Single-Cell Sequencing of Malignant Ascites Reveals Transcriptomic Remodeling of the Tumor Microenvironment during the Progression of Epithelial Ovarian Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subject
2.2. ScRNA-seq Data Processing
2.3. ScRNA-seq Data Analysis
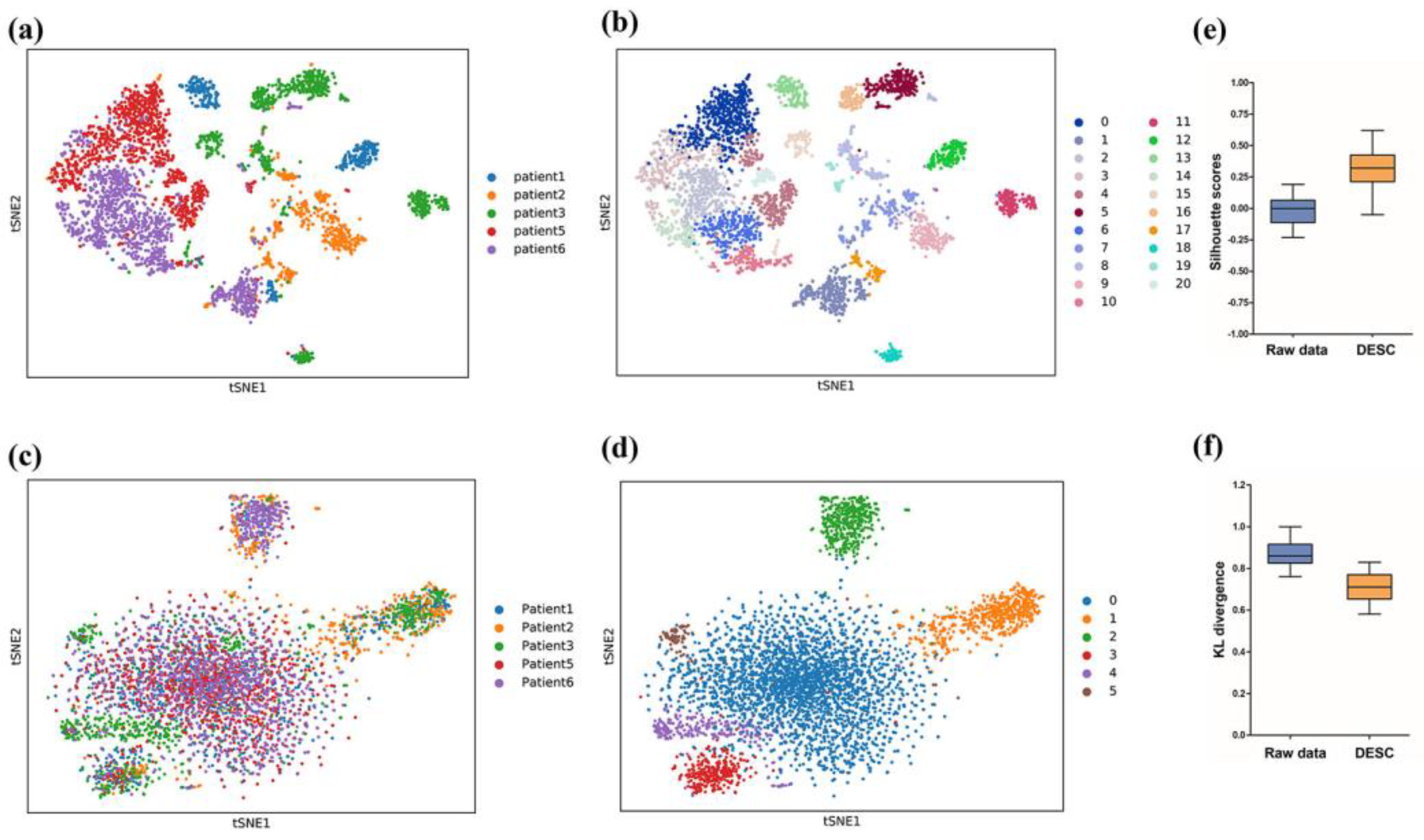
2.4. Batch Correction and Evaluation
2.5. Pseudotime Analysis
2.6. Pathway Analysis and Functional Annotation
3. Results
3.1. Batch Correction
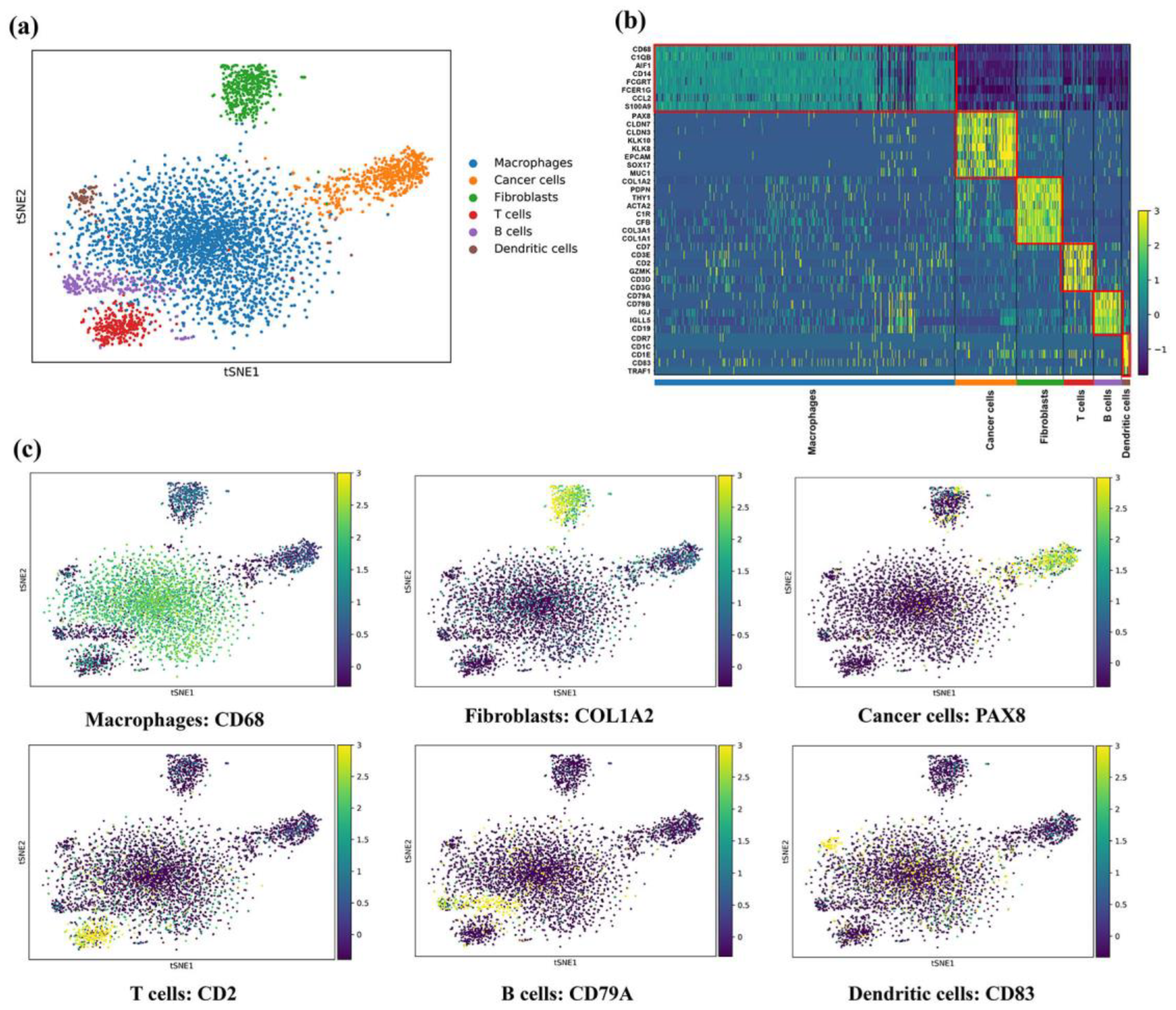
3.2. A Single-Cell Atlas in Ascites of EOC
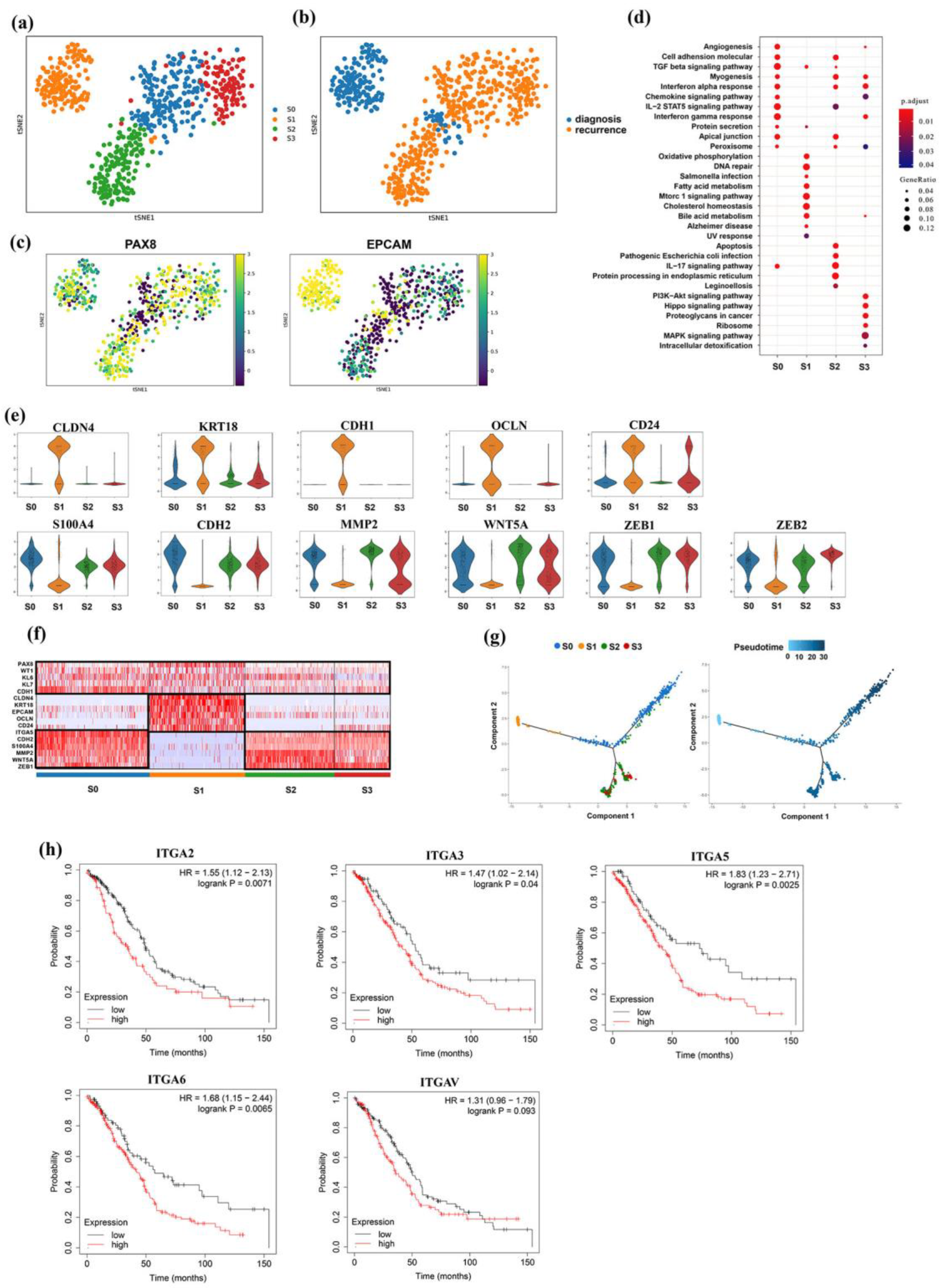
3.3. Intrinsic Tumor Cell Subpopulations of Ascites
3.4. Distinct Subgroup in Mesenchymal Cancer Cells
3.5. Identification of M2 Tumor-Associated Macrophages
3.6. Activated Cancer-Associated Fibroblasts Express Tumor Marker Genes
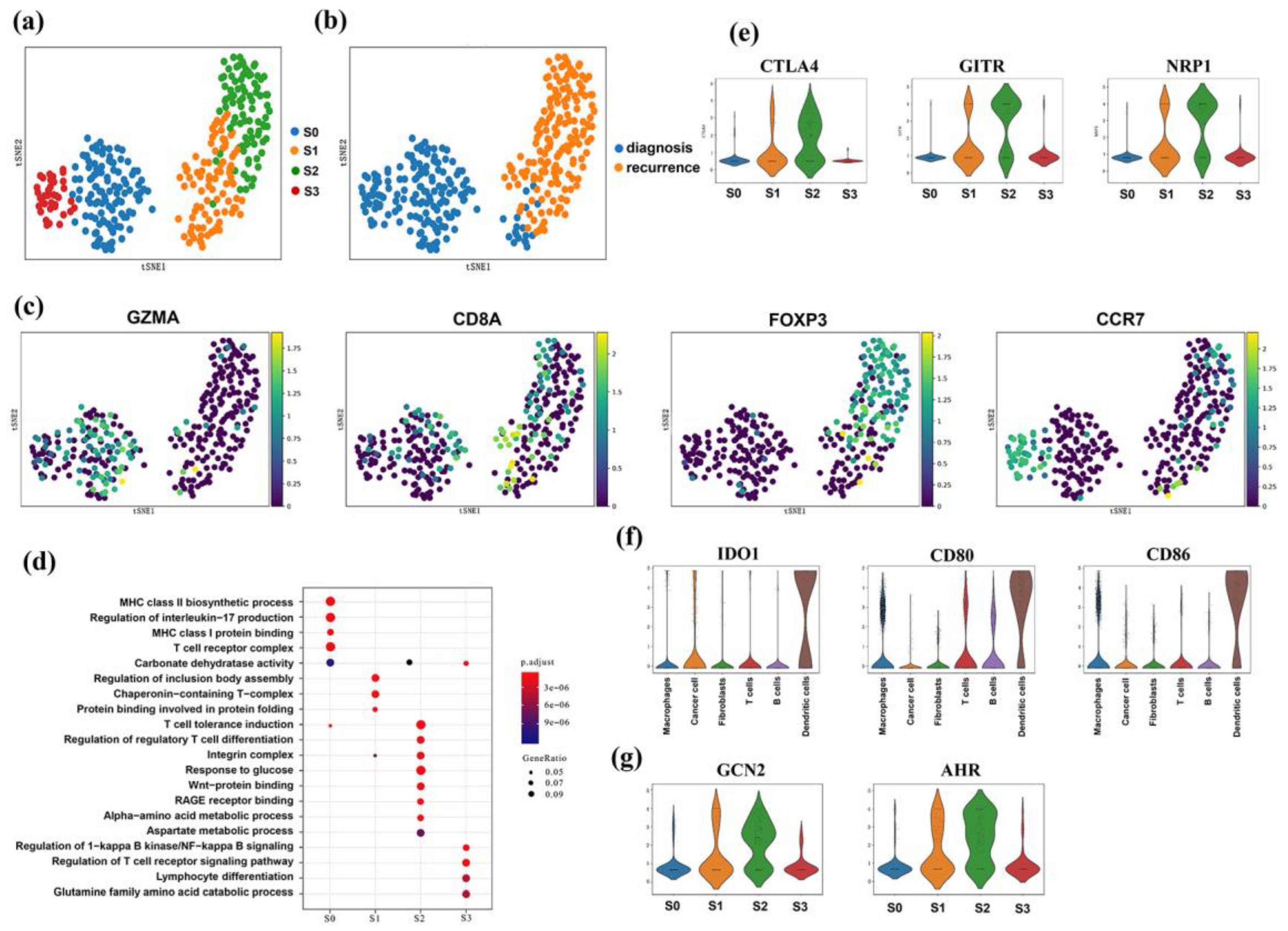
3.7. CD4+CD25+ T Regulatory Cells Showed Infiltration Features
3.8. Dendritic and Treg Cells Both Expressed IDO-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA A Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Kuroki, L.; Guntupalli, S.R. Treatment of epithelial ovarian cancer. BMJ 2020, 371, m3773. [Google Scholar] [CrossRef] [PubMed]
- Mcgranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Hass, R.; von der Ohe, J.; Ungefroren, H. Impact of the tumor microenvironment on tumor heterogeneity and consequences for cancer cell plasticity and stemness. Cancers 2020, 12, 3716. [Google Scholar] [CrossRef]
- Ford, C.E.; Werner, B.; Hacker, N.F.; Warton, K. The untapped potential of ascites in ovarian cancer research and treatment. Br. J. Cancer 2020, 123, 9–16. [Google Scholar] [CrossRef]
