Pathophysiological Role and Diagnostic Potential of R-Loops in Cancer and Beyond
Abstract
1. Introduction
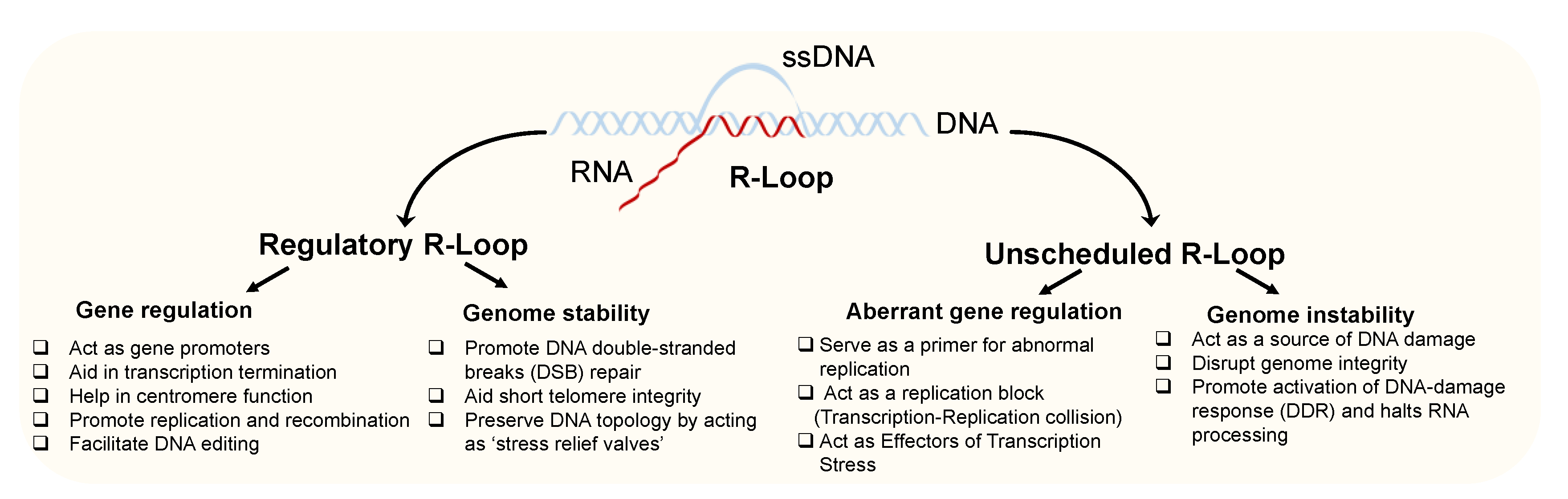
2. Roles of Regulatory R-Loops in Gene Regulation and Genome Stability
3. Role of Unscheduled R-Loops as a Source of DNA Damage and Genomic Instability
4. Role of R-Loop-Binding Proteins and (Co-)Transcriptional Mechanisms in R-Loop Formation, Resolution, and Prevention of Aberrant R-Loop Accumulation
| Protein | Function |
|---|---|
| Transcription initiation and capping | |
| Capping enzyme-Pol II complex [62] | Responsible for transcription initiation by modulating displacement of nascent RNA during transcription, thereby promoting R-loop formation |
| Transcription elongation | |
| Facilitates Chromatin Transcription (FACT) complex [17] | Helps in preventing R-loop accumulation-causing TRCs |
| Transcription termination, cleavage, and polyadenylation | |
| Cleavage and Polyadenylation (CPA) factors [15,29,69] (PCF11, CLP1, FIP1L1, CFT2, WDR33) | Suppresses R-loop formation and facilitates efficient mRNA cleavage, thereby preventing replication-stress-associated DNA damage |
| RNA processing and export | |
| Transcription and export complex (THO/TREX complex; Tho2/THOC2, Hpr1/THOC1, Mft1, Thp2, Sub2/UAP56) [13,73] | Inhibits aberrant R-loop formation and transcription-associated recombination |
| Splicing | |
| Serine- And Arginine-Rich Splicing Factor SRSF2 [16] | Prevents the formation of mutagenic R loop structures |
| RNA-Binding Protein With Serine Rich Domain 1 (RNPS1) [16] | Forms complex with ASF/SF2 to prevent transcriptional R-loops |
| R-loop degradation | |
| RNase H1/2 [10,74] | Prevents aberrant R-loop formation by timely removal of these hybrids |
| R-Loop-processing factors (DNA–RNA helicases) | |
| Senataxin (SETX) [75] | Binds to replication forks to protect its integrity across RNA-Polymerase-II-transcribed gene and unwinds unnecessary R-loops |
| Aquarius (AQR) [76] | Prevents R-loop formation by unwinding DNA–RNA hybrids |
| DExH-Box Helicase 9 (DHX9) [77] | Prevents R-loop formation by melting DNA–RNA hybrid with a 3′–5′ polarity |
| DExH-Box Helicase 11 (DHX11) [78] | Converts RNA G-Quadruplex structures into R-Loops to promote IgH class switch recombination |
| Werner Syndrome RecQ-Like Helicase (WRN) [64] | Protects the replication fork by preventing unscheduled R-loop formation |
