Phylogeographic Diversity Analysis of Bipolaris sorokiniana (Sacc.) Shoemaker Causing Spot Blotch Disease in Wheat and Barley
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Diseased Samples, Isolation of Bipolaris Isolates and Their Maintenance
2.2. DNA Extraction and PCR Amplification
2.3. Phylogenetic Analysis
2.4. Genetic Diversity and Haplotype Relationship
2.5. Demographic History and Divergence Time
3. Results
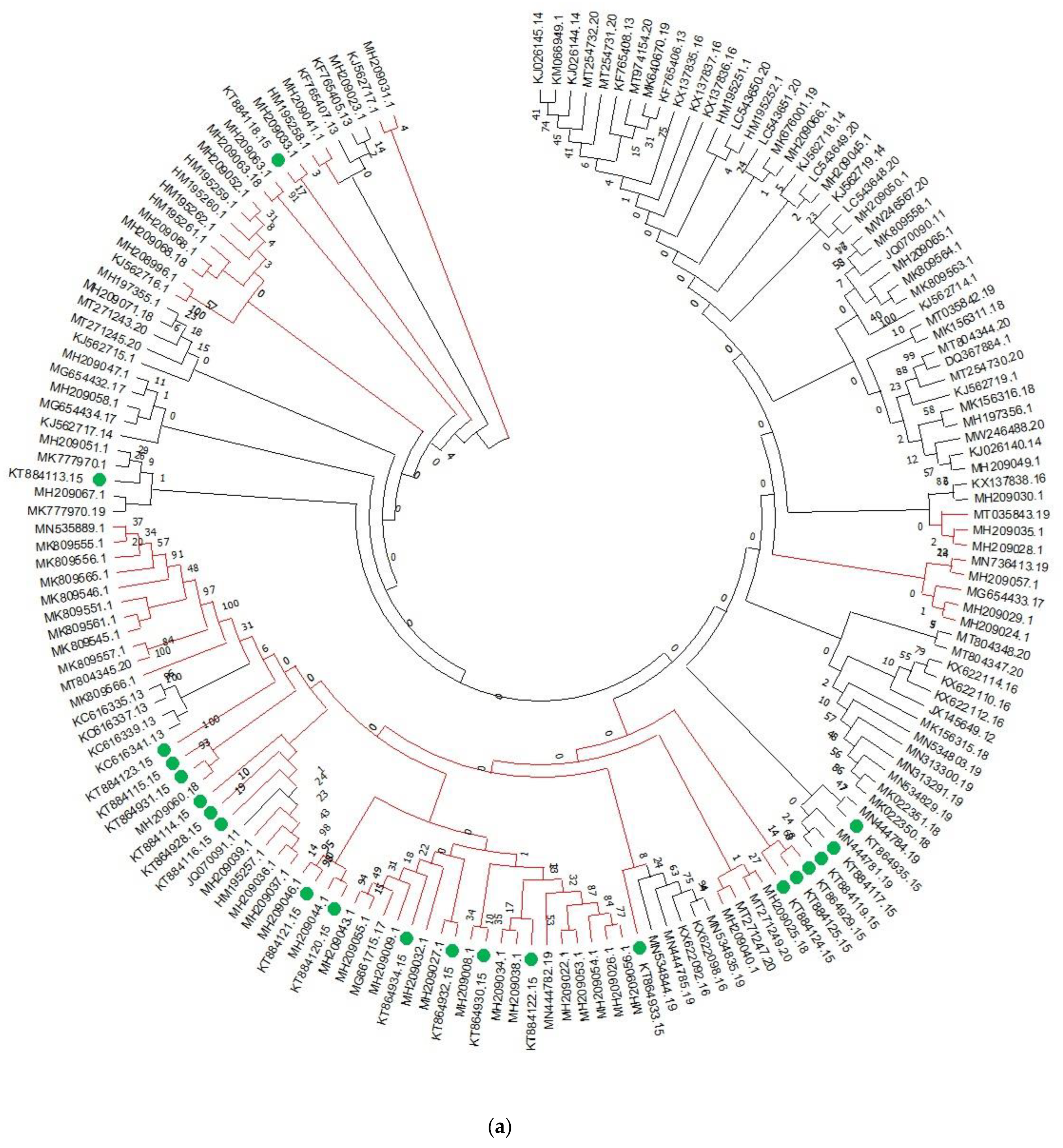
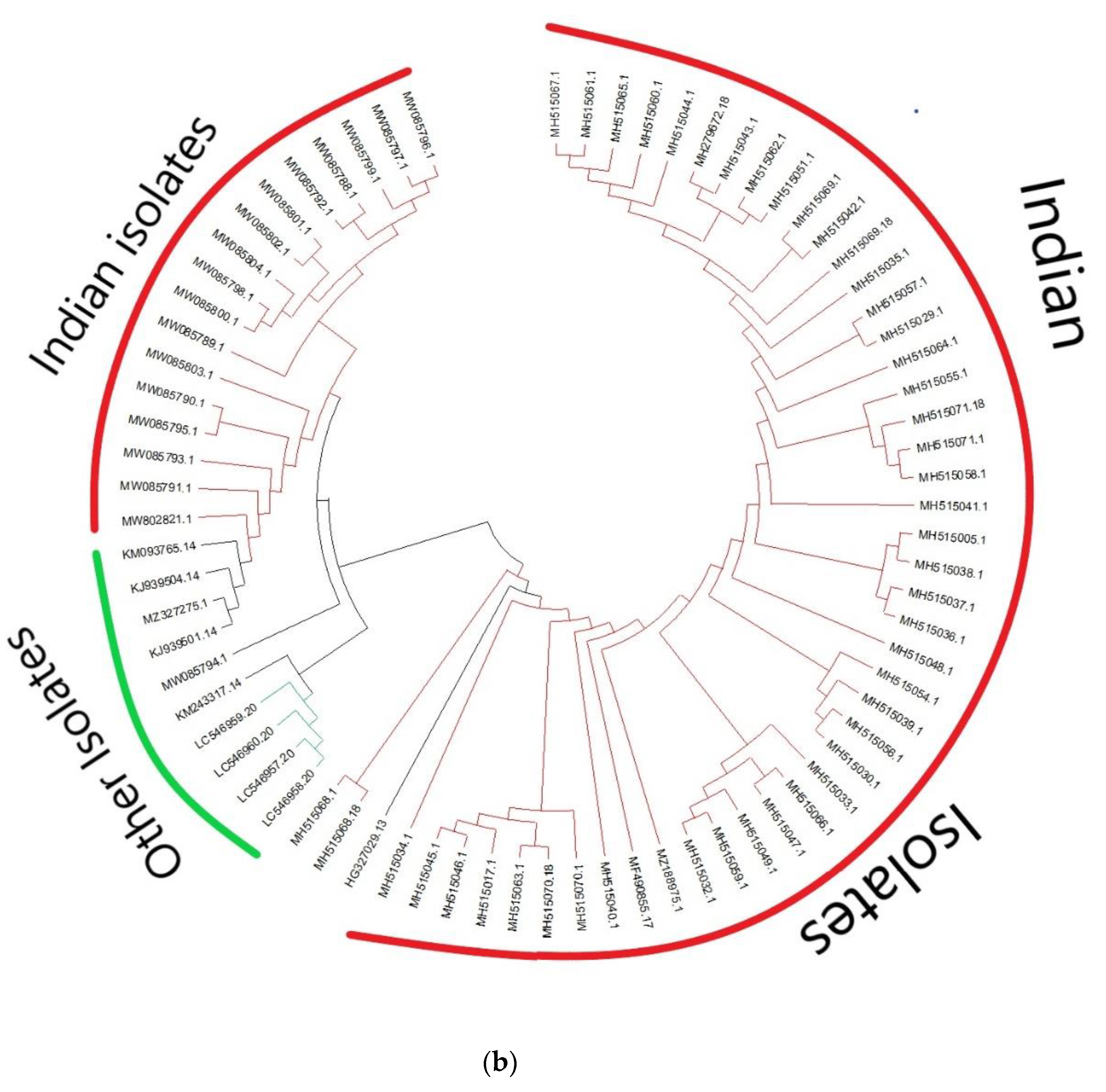
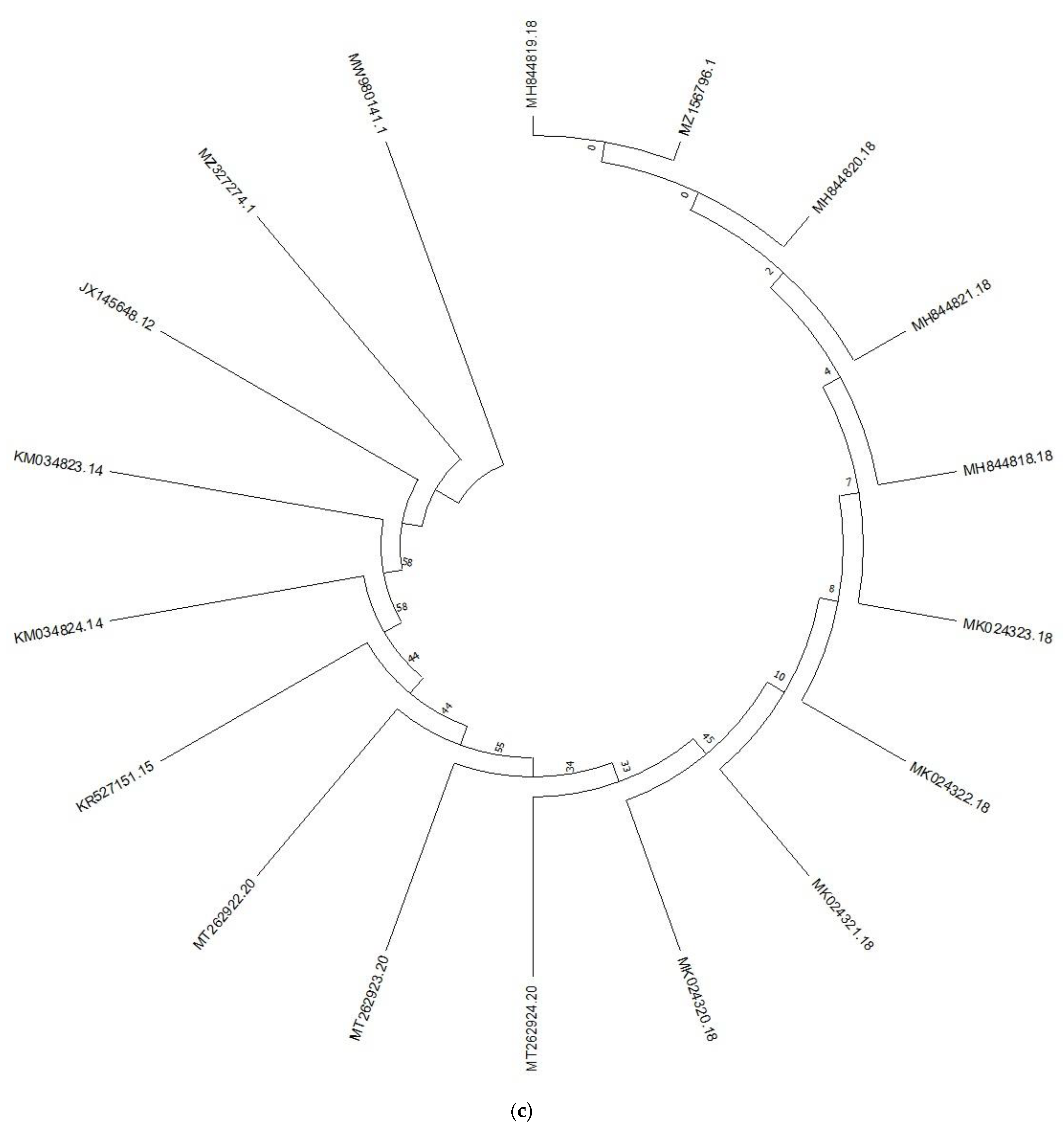
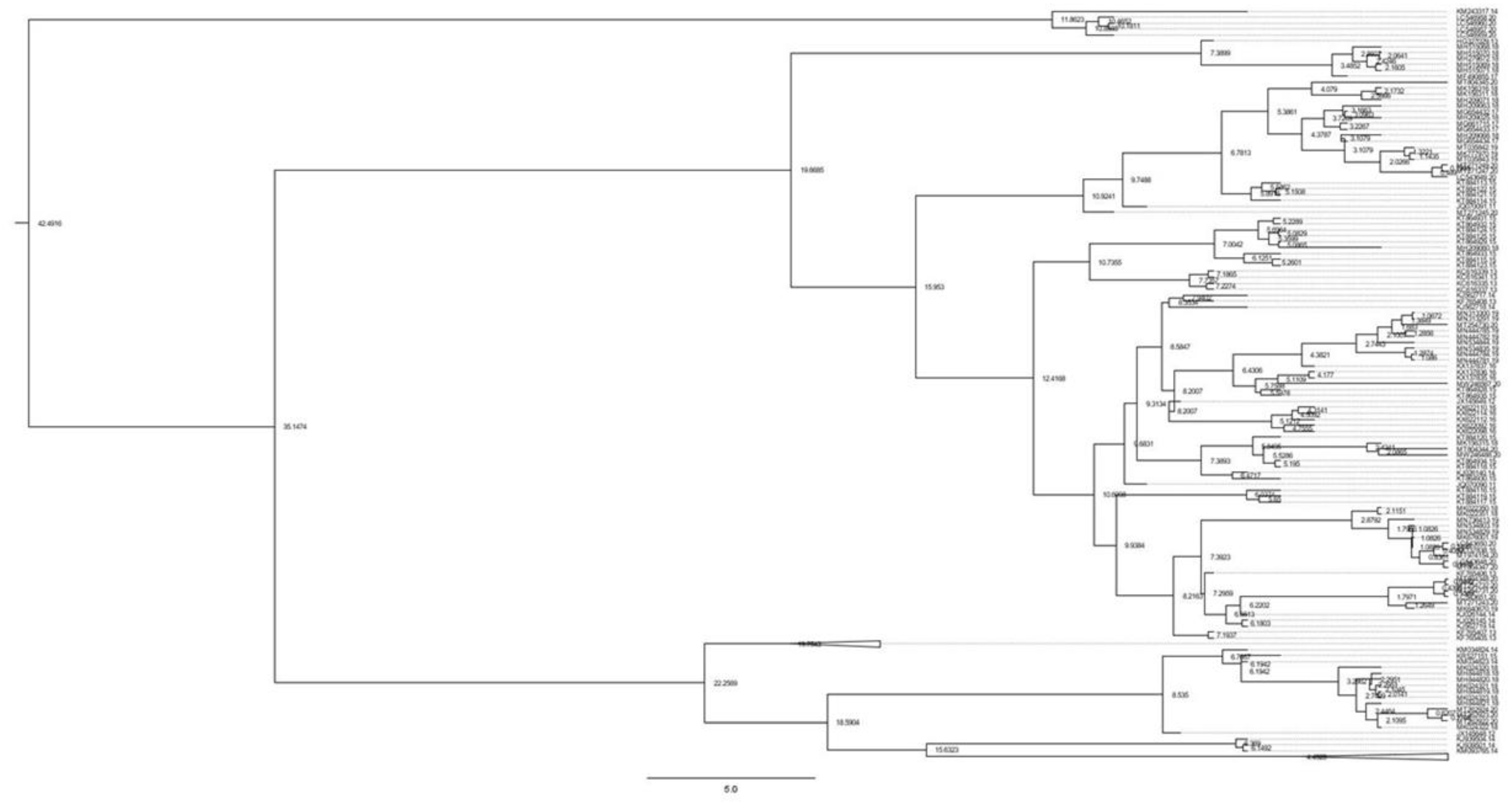
3.1. Analysis of Genetic Diversity and Phylogenetic Relationships
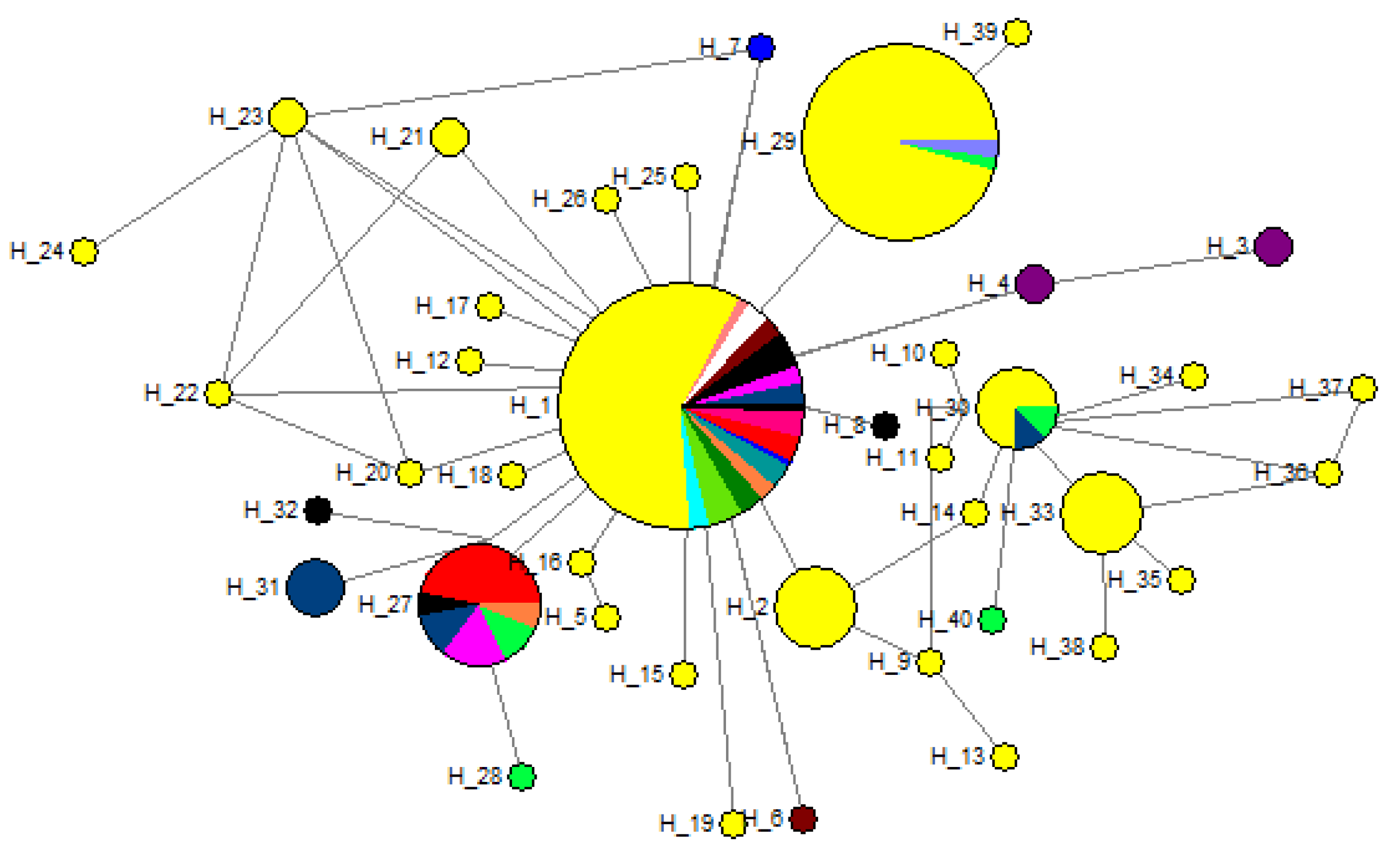
3.2. Haplotype Relationship
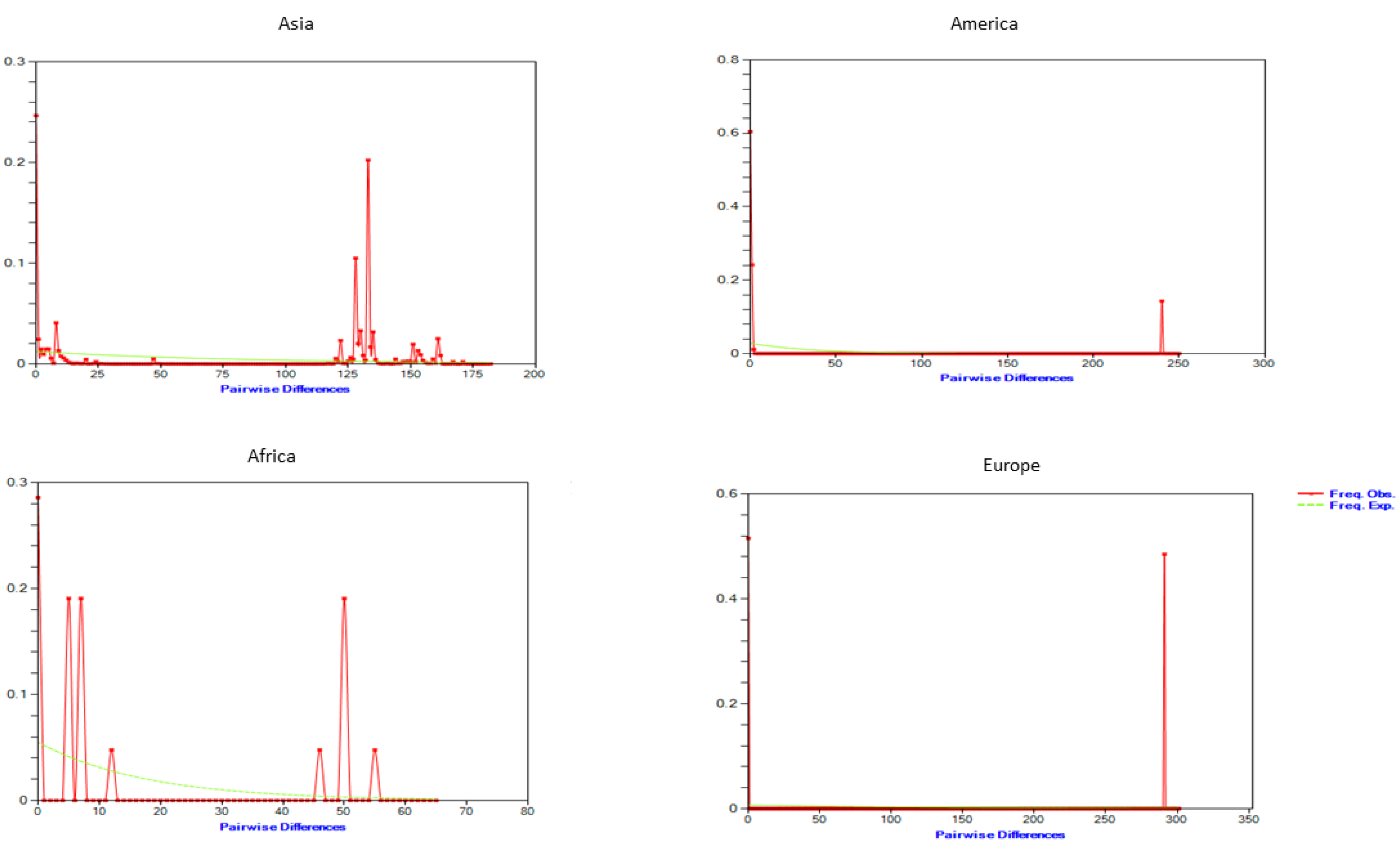
3.3. Population Demography Events and Divergence Time
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dutbayev, Y.; Kuldybayev, N.; Daugaliyeva, S.; Ismailova, E.; Sultanova, N.; Özer, G.; Slyamova, A.; Mukin, K.; Dababat, A.; Yessimbekova, M. Occurrence of spot blotch in spring barley caused by Bipolaris sorokiniana Shoem. in South-Eastern Kazakhstan. Sci. World J. 2022, 2022, 3602996. [Google Scholar] [CrossRef]
- Gupta, P.K.; Chand, R.; Vasistha, N.K.; Pandey, S.P.; Kumar, U.; Mishra, V.K.; Joshi, A.K. Spot blotch disease of wheat: The current status of research on genetics and breeding. Plant. Pathol. 2018, 67, 508–531. [Google Scholar] [CrossRef]
- Raemaekers, R.H. First occurrence in nature of Cochliobolussativus, the teleomorph of Bipolarissorokiniana. In Contribution to the Epidemiology of Bipolarissorokiniana Diseases and the Development of Rainfed Wheat, a New Crop in Zambia; Katholieke Universiteitte Leuven: Leuven, Belgium, 1991; pp. 70–85. [Google Scholar]