- Liang, H.; Yu, T.; Yue, H.; Hua, J.; Wang, C.; You, T.; Zhao, X.; Shan, H.; Rui, Y.; Yang, L. Lncrna ptar promotes emt and invasion-metastasis in serous ovarian cancer by competitively binding mir-101-3p to regulate zeb1 expression. Mol. Cancer 2018, 17, 119. [Google Scholar] [CrossRef]
- Chen, G.M.; Lavanya, K.; Ludwig, G.; Victor, K.; Zhaleh, S.; Gendoo, D.; Giovanni, P.; Birrer, M.J.; Benjamin, H.K.; Levi, W. Consensus on molecular subtypes of high-grade serous ovarian carcinoma. Clin. Cancer Res. 2018, 24, 5037–5047. [Google Scholar] [CrossRef]
- Wagner, A.; Regev, A.; Yosef, N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 2016, 34, 1145–1160. [Google Scholar] [CrossRef]
- Schelker, M.; Feau, S.; Du, J.; Ranu, N.; Klipp, E.; Macbeath, G.; Schoeberl, B.; Raue, A. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat. Commun. 2017, 8, 2032. [Google Scholar] [CrossRef]
- Izar, B.; Tirosh, I.; Stover, E.H.; Wakiro, I.; Regev, A. A single-cell landscape of high-grade serous ovarian cancer. Nat. Med. 2020, 26, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.N.; Ang, K.S.; Chevrier, M.; Zhang, X.; Lee, N.Y.S.; Goh, M.; Chen, J. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020, 21, 12. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, K.; Lyu, Y.; Pan, H.; Li, M. Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nat. Commun. 2020, 11, 2338. [Google Scholar] [CrossRef] [PubMed]
- Peter, R.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar]
- Wu, D.; Liu, L.; Ren, C.; Kong, D.; Zhang, P.; Jin, X.; Wang, T.; Zhang, G. Epithelial-mesenchymal interconversions and the regulatory function of the zeb family during the development and progression of ovarian cancer. Oncol. Lett. 2016, 11, 1463–1468. [Google Scholar] [CrossRef]