| DNA topology | |
| Topoisomerase I/IIIB [55,67,79] | Involved in maintaining R-loop resolution by interacting with RNA-splicing and DNA-processing factors |
| DNA repair and genome maintenance | |
| Ataxia Telangiectasia Mutated (ATM)/Ataxia Telangiectasia And Rad3 Related (ATR) Kinase [57] | DNA-damage response (DDR) kinases that become activated when R-loop-mediated DNA damage occurs |
| Breast Cancer Type 2 Susceptibility Protein (BRCA2/FANCD1) [41] | Binds to R-loops in response to dsDNA breaks to invite other DNA repair factors |
5. R-Loops Associated with Human Disease
- A.
- R-loops in Nucleotide Expansion Diseases
| Diseases | Genes Associated with R-Loops |
|---|---|
| Aging [8,103,104] | SETX |
| Alzheimer’s [8,103,104,105] | SETX, WW domain-containing oxidoreductase |
| Aicardi–Goutières syndrome (AGS) [106] | TREX1, RNASEH2 |
| AIDS-associated malignancies [107] | TREX complex |
| Amyotrophic lateral sclerosis (ALS) [30,93,94] | C9orf72 and ATXN2 (GGGCCC)n, SETX |
| Alternative lengthening of telomere (ALT)-dependent cancers [108] | TERRA complex |
| Ataxia with oculomotor apraxia (AOA2) [7,109,110] | SETX |
| Breast cancer [111,112,113,114] | BRAC1, BRAC2, Estrogen, SETX |
| Burkitt’s lymphoma [84] | c-MYC, TRD3-TOP3B |
| Colon cancer [115,116] | VIM |
| Myotonic dystrophy type 1 (DM1) [100] | DMPK |
| Embryonal tumors with multilayered rosettes (EMTR) [27] | C19MC |
| Eosinophilic leukemia [15] | FIP1 |
| Ewing’s sarcoma [117] | EWS-FLI, BRCA1 |
| Frontotemporal dementia (FTD) [94] | C9orf7 (GGGCCC)n |
| Fragile X syndrome type E (FRAXE) [101,102] | FRM2 (CCG)n |
| Friedreich ataxia (FRDA) or fragile X syndrome type A (FRAXA) [96,97,98,99] | FXN (GAA)n, FRM1 (CCG)n |
| Huntington’s disease (HD) [96,97,98,99] | HTT (CAG)n |
| Infertility [118] | SETX |
| Multiple myeloma [84,119] | c-MYC, TRD3-TOP3B, IFN |
| Myelodysplastic syndromes [120] | U2AF1 (S34F), SRSF2 |
| Polyglutamine-associated ataxias [95] | Multifactorial Nucleotide Expansion disorder |
| Parkinson’s disease [8] | SETX |
| Spinocerebellar ataxias (SCAs) [39] | ATXN1/2 (CAG)n |
| Immunodeficiency, centromere instability, and facial anomalies (ICF) syndrome [121] | TERRA |
| Wiskott–Aldrich syndrome (WAS), X-linked thrombocytopenia (XLT), and X-linked neutropenia [25] | XLT-WAS |
- B.
- R-loops in Neuronal Diseases
- C.
- R-loops in Cancer
- D.