- Sultana, S.; Adhikary, S.K.; Islam, M.; Rahman, S.M.M. Evaluation of Pathogenic Variability Based on Leaf Blotch Disease Development Components of Bipolarissorokiniana in Triticumaestivum and Agroclimatic Origin. Plant. Pathol. J. 2018, 34, 93–103. [Google Scholar] [CrossRef]
- Sharma, R.C.; Duveiller, E. Spot Blotch Continues to Cause Substantial Grain Yield Reductions under Resource-limited Farming Conditions. J. Phytopathol. 2006, 154, 482–488. [Google Scholar] [CrossRef]
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef]
- Verma, S.K.; Chaurasia, S.K.; Pankaj, Y.K.; Kumar, R. Study on the genetic variability and pathogenicity assessment among isolates of spot blotch causing fungi (Bipolaris sorokiniana) in wheat (Triticum aestivum, L.). Plant. Physiol. Rep. 2020, 25, 255–267. [Google Scholar] [CrossRef]
- Pandey, S.P.; Sharma SChand, R.; Shahi, P.; Josshi, A.K. Clonal variability and its relevance in generation of new pathotypes in the spot blotch pathogen, Bipolaris sorokiniana. Curr. Microbiol. 2008, 56, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Iftikhar, S.; Asad, S.; Munir, A.; Sultan, A.; Ahmad, I. Hosts of Bipolaris sorokiniana, the major pathogen of spot blotch of wheat in Pakistan. Pak. J. Bot. 2001, 41, 1433–1436. [Google Scholar]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Sweta; Prasad, L.C.; Sharma, S.; Kumar, S.; Prasad, R.; Pandey, S.P.; Chand, R.; Joshi, A.K. Identification of molecular marker and aggressiveness for different groups of Bipolaris sorokiniana isolates causing spot blotch disease in wheat (Triticum aestivum, L.). Curr. Microbiol. 2007, 55, 135–141. [Google Scholar] [CrossRef] [PubMed]