- Kan, T.; Wang, W.; Ip, P.P.; Zhou, S.; Yang, M. Single-cell emt-related transcriptional analysis revealed intra-cluster heterogeneity of tumor cell clusters in epithelial ovarian cancer ascites. Oncogene 2020, 39, 4227–4240. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Sica, A. Tumour-associated macrophages as a prototypic type ii polarised phagocyte population: Role in tumour progression. Eur. J. Cancer 2004, 40, 1660–1667. [Google Scholar] [CrossRef]
- Bartoschek, M.; Oskolkov, N.; Bocci, M.; Lövrot, J.; Larsson, C.; Sommarin, M.; Madsen, C.D.; Lindgren, D.; Pekar, G.; Karlsson, G. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat. Commun. 2018, 9, 5150. [Google Scholar] [CrossRef]
- Waisberg, D.R.; Parra, E.R.; Barbas-Filho, J.V.; Fernezlian, S.; Capelozzi, V.L. Increased fibroblast telomerase expression precedes myofibroblast α-smooth muscle actin expression in idiopathic pulmonary fibrosis. Clinics 2012, 67, 1039–1046. [Google Scholar] [CrossRef]
- Schulz, G.B.; Grimm, T.; Sers, C.; Riemer, P.; Horst, D. Prognostic value and association with epithelial-mesenchymal transition and molecular subtypes of the proteoglycan biglycan in advanced bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2019, 37, 530.e9–530.e18. [Google Scholar] [CrossRef]
- Pistore, C.; Giannoni, E.; Colangelo, T.; Rizzo, F.; Bonapace, I. DNA methylation variations are required for epithelial-to-mesenchymal transition induced by cancer-associated fibroblasts in prostate cancer cells. Oncogene 2017, 36, 5551–5566. [Google Scholar] [CrossRef] [PubMed]
- Villadangos, J.A.; Schnorrer, P. Intrinsic and cooperative antigen-presenting functions of dendritic-cell subsets in vivo. Nat. Rev. Immunol. 2007, 7, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.M.; Cardenas, C.; Tedja, R. The role of intra-tumoral heterogeneity and its clinical relevance in epithelial ovarian cancer recurrence and metastasis. Cancers 2019, 11, 1083. