRef]
- Torres, L.F.; Cogliati, B.; Otton, R. Green tea prevents NAFLD by modulation of miR-34a and miR-194 expression in a high-fat diet mouse model. Oxidative Med. Cell Longev. 2019, 2019, 4168380. [Google Scholar] [CrossRef]
- Nie, H.; Song, C.; Wang, D.; Cui, S.; Ren, T.; Cao, Z.; Liu, Q.; Chen, Z.; Chen, X.; Zhou, Y. MicroRNA-194 inhibition improves dietary-induced non-alcoholic fatty liver disease in mice through targeting on FXR. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2017, 1863, 3087. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Sato, Y.; Sassa, T.; Ohno, Y.; Kihara, A. Biochemical characterization of the very long-chain fatty acid elongase ELOVL7. FEBS Lett. 2011, 585, 3337–3341. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wan, D.; Zhang, Y.; Zhang, Y.; Long, C.; Chen, S.; He, L.; Tan, B.; Wu, X.; Yin, Y. Diurnal variations in polyunsaturated fatty acid contents and expression of genes involved in their de novo synthesis in pigs. Biochem. Biophys. Res. Commun. 2017, 483, 430–434. [Google Scholar] [CrossRef]
- Tang, F.; Yang, X.; Liu, D.; Zhang, X.; Huang, X.; He, X.; Shi, J.; Li, Z.; Wu, Z. Co-expression of fat1 and fat2 in transgenic pigs promotes synthesis of polyunsaturated fatty acids. Transgenic Res. 2019, 28, 369–379. [Google Scholar] [CrossRef]
- Crespo-Piazuelo, D.; Criado-Mesas, L.; Revilla, M.; Castelló, A.; Noguera, J.L.; Fernández, A.I.; Ballester, M.; Folch, J.M. Identification of strong candidate genes for backfat and intramuscular fatty acid composition in three crosses based on the Iberian pig. Sci. Rep. 2020, 10, 13962. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Hu, S.; Lai, T.; Wang, J.; Wang, L.; Lai, S. MiR-9-5p promotes rabbit preadipocyte differentiation by suppressing leptin gene expression. Lipids Health Dis. 2020, 19, 126. [Google Scholar] [CrossRef] [PubMed]
- Mentzel, C.; Anthon, C.; Jacobsen, M.J.; Karlskov-Mortensen, P.; Bruun, C.S.; Jorgensen, C.B.; Gorodkin, J.; Cirera, S.; Fredholm, M. Gender and obesity specific microRNA expression in adipose tissue from lean and obese pigs. PLoS ONE 2015, 10, e0131650. [Google Scholar] [CrossRef]
- Shamsi, F.; Zhang, H.; Tseng, Y.H. MicroRNA regulation of brown adipogenesis and thermogenic energy expenditure. Front. Endocrinol. 2017, 8, 205. [Google Scholar] [CrossRef]