- De Moura Nascimento, E.J.; van der Sand, S.T. Restriction analysis of the amplified ribosomal DNA spacers ITS1 and ITS2 of Bipolaris sorokiniana isolates. World J. Microbiol. Biotechnol. 2008, 24, 647–652. [Google Scholar] [CrossRef]
- Burdon, J.J.; Silk, J. Sources and patterns of diversity in plant-pathogenic fungi. Phytopathology 1997, 87, 664–669. [Google Scholar] [CrossRef] [PubMed]
- Tinline, R.D. Cochliobolus sativus: V. heterokayosis and parasexualtiy. Can. J. Bot. 1962, 40, 425–437. [Google Scholar] [CrossRef]
- Lahr, D.J.G.; Parfrey, L.W.; Mitchell, E.; Katz, L.; Lara, E. The chastity of amoebae: Re-evaluating evidence for sex in amoeboid organisms. Proc. Boil. Sci. 2011, 278, 2081–2090. [Google Scholar] [CrossRef]
- Van Ginkel, M.; Rajaram, S. Breeding for resistance to spot blotch in wheat: Global perspective. In Helminthosophism Diseases of Wheat: Spot Blotch and Tan Spot; Duveiller, E., Dubin, H.J., Reeves, J., McNab, A., Eds.; CIMMYT: Tolantongo, Mexico, 1998; pp. 162–170. [Google Scholar]
- Deli, T.; Kiel, C.; Schubart, C.D. Phylogeographic and evolutionary history analyses of the warty crab Eriphiaverrucosa (Decapoda, Brachyura, Eriphiidae) unveil genetic imprints of a late Pleistocene vicariant event across the Gibraltar Strait, erased by postglacial expansion and admixture among refugial lineages. BMC Evol. Biol. 2019, 19, 105. [Google Scholar] [CrossRef]
- Zhong, S.; Steffenson, B.J.; Martinez, J.P.; Ciuffetti, L.M. A Molecular genetic map and rlectrophoretic karyotype of the plant pathogenic fungus Cochliobolus sativus. Mol. Plant.-Microbe Interact. 2002, 15, 481–492. [Google Scholar] [CrossRef]
