Unraveling Signatures of Local Adaptation among Indigenous Groups from Mexico
Abstract
1. Introduction
2. Materials and Methods
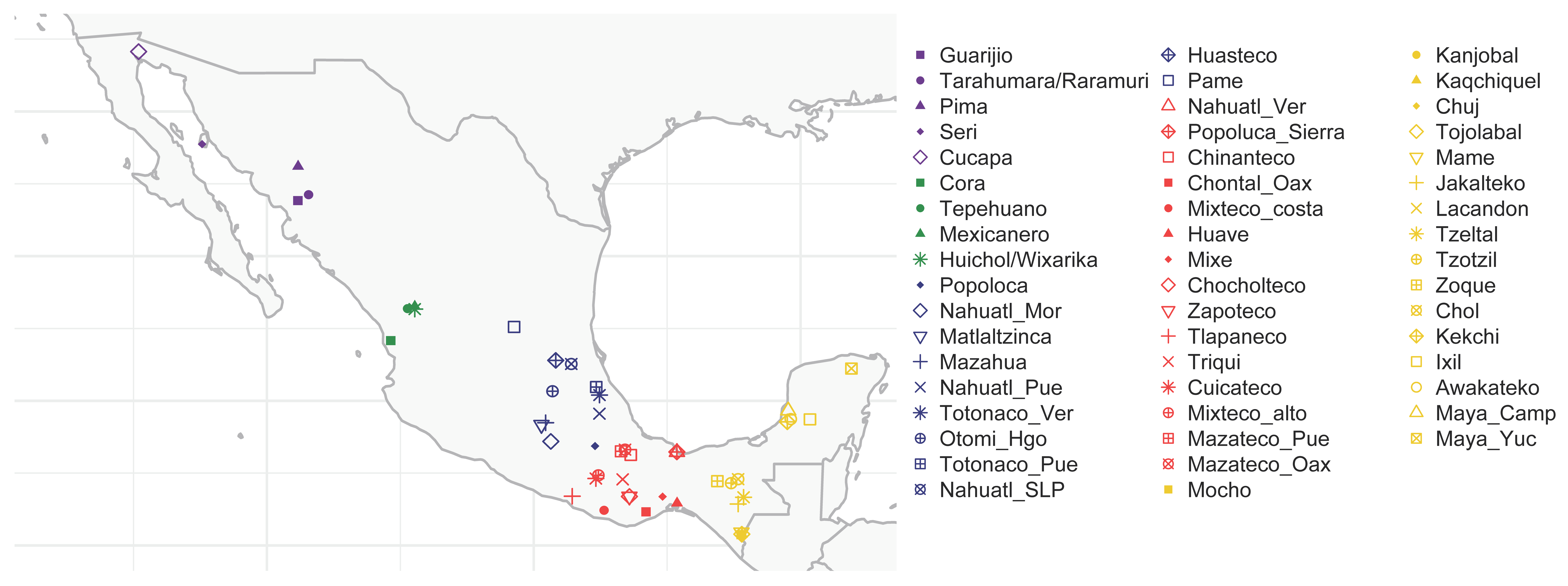
2.1. Samples
2.2. Genome-Wide Scan for Selection Signals
2.3. Enrichment Analysis
3. Results
3.1. Samples Description
3.2. Identification of Adaptation Signals
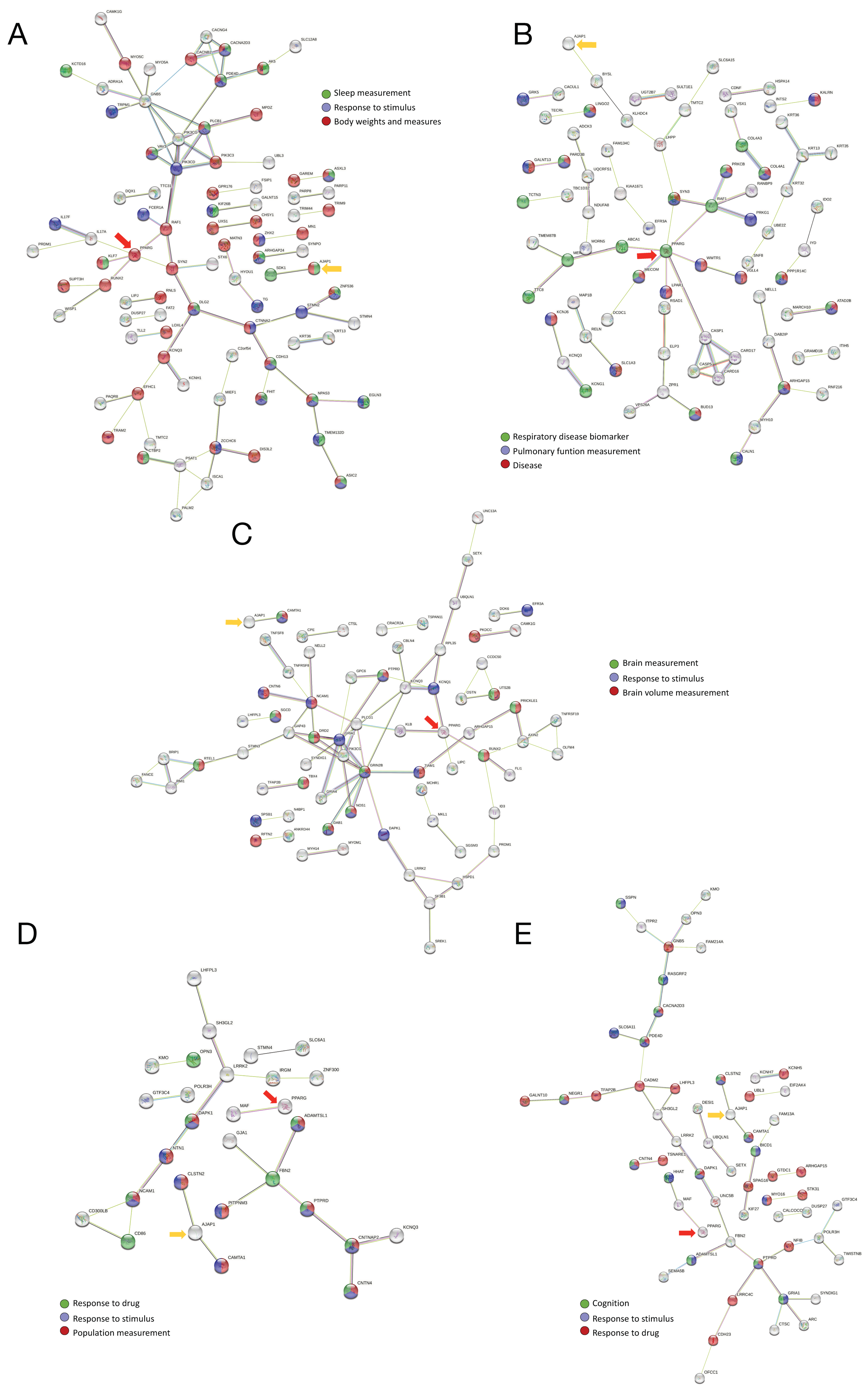
3.3. Protein–Protein Interaction (PPI) Network of Genes under Selection
3.4. Enrichment Pathway Analysis of the Network of Genes under Selection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Posth, C.; Nakatsuka, N.; Lazaridis, I.; Skoglund, P.; Mallick, S.; Lamnidis, T.C.; Rohland, N.; Nägele, K.; Adamski, N.; Bertolini, E.; et al. Reconstructing the Deep Population History of Central and South America. Cell 2018, 175, 1185–1197.e22. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mayar, J.V.; Vinner, L.; de Barros Damgaard, P.; de La Fuente, C.; Chan, J.; Spence, J.P.; Allentoft, M.E.; Vimala, T.; Racimo, F.; Pinotti, T.; et al. Early Human Dispersals within the Americas. Science 2018, 362, eaav2621. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, M.; Steinrücken, M.; Harris, K.; Schiffels, S.; Rasmussen, S.; DeGiorgio, M.; Albrechtsen, A.; Valdiosera, C.; Ávila-Arcos, M.C.; Malaspinas, A.S.; et al. Genomic Evidence for the Pleistocene and Recent Population History of Native Americans. Science 2015, 349, aab3884. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Villalobos, H.; Martínez-Sevilla, V.M.; Martínez-Cortés, G.; Aguilar-Velázquez, J.A.; Sosa-Macías, M.; Rubi-Castellanos, R.; González-Martín, A. Importance of the Geographic Barriers to Promote Gene Drift and Avoid Pre- and Post-Columbian Gene Flow in Mexican Native Groups: Evidence from Forensic STR Loci. Am. J. Phys. Anthropol. 2016, 160, 298–316. [Google Scholar] [CrossRef]
- Castro e Silva, M.A.; Ferraz, T.; Couto-Silva, C.M.; Lemes, R.B.; Nunes, K.; Comas, D.; Hünemeier, T. Population Histories and Genomic Diversity of South American Natives. Mol. Biol. Evol. 2022, 39, msab339. [Google Scholar] [CrossRef]
- Lindo, J.; Rogers, M.; Mallott, E.K.; Petzelt, B.; Mitchell, J.; Archer, D.; Cybulski, J.S.; Malhi, R.S.; DeGiorgio, M. Patterns of Genetic Coding Variation in a Native American Population before and after European Contact. Am. J. Hum. Genet. 2018, 102, 806–815. [Google Scholar] [CrossRef]