- Chen, Y.; Siegel, F.; Kipschull, S.; Haas, B.; Fröhlich, H.; Meister, G.; Pfeifer, A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat. Commun. 2013, 4, 1769. [Google Scholar] [CrossRef]
- Li, C.; He, H.; Zhu, M.; Zhao, S.; Li, X. Molecular characterisation of porcine miR-155 and its regulatory roles in the TLR3/TLR4 pathways. Dev. Comp. Immunol. 2013, 39, 110–116. [Google Scholar] [CrossRef]
- Ying, W.; Tseng, A.; Chang, R.C.-A.; Wang, H.; Lin, Y.-L.; Kanameni, S.; Brehm, T.; Morin, A.; Jones, B.; Splawn, T.; et al. miR-150 regulates obesity-associated insulin resistance by controlling B cell functions. Sci. Rep. 2016, 6, 20176. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Raza, S.H.A.; Ma, X.; Wang, J.; Wang, X.; Liang, C.; Yang, X.; Mei, C.; Suhail, S.M.; Zan, L. Bovine pre-adipocyte adipogenesis is regulated by bta-miR-150 through mTOR signaling. Front. Genet. 2021, 12, 636550. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Li, Y.; Mao, R.; Yang, H.; Zhang, Y.; Zhang, Y.; Guo, P.; Zhan, D.; Zhang, T. Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics 2020, 10, 4705–4719. [Google Scholar] [CrossRef]
- Yang, Z.; Bian, C.; Zhou, H.; Huang, S.; Wang, S.; Liao, L.; Zhao, R.C. MicroRNA hsa-miR-138 inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells through adenovirus EID-1. Stem Cells Dev. 2011, 20, 259–267. [Google Scholar] [CrossRef]
- Pahlavani, M.; Wijayatunga, N.; Kalupahana, N.S.; Ramalingam, L.; Gunaratne, P.H.; Coarfa, C.; Rajapakshe, K.; Kottapalli, P.; Moustaid-Moussa, N. Transcriptomic and microRNA analyses of gene networks regulated by eicosapentaenoic acid in brown adipose tissue of diet-induced obese mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wang, Y.; Ma, L.; Zhong, Z.; Yang, X.; Tao, X.; Chen, X.; He, Z.; Yang, Y.; Zeng, K.; et al. Comparison of microRNAs in adipose and muscle tissue from seven indigenous Chinese breeds and Yorkshire pigs. Anim. Genet. 2019, 50, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Jin, L.; Han, L.; Yuan, Y.; Mu, Q.; Wang, H.; Yang, J.; Ning, G.; Zhou, D.; Zhang, Z. miR-129-5p inhibits adipogenesis through autophagy and may be a potential biomarker for obesity. Int. J. Endocrinol. 2019, 2019, 5069578. [Google Scholar] [CrossRef]
- Lv, S.; Ma, M.; Sun, Y.; Wang, X.; Qimuge, N.; Qin, J.; Pang, W. MicroRNA-129-5p inhibits 3T3-L1 preadipocyte proliferation by targeting G3BP1. Anim. Cells Syst. 2017, 21, 269–277. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, X.; Huang, W.; Miao, X. Identification and characterization of differentially expressed miRNAs in subcutaneous adipose between Wagyu and Holstein cattle. Sci. Rep. 2017, 7, 44026. [Google Scholar] [CrossRef]