- Lu, S.; Platz, G.J.; Edwards, M.C.; Friesen, T.L. Mating type locus-specific polymerase chain reaction markers for differentiation of Pyrenophorateres f. teres and P. teres f. maculata, the causal agents of barley net blotch. Phytopathology 2010, 100, 1298–1306. [Google Scholar]
- Aggarwal, R.; Singh, V.B.; Shukla, R.; Gurjar, M.S.; Gupta, S.; Sharma, T.R. URP-based DNA fingerprinting of Bipolaris sorokiniana isolates causing ppot blotch of Wheat. J. Phytopathol. 2010, 158, 210–216. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Senwanna, C.; Cheewangkoon, R.; Crous, P.W. New species and records of Bipolaris and Curvularia from Thailand. Mycosphere 2017, 8, 1556–1574. [Google Scholar] [CrossRef]
- Bhattacharjee, P.; Sen, A.; Chakraborty, U. Chakraborty BRAPD analysis and rDNA gene sequence-based phylogeny of Bipolaris sorokiniana, a spot blotch pathogen of sorghum. NBU J. Plant. Sci. 2019, 11, 101–114. [Google Scholar] [CrossRef]
- Meng, Y.; Wang, J.; Bai, B.; Wang, L.; Yao, L.; Ma, Z.; Si, E.; Li, B.; Ma, X.; Shang, X.; et al. Genome sequence resource for pathogen Bipolaris sorokiniana Shoemaker GN1 causing spot blotch of barley (Hordeum vulgare L.). Plant Dis. 2020, 104, 1574–1577. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.B.; Spadari, C.C.; Feltrin, T.; Frazzon, A.P.G.; Germani, J.C.; Van Der Sand, S.T. Genetic variability of Bipolaris sorokiniana isolates using URP-PCR. Trop. Plant. Pathol. 2014, 39, 163–171. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, D.; Gong, G.; Qi, X.; Ye, K.; Zhou, Y.; Liu, N.; Chang, X. Spot blotch on volunteer wheat plants in Sichuan, China. J. Plant. Pathol. 2015, 97, 173–176. [Google Scholar] [CrossRef]