- R-loops in other diseases
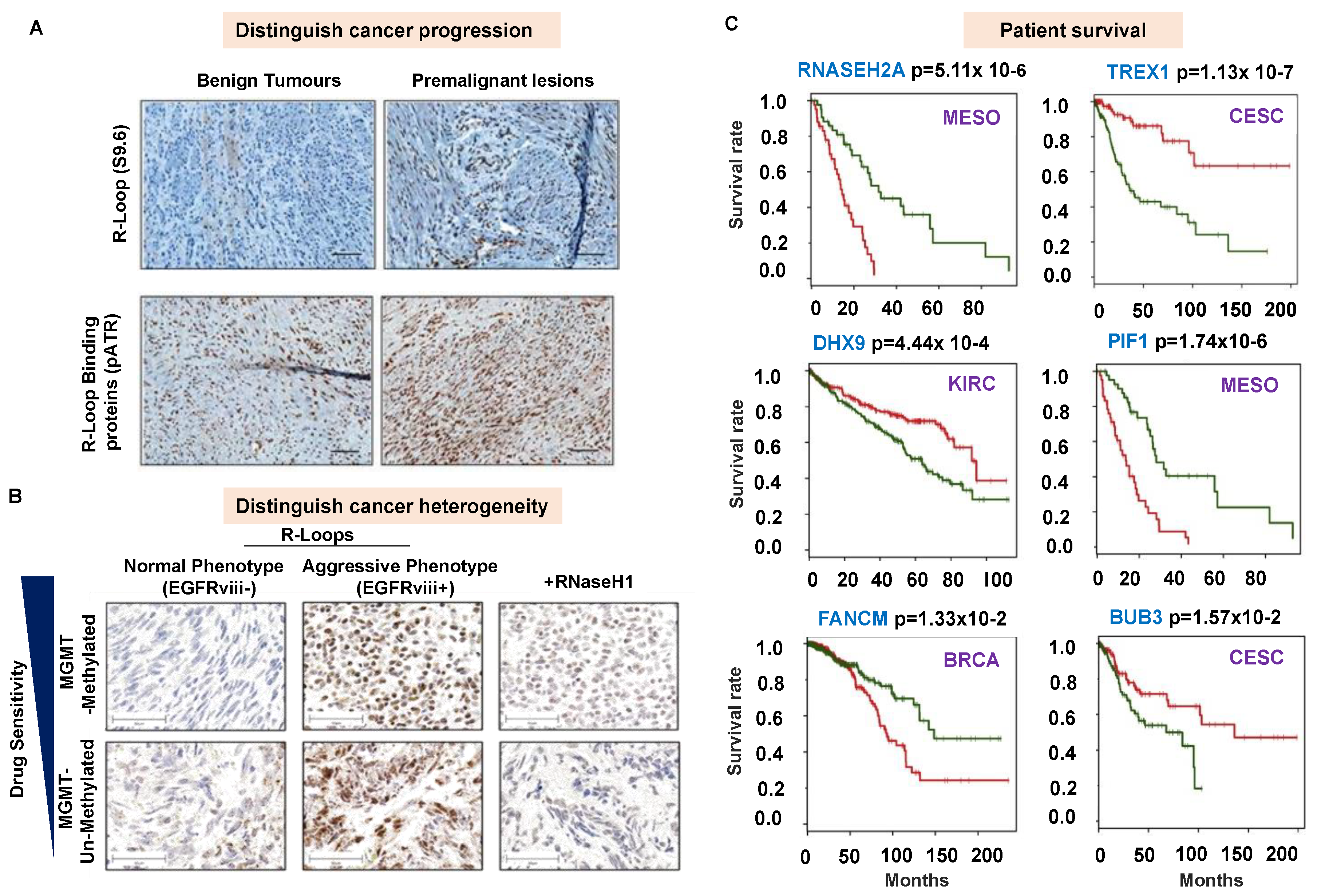
6. R-Loops as a Diagnostic Biomarker?
7. Emerging Technologies for Detecting R-Loops
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santos-Pereira, J.M.; Aguilera, A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015, 16, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.; Chédin, F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell 2016, 63, 167–178. [Google Scholar] [CrossRef]
- Allison, D.F.; Wang, G.G. R-loops: Formation, function, and relevance to cell stress. Cell Stress 2019, 3, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, Y.A.; Fernando, C.M.; Tran, E.J. The balancing act of R-loop biology: The good, the bad, and the ugly. J. Biol. Chem. 2020, 295, 905–913. [Google Scholar] [CrossRef]
- Aguilera, A.; García-Muse, T. R loops: From transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Groh, M.; Gromak, N. Out of balance: R-loops in human disease. PLoS Genet. 2014, 10, e1004630. [Google Scholar] [CrossRef] [PubMed]
- Mackay, R.P.; Xu, Q.; Weinberger, P.M. R-Loop Physiology and Pathology: A Brief Review. DNA Cell Biol. 2020, 39, 1914–1925. [Google Scholar] [CrossRef]
- Skourti-Stathaki, K.; Proudfoot, N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Mosler, T.; Conte, F.; Longo, G.M.C.; Mikicic, I.; Kreim, N.; Möckel, M.M.; Petrosino, G.; Flach, J.; Barau, J.; Luke, B.; et al. R-loop proximity proteomics identifies a role of DDX41 in transcription-associated genomic instability. Nat. Commun. 2021, 12, 7314. [Google Scholar] [CrossRef] [PubMed]
- Lam, F.C.; Kong, Y.W.; Huang, Q.; Vu Han, T.L.; Maffa, A.D.; Kasper, E.M.; Yaffe, M.B. BRD4 prevents the accumulation of R-loops and protects against transcription-replication collision events and DNA damage. Nat. Commun. 2020, 11, 4083. [Google Scholar] [CrossRef] [PubMed]
- Huertas, P.; Aguilera, A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 2003, 12, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Proudfoot, N.J.; Gromak, N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 2011, 42, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Stirling, P.C.; Chan, Y.A.; Minaker, S.W.; Aristizabal, M.J.; Barrett, I.; Sipahimalani, P.; Kobor, M.S.; Hieter, P. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012, 26, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Manley, J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 2005, 122, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Moyano, E.; Mergui, X.; García-Rubio, M.L.; Barroso, S.; Aguilera, A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 2014, 28, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Wahba, L.; Amon, J.D.; Koshland, D.; Vuica-Ross, M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA: DNA hybrids from generating genome instability. Mol. Cell 2011, 44, 978–988. [Google Scholar] [CrossRef]
- Richard, P.; Manley, J.L. R Loops and Links to Human Disease. J. Mol. Biol. 2017, 429, 3168–3180. [Google Scholar] [CrossRef]