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Hong, S.; Dong, L.; Cheng, B.; Lin, L.; Zhao, B.; Chen, Y.-G.; Chen, X. Dynamic sialylation in transforming growth factor-β (tgf-β)-induced epithelial to mesenchymal transition. J. Biol. Chem. 2015, 290, 12000–12013. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Li, H.; Zhu, H.; Xiao, X.; Ma, Y. Genistein inhibits estradiol-and environmental endocrine disruptor-induced growth effects on neuroblastoma cells in vitro. Oncol. Lett. 2013, 5, 1583–1586. [Google Scholar] [CrossRef]
- Singla, M.; Kumar, A.; Bal, A.; Sarkar, S.; Bhattacharyya, S. Epithelial to mesenchymal transition induces stem cell like phenotype in renal cell carcinoma cells. Cancer Cell Int. 2018, 18, 57. [Google Scholar] [CrossRef]
- Yshii, L.; Pignolet, B.; Mauré, E.; Pierau, M.; Brunner-Weinzierl, M. Ifn-γ is a therapeutic target in paraneoplastic cerebellar degeneration. JCI Insight 2019, 4, e127001. [Google Scholar]
- Gubbels, J.A.; Felder, M.; Horibata, S.; Belisle, J.A.; Kapur, A.; Holden, H.; Petrie, S.; Mign Ea Ult, M.; Rancourt, C.; Connor, J.P. Muc16 provides immune protection by inhibiting synapse formation between nk and ovarian tumor cells. Mol. Cancer 2010, 9, 11. [Google Scholar] [CrossRef]
- Lane, D.; Matte, I.; Laplante, C.; Garde-Granger, P.; Rancourt, C.; Piché, A. Osteoprotegerin (opg) activates integrin, focal adhesion kinase (fak), and akt signaling in ovarian cancer cells to attenuate trail-induced apoptosis. J. Ovarian Res. 2013, 6, 82. [Google Scholar] [CrossRef][Green Version]
- Lu, L.; Katsaros, D.; Wiley, A.; Longrais, I.A.R.D.L.; Yu, H. Expression of mdr1 in epithelial ovarian cancer and its association with disease progression. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2007, 16, 395–403. [Google Scholar] [CrossRef]