- Kang, R.; Hu, Y.; Wang, L.; Xie, S.; Li, Y.; Yuan, H.; Li, H. Pathogenicity variation and DNA polymorphism of Bipolaris sorokiniana infecting winter wheat in the Huanghuai foold plain of China. Plant. Pathol. 2021, 70, 87–89. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Dong, Y.; Jayawardena, R.S.; Jeewon, R.; Phukhamsakda, C.; Bundhun, D.; Hyde, K.D.; Sheng, J. A polyphasic approach to delineate species in Bipolaris. Fungal Divers. 2020, 102, 225–256. [Google Scholar] [CrossRef]
- Sharma, P.N.; Kaur, M.; Sharma, O.P.; Pathania, A. Morphological, pathological and molecular variability in Colletotrichum capsici, the cause of fruit rot of chillies in the Subtropical region of North-western India. J. Phytopathol. 2005, 153, 232–237. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, S.; Pandey, B.; Muthusamy, S.; Kumar, S.; Saharan, M.; Kumar, S.; Singroha, G.; Sharma, I.; Singh, G. Development and validation of microsatellite markers for Karnal bunt (Tilletia indica) and loose smut (Ustilago segetum tritici) of wheat from related fungal species. J. Phytopathol. 2018, 166, 729–738. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–955. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.R. Genetic evidence for a Pleistocene population expansion. Evolution 1995, 49, 608–615. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. Molecular Evolution, Phylogenetics and Epidemiology. FigTree v1.3.1. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 October 2022).