- Wells, J.P.; White, J.; Stirling, P.C. R Loops and Their Composite Cancer Connections. Trends Cancer 2019, 5, 619–631. [Google Scholar] [CrossRef]
- Rakshit, S.; Sunny, J.S.; George, M.; Hanna, L.E.; Sarkar, K. R-loop modulated epigenetic regulation in T helper cells mechanistically associates coronary artery disease and non-small cell lung cancer. Transl. Oncol. 2021, 14, 101189. [Google Scholar] [CrossRef] [PubMed]
- Barchi, M.; Bielli, P.; Dolci, S.; Rossi, P.; Grimaldi, P. Non-Coding RNAs and Splicing Activity in Testicular Germ Cell Tumors. Life 2021, 11, 736. [Google Scholar] [CrossRef]
- Cuartas, J.; Gangwani, L. R-loop Mediated DNA Damage and Impaired DNA Repair in Spinal Muscular Atrophy. Front. Cell. Neurosci. 2022, 16, 826608. [Google Scholar] [CrossRef]
- Perego, M.G.L.; Taiana, M.; Bresolin, N.; Comi, G.P.; Corti, S. R-Loops in Motor Neuron Diseases. Mol. Neurobiol. 2019, 56, 2579–2589. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, K.; Han, S.S.; Wen, K.K.; Ochs, H.D.; Dupré, L.; Seidman, M.M.; Vyas, Y.M. R-loops cause genomic instability in T helper lymphocytes from patients with Wiskott-Aldrich syndrome. J. Allergy Clin. Immunol. 2018, 142, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gao, X.; Zhou, Z.; Kang, S.W.; Yang, Y.; Liu, H.; Zhang, C.; Wen, Z.; Rao, X.; Wang, D.; et al. San1 deficiency leads to cardiomyopathy due to excessive R-loop-associated DNA damage and cardiomyocyte hypoplasia. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166237. [Google Scholar] [CrossRef] [PubMed]
- Lambo, S.; Gröbner, S.N.; Rausch, T.; Waszak, S.M.; Schmidt, C.; Gorthi, A.; Romero, J.C.; Mauermann, M.; Brabetz, S.; Krausert, S.; et al. The molecular landscape of ETMR at diagnosis and relapse. Nature 2019, 576, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Boros-Oláh, B.; Dobos, N.; Hornyák, L.; Szabó, Z.; Karányi, Z.; Halmos, G.; Roszik, J.; Székvölgyi, L. Drugging the R-loop interactome: RNA-DNA hybrid binding proteins as targets for cancer therapy. DNA Repair 2019, 84, 102642. [Google Scholar] [CrossRef]
- Spada, S.; Luke, B.; Danckwardt, S. The Bidirectional Link Between RNA Cleavage and Polyadenylation and Genome Stability: Recent Insights From a Systematic Screen. Front. Genet. 2022, 13, 854907. [Google Scholar] [CrossRef] [PubMed]
- Grunseich, C.; Wang, I.X.; Watts, J.A.; Burdick, J.T.; Guber, R.D.; Zhu, Z.; Bruzel, A.; Lanman, T.; Chen, K.; Schindler, A.B.; et al. Senataxin Mutation Reveals How R-Loops Promote Transcription by Blocking DNA Methylation at Gene Promoters. Mol. Cell 2018, 69, 426–437.e7. [Google Scholar] [CrossRef] [PubMed]
- Ginno, P.A.; Lott, P.L.; Christensen, H.C.; Korf, I.; Chédin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 2012, 45, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e4. [Google Scholar] [CrossRef]
- Chen, P.B.; Chen, H.V.; Acharya, D.; Rando, O.J.; Fazzio, T.G. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 2015, 22, 999–1007. [Google Scholar] [CrossRef]