- Lindo, J.; Huerta-Sánchez, E.; Nakagome, S.; Rasmussen, M.; Petzelt, B.; Mitchell, J.; Cybulski, J.S.; Willerslev, E.; Degiorgio, M.; Malhi, R.S. A Time Transect of Exomes from a Native American Population before and after European Contact. Nat. Commun. 2016, 7, 13175. [Google Scholar] [CrossRef]
- García-Ortiz, H.; Barajas-Olmos, F.; Contreras-Cubas, C.; Cid-Soto, M.Á.; Córdova, E.J.; Centeno-Cruz, F.; Mendoza-Caamal, E.; Cicerón-Arellano, I.; Flores-Huacuja, M.; Baca, P.; et al. The Genomic Landscape of Mexican Indigenous Populations Brings Insights into the Peopling of the Americas. Nat. Commun. 2021, 12, 5942. [Google Scholar] [CrossRef]
- Ávila-Arcos, M.C.; McManus, K.F.; Sandoval, K.; Rodríguez-Rodríguez, J.E.; Villa-Islas, V.; Martin, A.R.; Luisi, P.; Peñaloza-Espinosa, R.I.; Eng, C.; Huntsman, S.; et al. Population History and Gene Divergence in Native Mexicans Inferred from 76 Human Exomes. Mol. Biol. Evol. 2020, 37, 994–1006. [Google Scholar] [CrossRef]
- Amorim, C.E.G.; Nunes, K.; Meyer, D.; Comas, D.; Bortolini, M.C.; Salzano, F.M.; Hünemeier, T. Genetic Signature of Natural Selection in First Americans. Proc. Natl. Acad. Sci. USA 2017, 114, 2195–2199. [Google Scholar] [CrossRef]
- Mendoza-Revilla, J.; Chacón-Duque, J.C.; Fuentes-Guajardo, M.; Ormond, L.; Wang, K.; Hurtado, M.; Villegas, V.; Granja, V.; Acuña-Alonzo, V.; Jaramillo, C.; et al. Disentangling Signatures of Selection before and after European Colonization in Latin Americans. Mol. Biol. Evol. 2022, 39, msac076. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, A.W.; Mata-Míguez, J.; Miró-Herrans, A.; Briggs-Cloud, M.; Sylestine, A.; Barajas-Olmos, F.; Garcia-Ortiz, H.; Rzhetskaya, M.; Orozco, L.; Raff, J.A.; et al. Comparing Signals of Natural Selection between Three Indigenous North American Populations. Proc. Natl. Acad. Sci. USA 2019, 116, 9312–9317. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Hansen, M.E.B.; Lo, Y.; Tishkoff, S.A. Going Global by Adapting Local: A Review of Recent Human Adaptation. Science 2016, 354, 54–59. [Google Scholar] [CrossRef]
- Benton, M.L.; Abraham, A.; LaBella, A.L.; Abbot, P.; Rokas, A.; Capra, J.A. The Influence of Evolutionary History on Human Health and Disease. Nat. Rev. Genet. 2021, 22, 269–283. [Google Scholar] [CrossRef]