- Zhang, W.; Manawasinghe, I.S.; Zhao, W.; Xu, J.; Brooks, S.; Zhao, X.; Hyde, K.D.; Chethana, K.W.T.; Liu, J.; Li, X.; et al. Multiple gene genealogy reveals high genetic diversity and evidence for multiple origins of Chinese Plasmopara viticola population. Sci. Rep. 2017, 7, 17304. [Google Scholar] [CrossRef]
- Stumpf, M.P. Haplotype diversity and SNP frequency dependence in the description of genetic variation. Eur. J. Hum. Genet. 2004, 12, 469–477. [Google Scholar] [CrossRef]
- Dietzel, K.; Valle, D.; Fierer, N.; U’Ren, J.M.; Barberán, A. Geographical distribution of fungal plant pathogens in Dust across the United States. Front. Ecol. Evol. 2019, 7, 304. [Google Scholar] [CrossRef]
- Zaffarano, P.L.; McDonald, B.A.; Linde, C.C. Rapid speciation following recent host shifts in the plant pathogenic fungus rhynchosporium. Evolution 2008, 62, 1418–1436. [Google Scholar] [CrossRef]
- Katoch, A.; Prabhakar, C.S.; Sharma, P.N. Metageographic population analysis of Colletotrichumtruncatum associated with chili fruit rot and other hosts using ITS region nucleotide sequences. J. Plant. Biochem. Biotechnol. 2016, 25, 64–72. [Google Scholar] [CrossRef]
- Prabhakar, C.S.; Mehta, P.K.; Sood, P.; Singh, S.K.; Sharma, P.; Sharma, P.N. Population genetic structure of the melon fly, Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae) based on mitochondrial cytochrome oxidase (COI) gene sequences. Genetica 2012, 140, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wei, H.; Taheri, M.; Khormali, F.; Danukalova, G.; Chen, F. Early Pleistocene climate in western arid central Asia inferred from loess-palaeosol sequences. Sci. Rep. 2016, 6, 20560. [Google Scholar] [CrossRef] [PubMed]