- Nagle, C.M.; Chenevix-Trench, G.; Spurdle, A.B.; Webb, P.M. The role of glutathione-s-transferase polymorphisms in ovarian cancer survival. Eur. J. Cancer 2007, 43, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Yao, L.; Han, Y.; Hao, P.; Lu, P. Quantitative phosphoproteomics reveals system-wide phosphorylation network altered by spry in mouse mammary stromal fibroblasts. Int. J. Mol. Sci. 2019, 20, 5400. [Google Scholar] [CrossRef] [PubMed]
- Berzaghi, R.; Ahktar, M.A.; Islam, A.; Pedersen, B.D.; Hellevik, T.; Martinez-Zubiaurre, I. Fibroblast-mediated immunoregulation of macrophage function is maintained after irradiation. Cancers 2019, 11, 689. [Google Scholar] [CrossRef]
- Julia, E.; Elena, C.; Parisa, A.; Alexandra, M.; Enni, M. Analysis of gene expression signatures in cancer-associated stroma from canine mammary tumours reveals molecular homology to human breast carcinomas. Int. J. Mol. Sci. 2017, 18, 1101. [Google Scholar]
- Shuai, X.; Wei-min, L.; Tong, Y.L.; Dong, N.; Sheng, Z.Y.; Yao, Y.M. Expression of il-37 contributes to the immunosuppressive property of human cd4+cd25+ regulatory t cells. Sci. Rep. 2015, 5, 14478. [Google Scholar] [CrossRef]
- Larmonier, N.; Marron, M.; Zeng, Y.; Cantrell, J.; Romanoski, A.; Sepassi, M.; Thompson, S.; Chen, X.; Andreansky, S.; Katsanis, E. Tumor-derived cd4+cd25+ regulatory t cell suppression of dendritic cell function involves tgf-β and il-10. Cancer Immunol. Immunother. 2007, 56, 48–59. [Google Scholar] [CrossRef]
- Chang, W.C.; Li, C.H.; Huang, S.C.; Chang, D.Y.; Chou, L.Y.; Sheu, B.C. Clinical significance of regulatory t cells and cd8+ effector populations in patients with human endometrial carcinoma. Cancer 2010, 116, 5777–5788. [Google Scholar] [CrossRef]