- Delaneau, O.; Zagury, J.-F.; Robinson, M.R.; Marchini, J.L.; Dermitzakis, E.T. Accurate, Scalable and Integrative Haplotype Estimation. Nat. Commun. 2019, 10, 5436. [Google Scholar] [CrossRef]
- 1000 Genomes Project Consortium. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Cadzow, M.; Boocock, J.; Nguyen, H.T.; Wilcox, P.; Merriman, T.R.; Black, M.A. A Bioinformatics Workflow for Detecting Signatures of Selection in Genomic Data. Front. Genet. 2014, 5, 293. [Google Scholar] [CrossRef]
- Maclean, C.A.; Chue Hong, N.P.; Prendergast, J.G.D. Hapbin: An Efficient Program for Performing Haplotype-Based Scans for Positive Selection in Large Genomic Datasets. Mol. Biol. Evol. 2015, 32, 3027–3029. [Google Scholar] [CrossRef]
- Yi, X.; Liang, Y.; Huerta-Sanchez, E.; Jin, X.; Cuo, Z.X.P.; Pool, J.E.; Xu, X.; Jiang, H.; Vinckenbosch, N.; Korneliussen, T.S.; et al. Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude. Science 2010, 329, 75–78. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, T.; Salmela, E.; Andersen, P.M.; Dahlman-Wright, K.; Sistonen, P.; Savontaus, M.-L.; Schreiber, S.; Lahermo, P.; Kere, J. Genomic Landscape of Positive Natural Selection in Northern European Populations. Eur. J. Hum. Genet. 2010, 18, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, S.; Mathieson, I. FADS1 and the Timing of Human Adaptation to Agriculture. Mol. Biol. Evol. 2018, 35, 2957–2970. [Google Scholar] [CrossRef]
- Song, W.; Shi, Y.; Wang, W.; Pan, W.; Qian, W.; Yu, S.; Zhao, M.; Lin, G.N. A Selection Pressure Landscape for 870 Human Polygenic Traits. Nat. Hum. Behav. 2021, 5, 1731–1743. [Google Scholar] [CrossRef]
- Yasumizu, Y.; Sakaue, S.; Konuma, T.; Suzuki, K.; Matsuda, K.; Murakami, Y.; Kubo, M.; Palamara, P.F.; Kamatani, Y.; Okada, Y.; et al. Genome-Wide Natural Selection Signatures Are Linked to Genetic Risk of Modern Phenotypes in the Japanese Population. Mol. Biol. Evol. 2020, 37, 1306–1316. [Google Scholar] [CrossRef]
- Rühlemann, M.C.; Hermes, B.M.; Bang, C.; Doms, S.; Moitinho-Silva, L.; Thingholm, L.B.; Frost, F.; Degenhardt, F.; Wittig, M.; Kässens, J.; et al. Genome-Wide Association Study in 8956 German Individuals Identifies Influence of ABO Histo-Blood Groups on Gut Microbiome. Nat. Genet. 2021, 53, 147–155. [Google Scholar] [CrossRef]