- Arab, K.; Karaulanov, E.; Musheev, M.; Trnka, P.; Schäfer, A.; Grummt, I.; Niehrs, C. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 2019, 51, 217–223. [Google Scholar] [CrossRef]
- Skourti-Stathaki, K.; Kamieniarz-Gdula, K.; Proudfoot, N.J. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 2014, 516, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.T.; Coulson, R.L.; Gonzales, M.L.; Crary, F.K.; Wong, S.S.; Adams, S.; Ach, R.A.; Tsang, P.; Yamada, N.A.; Yasui, D.H.; et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc. Natl. Acad. Sci. USA 2013, 110, 13938–13943. [Google Scholar] [CrossRef]
- Nakama, M.; Kawakami, K.; Kajitani, T.; Urano, T.; Murakami, Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 2012, 17, 218–233. [Google Scholar] [CrossRef]
- Castellano-Pozo, M.; Santos-Pereira, J.M.; Rondón, A.G.; Barroso, S.; Andújar, E.; Pérez-Alegre, M.; García-Muse, T.; Aguilera, A. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell 2013, 52, 583–590. [Google Scholar] [CrossRef]
- Yu, K.; Chedin, F.; Hsieh, C.L.; Wilson, T.E.; Lieber, M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003, 4, 442–451. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- D’Alessandro, G.; Whelan, D.R.; Howard, S.M.; Vitelli, V.; Renaudin, X.; Adamowicz, M.; Iannelli, F.; Jones-Weinert, C.W.; Lee, M.; Matti, V.; et al. BRCA2 controls DNA: RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 2018, 9, 5376. [Google Scholar] [CrossRef]
- Cohen, S.; Puget, N.; Lin, Y.L.; Clouaire, T.; Aguirrebengoa, M.; Rocher, V.; Pasero, P.; Canitrot, Y.; Legube, G. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 2018, 9, 533. [Google Scholar] [CrossRef]
- Li, L.; Germain, D.R.; Poon, H.Y.; Hildebrandt, M.R.; Monckton, E.A.; McDonald, D.; Hendzel, M.J.; Godbout, R. DEAD Box 1 Facilitates Removal of RNA and Homologous Recombination at DNA Double-Strand Breaks. Mol. Cell. Biol. 2016, 36, 2794–2810. [Google Scholar] [CrossRef]
- Britton, S.; Dernoncourt, E.; Delteil, C.; Froment, C.; Schiltz, O.; Salles, B.; Frit, P.; Calsou, P. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014, 42, 9047–9062. [Google Scholar] [CrossRef] [PubMed]
- Ohle, C.; Tesorero, R.; Schermann, G.; Dobrev, N.; Sinning, I.; Fischer, T. Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell 2016, 167, 1001–1013.e7. [Google Scholar] [CrossRef]
- Yasuhara, T.; Kato, R.; Hagiwara, Y.; Shiotani, B.; Yamauchi, M.; Nakada, S.; Shibata, A.; Miyagawa, K. Human Rad52 Promotes XPG-Mediated R-loop Processing to Initiate Transcription-Associated Homologous Recombination Repair. Cell 2018, 175, 558–570.e11. [Google Scholar] [CrossRef]
- Balk, B.; Maicher, A.; Dees, M.; Klermund, J.; Luke-Glaser, S.; Bender, K.; Luke, B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013, 20, 1199–1205. [Google Scholar] [CrossRef]
- Arora, R.; Azzalin, C.M. Telomere elongation chooses TERRA ALTernatives. RNA Biol. 2015, 12, 938–941. [Google Scholar] [CrossRef]