- Metz, R.; Rust, S.; Duhadaway, J.B.; Mautino, M.R.; Prendergast, G.C. Ido inhibits a tryptophan sufficiency signal that stimulates mtor: A novel ido effector pathway targeted by d-1-methyl-tryptophan. OncoImmunology 2012, 1, 1460–1468. [Google Scholar] [CrossRef]
- Muller, A.J.; Duhadaway, J.B.; Donover, P.S.; Sutanto-Ward, E.; Prendergast, G.C. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene bin1, potentiates cancer chemotherapy. Nat. Med. 2005, 11, 312. [Google Scholar] [CrossRef]
- Opitz, C.A.; Litzenburger, U.M.; Sahm, F.; Ott, M.; Tritschler, I.; Trump, S.; Schumacher, T.; Jestaedt, L.; Schrenk, D.; Weller, M.; et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011, 478, 197. [Google Scholar] [CrossRef]
- Qin, Y.; Chen, K.; Gu, W.; Dong, X.; Lei, R.; Chang, Y.; Xue, B.; Xia, S.; Li, Z.; Zhang, J. Small size fullerenol nanoparticles suppress lung metastasis of breast cancer cell by disrupting actin dynamics. J. Nanobiotechnol. 2018, 16, 54. [Google Scholar] [CrossRef] [PubMed]


| Patient ID | Platform | BRCA Status | Treatment Status of Sample | Clinical Status at Time of Sampling |
|---|---|---|---|---|
| Patient 1 | 10X Genomics | WT | On-treatment | Recurrent |
| Patient 2 | 10X Genomics | WT | On-treatment | Recurrent |
| Patient 3 | 10X Genomics | N/A | Treatment-naïve | Diagnosis |
| Patient 5 | 10X Genomics | WT | Treatment-naïve | Diagnosis |
| Patient 6 | 10X Genomics | N/A | Treatment-naïve | Diagnosis |
| Cell Types | Markers | Number and Ratio in All Samples | Number and Ratio in Diagnosis Samples | Number and Ratio in Recurrent Samples |
|---|---|---|---|---|
| Macrophages | CD68, C1QB, CD14 | 2104, 44.96% | 1282, 48.16% | 822, 40.73% |