- Ishida, S.; Kato, K.; Tanaka, M.; Odamaki, T.; Kubo, R.; Mitsuyama, E.; Xiao, J.; Yamaguchi, R.; Uematsu, S.; Imoto, S.; et al. Genome-Wide Association Studies and Heritability Analysis Reveal the Involvement of Host Genetics in the Japanese Gut Microbiota. Commun. Biol. 2020, 3, 686. [Google Scholar] [CrossRef]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-Scale Association Analyses Identify Host Factors Influencing Human Gut Microbiome Composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef]
- Reich, D.; Thangaraj, K.; Patterson, N.; Price, A.L.; Singh, L. Reconstructing Indian Population History. Nature 2009, 461, 489–494. [Google Scholar] [CrossRef]
- Ragsdale, C.S.; Edgar, H.J.H. Cultural Interaction and Biological Distance in Postclassic Period Mexico. Am. J. Phys. Anthropol. 2015, 157, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.V.P.L.; Velázquez-Cruz, R.; Baca, V.; Lima, G.; Granados, J.; Orozco, L.; Alarcón-Riquelme, M.E. Genetic Association of IRF5 with SLE in Mexicans: Higher Frequency of the Risk Haplotype and Its Homozygozity than Europeans. Hum. Genet. 2007, 121, 721–727. [Google Scholar] [CrossRef] [PubMed]
- O’Fallon, B.D.; Fehren-Schmitz, L. Native Americans Experienced a Strong Population Bottleneck Coincident with European Contact. Proc. Natl. Acad. Sci. USA 2011, 108, 20444–20448. [Google Scholar] [CrossRef]
- Vågene, Å.J.; Herbig, A.; Campana, M.G.; Robles García, N.M.; Warinner, C.; Sabin, S.; Spyrou, M.A.; Andrades Valtueña, A.; Huson, D.; Tuross, N.; et al. Salmonella Enterica Genomes from Victims of a Major Sixteenth-Century Epidemic in Mexico. Nat. Ecol. Evol. 2018, 2, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Rees, J.S.; Castellano, S.; Andrés, A.M. The Genomics of Human Local Adaptation. Trends Genet. 2020, 36, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Jeong, C.; di Rienzo, A. Adaptations to Local Environments in Modern Human Populations. Curr. Opin. Genet. Dev. 2014, 29, 1–8. [Google Scholar] [CrossRef]
- Minster, R.L.; Hawley, N.L.; Su, C.-T.; Sun, G.; Kershaw, E.E.; Cheng, H.; Buhule, O.D.; Lin, J.; Reupena, M.S.; Viali, S.; et al. A Thrifty Variant in CREBRF Strongly Influences Body Mass Index in Samoans. Nat. Genet. 2016, 48, 1049–1054. [Google Scholar] [CrossRef]