- Hilton, C.; Neville, M.J.; Wittemans, L.B.; Todorcevic, M.; Pinnick, K.E.; Pulit, S.L.; Kulyté, A.; Dahlman, I.; Wareham, N.J.; Lotta, L.A.; et al. MicroRNA-196a links human body fat distribution to adipose tissue extracellular matrix composition. EBioMedicine 2019, 44, 467–475. [Google Scholar] [CrossRef]
- Liu, L.; Qian, K.; Wang, C. Discovery of porcine miRNA-196a/b may influence porcine adipogenesis in longissimus dorsi muscle by miRNA sequencing. Anim. Genet. 2017, 48, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Hou, Y.; Ling, Z.; Chen, Q.; Xu, T.; Liu, L.; Yu, N.; Ni, W.; Ding, X.; Zhang, X.; et al. Identification of candidate genes and regulatory competitive endogenous RNA (ceRNA) networks underlying intramuscular fat content in yorkshire pigs with extreme fat deposition phenotypes. Int. J. Mol. Sci. 2022, 23, 12596. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Liu, S.; Qiu, Y.; Li, G.; Li, Y.; Li, M.; Yang, G. Expression profiles and biological roles of miR-196a in swine. Genes 2016, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Liu, T.; Yousuf, S.; Zhang, X.; Huang, W.; Li, A.; Xie, L.; Miao, X. Identification of potential miRNA-mRNA regulatory network and the key miRNAs in intramuscular and subcutaneous adipose. Front. Vet. Sci. 2022, 9, 976603. [Google Scholar] [CrossRef]
- Chen, T.; Cui, J.; Ma, L.; Zeng, Y.; Chen, W. The Effect of MicroRNA-331-3p on preadipocytes proliferation and differentiation and fatty acid accumulation in Laiwu pigs. Biomed. Res. Int. 2019, 2019, 9287804. [Google Scholar] [CrossRef]
- Steinberg, G.R.; Kemp, B.E. Adiponectin: Starving for attention. Cell Metab. 2007, 6, 3–4. [Google Scholar] [CrossRef]
- Fang, H.; Judd, R.L. Adiponectin regulation and function. Compr. Physiol. 2018, 8, 1031–1063. [Google Scholar]
- Bardag-Gorce, F.; Diaz, A.; Niihara, R.; Stark, J.; Cortez, D.; Lee, A.; Hoft, R.; Niihara, Y. Aldehyde dehydrogenases expression in corneal epithelial cells with limbal stem cell deficiency. Int. J. Mol. Sci. 2022, 23, 4032. [Google Scholar] [CrossRef]
- Yamazoe, Y.; Tohkin, M. Development of template systems for ligand interactions of CYP3A5 and CYP3A7 and their distinctions from CYP3A4 template. Drug Metab. Pharmacokinet. 2021, 38, 19. [Google Scholar] [CrossRef]