- Lauer, T.; Vlaminck, S.; Frechen, M.; Rolf, C.; Kehl, M.; Sharifi, J.; Lehndorff, E.; Khormali, F. The Agh Band loess-palaeosol sequence–A terrestrial archive for climatic shifts during the last and penultimate glacial–interglacial cycles in a semiarid region in northern Iran. Quat. Int. 2017, 429, 13–30. [Google Scholar] [CrossRef]
- Zhang, H.X.; Zhang, M.L. Genetic structure of the Delphinium navicul are species group tracks Pleistocene climatic oscillations in the Tianshan Mountains, arid Central Asia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2012, 5, 353–355. [Google Scholar]
| Population | N | Hd | π | S | K | Fu’ Fs | Tajima’s D | R2 |
|---|---|---|---|---|---|---|---|---|
| India | 161 | 0.74 ± 0.028 | 0.34 ± 0.01 | 273 | 96.5 | 44.4 | 0.278 | 0.175 |
| Japan | 11 | 0.76 ± 0.08 | 0.45 ± 0.06 | 208 | 101.78 | 22.85 | −0.05 | 0.167 |
| Syria | 4 | 1 ± 0.177 | 0.009 ± 0.002 | 1 | 0.66 | −0.361 | 0.389 | 0.182 |
| USA | 4 | 1 ± 0.17 | 0.28 ± 0.152 | 128 | 64 | 2.988 | −0.88 *** | 0.43 |
| Argentina | 6 | 1 ± 0.096 | 0.5 ± 0.107 | 202 | 114.5 | 2.78 | −0.26 | 0.282 |
| Jordan | 4 | 0.5 ± 0.265 | 0.01 ± 0.006 | 2 | 1 | 4.419 | −0.84 | 0.433 |
| Egypt | 3 | 1 ± 0.272 | 0.0896 ± 0.03 | 2 | 1.33 | - | - | 0.362 |
| Turkey | 8 | 0.57 ± 0.09 | 0.33 ± 0.05 | 291 | 166 | 27.43 | 2.64 *** | 0.285 |
| China | 9 | 0.833 ± 0.127 | 0.240 ± 0.110 | 238 | 69.63 | 7.4 | −1.796 * | 0.215 |
| Azerbaijan | 10 | 0.64 ± 0.10 | 0.30 ± 0.0 | 306 | 163 | 27.17 | 2.43 ** | 0.264 |
| Mexico | 4 | 0.5 ± 0.265 | 0.0019 ± 0. | 2 | 1 | 1.099 | −0.709 | 0.433 |
| Russia | 5 | 1 ± 0.126 | 0.045 ± 0.012 | 51 | 27.2 | 0.874 | −0.97 | 0.218 |
| Kazakhstan | 6 | 0.6 ± 0.12 | 0.34 ± 0.07 | 282 | 169 | 20.9 | 2.4 | 0.3 |