| Cancer cells | PAX8, CLDN7, KLK8 | 806, 17.22% | 278, 10.44% | 528, 26.16% |
| Fibroblasts | COL1A2, PDPN, COL1A1, | 763, 16.30% | 420, 15.78% | 343, 17.00% |
| T cells | CD2, CD7, CD3E | 446, 9.53% | 225, 8.45% | 221, 10.95% |
| B cells | CD79A, CD19, CD79B | 425, 9.08% | 351, 13.19% | 74, 3.67% |
| Dendritic cells | CDR7, CD83, CD1C | 136, 2.91% | 106, 3.98% | 30, 1.49% |
| Total | 4680, 100% | 2662, 100% | 2018, 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, W.; Wang, D.; Zhang, L.; Wang, X.; He, J.; Cao, L.; Li, K.; Xie, H. Single-Cell Sequencing of Malignant Ascites Reveals Transcriptomic Remodeling of the Tumor Microenvironment during the Progression of Epithelial Ovarian Cancer. Genes 2022, 13, 2276. https://doi.org/10.3390/genes13122276
Li Y, Wang W, Wang D, Zhang L, Wang X, He J, Cao L, Li K, Xie H. Single-Cell Sequencing of Malignant Ascites Reveals Transcriptomic Remodeling of the Tumor Microenvironment during the Progression of Epithelial Ovarian Cancer. Genes. 2022; 13(12):2276. https://doi.org/10.3390/genes13122276
Chicago/Turabian StyleLi, Yiqun, Wenjie Wang, Danyun Wang, Liuchao Zhang, Xizhi Wang, Jia He, Lei Cao, Kang Li, and Hongyu Xie. 2022. "Single-Cell Sequencing of Malignant Ascites Reveals Transcriptomic Remodeling of the Tumor Microenvironment during the Progression of Epithelial Ovarian Cancer" Genes 13, no. 12: 2276. https://doi.org/10.3390/genes13122276
APA StyleLi, Y., Wang, W., Wang, D., Zhang, L., Wang, X., He, J., Cao, L., Li, K., & Xie, H. (2022). Single-Cell Sequencing of Malignant Ascites Reveals Transcriptomic Remodeling of the Tumor Microenvironment during the Progression of Epithelial Ovarian Cancer. Genes, 13(12), 2276. https://doi.org/10.3390/genes13122276