- Landrier, J.; Kasiri, E.; Karkeni, E.; Mihály, J.; Béke, G.; Weiss, K.; Lucas, R.; Aydemir, G.; Salles, J.; Walrand, S.; et al. Reduced adiponectin expression after high-fat diet is associated with selective up-regulation of ALDH1A1 and further retinoic acid receptor signaling in adipose tissue. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2017, 31, 203–211. [Google Scholar] [CrossRef]
- Xue, H.; Lu, J.; Yuan, R.; Liu, J.; Liu, Y.; Wu, K.; Wu, J.; Du, J.; Shen, B. Knockdown of CLIC4 enhances ATP-induced HN4 cell apoptosis through mitochondrial and endoplasmic reticulum pathways. Cell Biosci. 2016, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Negri, S.; Scolari, F.; Vismara, M.; Brunetti, V.; Faris, P.; Terribile, G.; Sancini, G.; Berra-Romani, R.; Moccia, F. GABA(A) and GABA(B) receptors mediate GABA-induced intracellular Ca(2+) signals in human brain microvascular endothelial cells. Cells 2022, 11, 3860. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Xu, Y.; Hou, Y.; Qi, X.; Zhou, L.; Liu, H.; Luan, Y.; Jing, L.; Miao, Y.; Zhao, S.; et al. Proteomic analysis indicates that mitochondrial energy metabolism in skeletal muscle tissue is negatively correlated with feed efficiency in pigs. Sci. Rep. 2017, 7, 45291. [Google Scholar] [CrossRef] [PubMed]
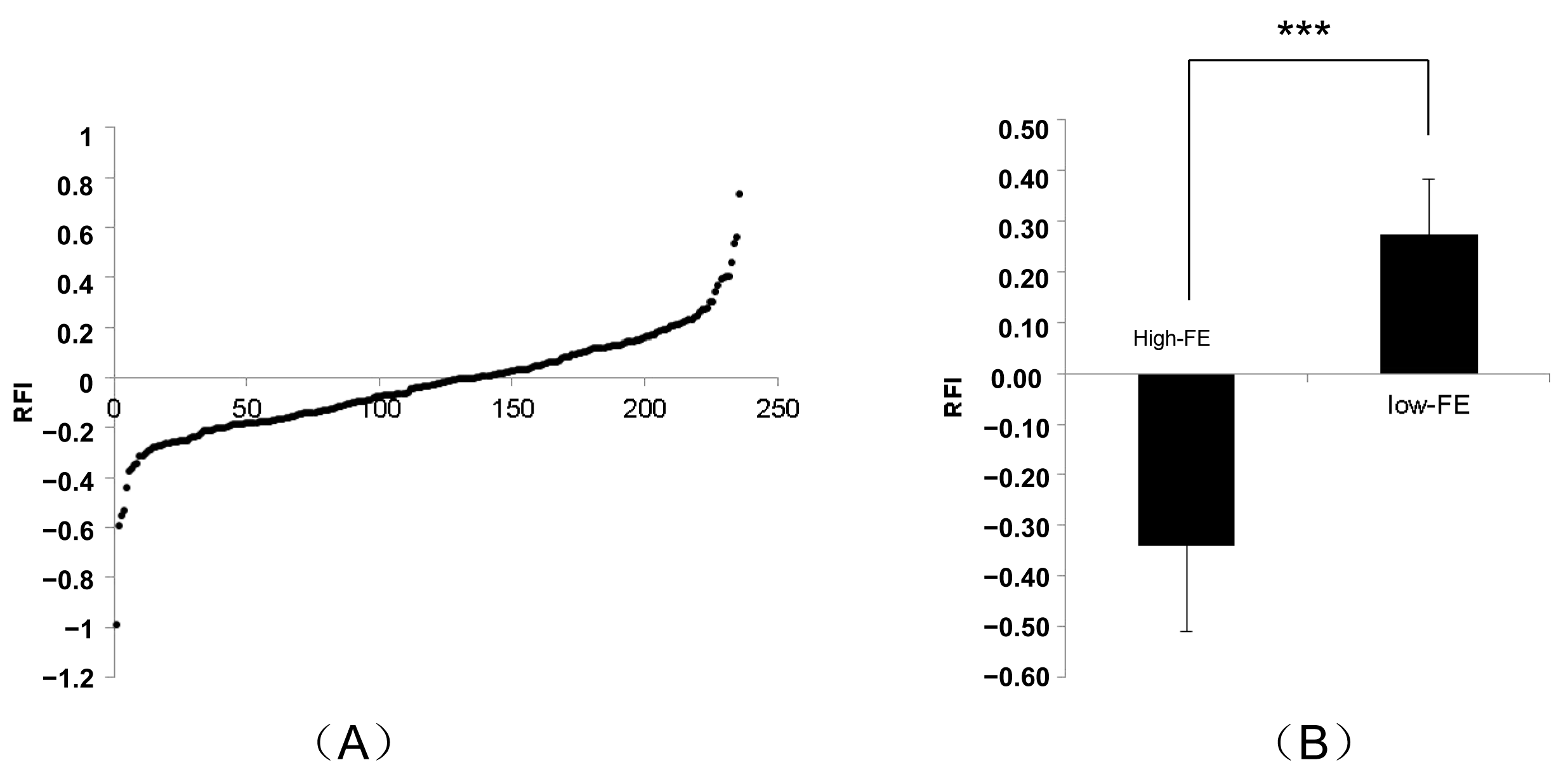

| High FE (n = 50) | Low FE (n = 50) | p-Value | |
|---|---|---|---|
| FCR | 2.23 ± 0.20 | 2.92 ± 0.31 | 9.60694 × 10−24 |
| RFI (kg/day) | −0.28 ± 0.14 | 0.24 ± 0.13 | 2.228 × 10−36 |
| FI | 1.78 ± 0.29 | 2.35 ± 0.28 | 3.02393 × 10−16 |
| ADG | 0.80 ± 0.14 | 0.82 ± 0.14 | 0.67 |
| Initial BW (kg) | 38.87 ± 2.12 | 39.27 ± 2.94 | 0.37 |
| Final BW (kg) | 90.13 ± 3.41 | 90.84 ± 4.27 | 0.24 |
| AMBW | 21.98 ± 0.82 | 22.19 ± 0.62 | 0.45 |
| ABF (mm) | 18.68 ± 2.74 | 21.49 ± 2.23 | 1.0061 × 10−5 |
| LMA (cm2) | 45.75 ± 6.22 | 44.78 ± 6.87 | 0.46 |
| Reads | High FE-1 | High FE-2 | High FE-3 | Low FE-1 | Low FE-2 | Low FE-3 |
|---|---|---|---|---|---|---|
| Total reads | 20,197,804 | 12,835,372 | 20,920,937 | 26,772,081 | 17,184,327 | 23,376,133 |
| Clean reads | 18,424,875 | 10,611,213 | 18,687,418 | 22,321,137 | 15,274,616 | 20,018,461 |
| Qualified% | 0.912 | 0.827 | 0.893 | 0.834 | 0.889 | 0.856 |
| mapped | 4,251,042 | 2,045,141 | 4,265,705 | 6,004,627 | 5,288,460 | 4,190,707 |