- Maguolo, A.; Zusi, C.; Corradi, M.; Emiliani, F.; Fornari, E.; Morandi, A.; Maffeis, C. FADS Cluster Variants Are Associated with Insulin Sensitivity in Children and Adolescents with Overweight and Obesity. Eur. J. Clin. Nutr. 2022, 76, 1776–1778. [Google Scholar] [CrossRef]
- Lecarpentier, Y.; Claes, V.; Vallée, A.; Hébert, J.L. Interactions between PPAR Gamma and the Canonical Wnt/Beta-Catenin Pathway in Type 2 Diabetes and Colon Cancer. PPAR Res. 2017, 2017, 5879090. [Google Scholar] [CrossRef]
- Rosas-Plaza, S.; Hernández-Terán, A.; Navarro-Díaz, M.; Escalante, A.E.; Morales-Espinosa, R.; Cerritos, R. Human Gut Microbiome across Different Lifestyles: From Hunter-Gatherers to Urban Populations. Front. Microbiol. 2022, 13, 843170. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Ortiz, H.; Barajas-Olmos, F.; Contreras-Cubas, C.; Reynolds, A.W.; Flores-Huacuja, M.; Snow, M.; Ramos-Madrigal, J.; Mendoza-Caamal, E.; Baca, P.; López-Escobar, T.A.; et al. Unraveling Signatures of Local Adaptation among Indigenous Groups from Mexico. Genes 2022, 13, 2251. https://doi.org/10.3390/genes13122251
García-Ortiz H, Barajas-Olmos F, Contreras-Cubas C, Reynolds AW, Flores-Huacuja M, Snow M, Ramos-Madrigal J, Mendoza-Caamal E, Baca P, López-Escobar TA, et al. Unraveling Signatures of Local Adaptation among Indigenous Groups from Mexico. Genes. 2022; 13(12):2251. https://doi.org/10.3390/genes13122251
Chicago/Turabian StyleGarcía-Ortiz, Humberto, Francisco Barajas-Olmos, Cecilia Contreras-Cubas, Austin W. Reynolds, Marlen Flores-Huacuja, Meradeth Snow, Jazmín Ramos-Madrigal, Elvia Mendoza-Caamal, Paulina Baca, Tomás A. López-Escobar, and et al. 2022. "Unraveling Signatures of Local Adaptation among Indigenous Groups from Mexico" Genes 13, no. 12: 2251. https://doi.org/10.3390/genes13122251
APA StyleGarcía-Ortiz, H., Barajas-Olmos, F., Contreras-Cubas, C., Reynolds, A. W., Flores-Huacuja, M., Snow, M., Ramos-Madrigal, J., Mendoza-Caamal, E., Baca, P., López-Escobar, T. A., Bolnick, D. A., Flores-Martínez, S. E., Ortiz-Lopez, R., Kostic, A. D., Villafan-Bernal, J. R., Galaviz-Hernández, C., Centeno-Cruz, F., García-Zapién, A. G., Monge-Cázares, T., ... Orozco, L. (2022). Unraveling Signatures of Local Adaptation among Indigenous Groups from Mexico. Genes, 13(12), 2251. https://doi.org/10.3390/genes13122251