| Morocco | 4 | 0.8 ± 0.22 | 0.006 ± 0.002 | 6 | 3.16 | 0.811 | −0.314 | 0.365 |
| Total | 239 | 0.7 ± 0.027 | 0.33 ± 0.01 | 225 | 75.5 | 60.02 | −0.105 | 0.3 |
| F-Statistic | ||||
|---|---|---|---|---|
| Groups | Variation% | Փst | Փsc | Փct |
| Among populations | 5.84681 | 0.05847 | - | - |
| within populations | 94.15319 | |||
| Փst | Փsc | Փct | ||
| Among groups | 7.44 | 0.182 | 0.233 | −0.066 |
| Among populations within groups | 27.77 | |||
| Within population within groups | 91.28 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, P.; Mishra, S.; Singroha, G.; Kumar, R.S.; Singh, S.K.; Singh, G.P. Phylogeographic Diversity Analysis of Bipolaris sorokiniana (Sacc.) Shoemaker Causing Spot Blotch Disease in Wheat and Barley. Genes 2022, 13, 2206. https://doi.org/10.3390/genes13122206
Sharma P, Mishra S, Singroha G, Kumar RS, Singh SK, Singh GP. Phylogeographic Diversity Analysis of Bipolaris sorokiniana (Sacc.) Shoemaker Causing Spot Blotch Disease in Wheat and Barley. Genes. 2022; 13(12):2206. https://doi.org/10.3390/genes13122206
Chicago/Turabian StyleSharma, Pradeep, Shefali Mishra, Garima Singroha, Rajan Selva Kumar, Sanjay Kumar Singh, and Gyanendra Pratap Singh. 2022. "Phylogeographic Diversity Analysis of Bipolaris sorokiniana (Sacc.) Shoemaker Causing Spot Blotch Disease in Wheat and Barley" Genes 13, no. 12: 2206. https://doi.org/10.3390/genes13122206
APA StyleSharma, P., Mishra, S., Singroha, G., Kumar, R. S., Singh, S. K., & Singh, G. P. (2022). Phylogeographic Diversity Analysis of Bipolaris sorokiniana (Sacc.) Shoemaker Causing Spot Blotch Disease in Wheat and Barley. Genes, 13(12), 2206. https://doi.org/10.3390/genes13122206