| unmapped | 14,173,833 | 8,566,072 | 14,421,713 | 16,316,510 | 9,986,156 | 15,827,754 |
| mapped% | 0.231 | 0.193 | 0.228 | 0.269 | 0.346 | 0.209 |
| unmapped% | 0.769 | 0.807 | 0.772 | 0.731 | 0.654 | 0.791 |
| miRNA | Ref miRNA (Human) | Fold Change (High/Low) | p-Value | Mature Sequences |
|---|---|---|---|---|
| ssc-miR-122-5p | miR-122-5p | −3.50 | 1.64483 × 10−5 | uggagugugacaaugguguuu |
| ssc-miR-192 | miR-192-5p | −2.98 | 0.000328674 | cugaccuaugaauugacagccag |
| ssc-miR-194a-5p | miR-194-5p | −2.76 | 0.000665162 | uguaacagcaacuccaugugga |
| ssc-miR-10386 | −3.18 | 0.000765051 | gucguccucucccucccuccu | |
| ssc-miR-155-5p | miR-155-5p | 2.29 | 0.001718442 | uuaaugcuaauugugauaggggu |
| ssc-miR-150 | miR-150-5p | 2.71 | 0.001962553 | ucucccaacccuuguaccagug |
| ssc-miR-194b-5p | miR-194-5p | −2.43 | 0.00437337 | uguaacagcgacuccaugugga |
| ssc-miR-582-5p | miR-582-5p | 3.55 | 0.015997654 | uacaguuguucaaccaguuacu |
| ssc-miR-331-5p | miR-331-3p | 2.08 | 0.019987215 | gccccugggccuauccuagaac |
| ssc-miR-136-5p | miR-136-3p | −3.32 | 0.020242907 | caucaucgucucaaaugagucu |
| ssc-miR-129a-5p | miR-129-5p | −2.02 | 0.020370961 | cuuuuugcggucugggcuugc |
| ssc-miR-9 | miR-9-5p | −2.22 | 0.023102635 | ucuuugguuaucuagcuguauga |
| ssc-miR-138 | miR-138-5p | −2.27 | 0.039803139 | agcugguguugugaaucaggccgu |
| ssc-miR-196a | miR-196a-5p | 2.07 | 0.04389995 | uagguaguuucauguuguuggg |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, M.; Ren, Z.; Miao, Y. Identification of Differentially Expressed miRNAs in Porcine Adipose Tissues and Evaluation of Their Effects on Feed Efficiency. Genes 2022, 13, 2406. https://doi.org/10.3390/genes13122406
Liao M, Ren Z, Miao Y. Identification of Differentially Expressed miRNAs in Porcine Adipose Tissues and Evaluation of Their Effects on Feed Efficiency. Genes. 2022; 13(12):2406. https://doi.org/10.3390/genes13122406
Chicago/Turabian StyleLiao, Mingxing, Zhuqing Ren, and Yuanxin Miao. 2022. "Identification of Differentially Expressed miRNAs in Porcine Adipose Tissues and Evaluation of Their Effects on Feed Efficiency" Genes 13, no. 12: 2406. https://doi.org/10.3390/genes13122406
APA StyleLiao, M., Ren, Z., & Miao, Y. (2022). Identification of Differentially Expressed miRNAs in Porcine Adipose Tissues and Evaluation of Their Effects on Feed Efficiency. Genes, 13(12), 2406. https://doi.org/10.3390/genes13122406