- Pfeiffer, V.; Crittin, J.; Grolimund, L.; Lingner, J. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013, 32, 2861–2871. [Google Scholar] [CrossRef]
- Graf, M.; Bonetti, D.; Lockhart, A.; Serhal, K.; Kellner, V.; Maicher, A.; Jolivet, P.; Teixeira, M.T.; Luke, B. Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell 2017, 170, 72–85.e14. [Google Scholar] [CrossRef]
- Stolz, R.; Sulthana, S.; Hartono, S.R.; Malig, M.; Benham, C.J.; Chedin, F. Interplay between DNA sequence and negative superhelicity drives R-loop structures. Proc. Natl. Acad. Sci. USA 2019, 116, 6260–6269. [Google Scholar] [CrossRef]
- Huang, F.T.; Yu, K.; Balter, B.B.; Selsing, E.; Oruc, Z.; Khamlichi, A.A.; Hsieh, C.L.; Lieber, M.R. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol. Cell. Biol. 2007, 27, 5921–5932. [Google Scholar] [CrossRef]
- Domínguez-Sánchez, M.S.; Barroso, S.; Gómez-González, B.; Luna, R.; Aguilera, A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet. 2011, 7, e1002386. [Google Scholar] [CrossRef] [PubMed]
- Kotsantis, P.; Silva, L.M.; Irmscher, S.; Jones, R.M.; Folkes, L.; Gromak, N.; Petermann, E. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 2016, 7, 13087. [Google Scholar] [CrossRef]
- Tuduri, S.; Crabbé, L.; Conti, C.; Tourrière, H.; Holtgreve-Grez, H.; Jauch, A.; Pantesco, V.; de Vos, J.; Thomas, A.; Theillet, C.; et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009, 11, 1315–1324. [Google Scholar] [CrossRef]
- El Hage, A.; French, S.L.; Beyer, A.L.; Tollervey, D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010, 24, 1546–1558. [Google Scholar] [CrossRef]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Marabitti, V.; Lillo, G.; Malacaria, E.; Palermo, V.; Sanchez, M.; Pichierri, P.; Franchitto, A. ATM pathway activation limits R-loop-associated genomic instability in Werner syndrome cells. Nucleic Acids Res. 2019, 47, 3485–3502. [Google Scholar] [CrossRef]
- Di Domenico, E.G.; Romano, E.; Del Porto, P.; Ascenzioni, F. Multifunctional role of ATM/Tel1 kinase in genome stability: From the DNA damage response to telomere maintenance. Biomed. Res. Int. 2014, 2014, 787404. [Google Scholar] [CrossRef]
- Friedel, A.M.; Pike, B.L.; Gasser, S.M. ATR/Mec1: Coordinating fork stability and repair. Curr. Opin. Cell Biol. 2009, 21, 237–244. [Google Scholar] [CrossRef]
- Danckwardt, S.; Hentze, M.W.; Kulozik, A.E. 3′ end mRNA processing: Molecular mechanisms and implications for health and disease. EMBO J. 2008, 27, 482–498. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Chu, C.; Shatkin, A.J.; Manley, J.L. Human capping enzyme promotes formation of transcriptional R loops in vitro. Proc. Natl. Acad. Sci. USA 2007, 104, 17620–17625. [Google Scholar] [CrossRef]
- Mason, P.B.; Struhl, K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell Biol. 2003, 23, 8323–8333. [Google Scholar] [CrossRef]
- Marabitti, V.; Valenzisi, P.; Lillo, G.; Malacaria, E.; Palermo, V.; Pichierri, P.; Franchitto, A. R-Loop-Associated Genomic Instability and Implication of WRN and WRNIP1. Int. J. Mol. Sci. 2022, 23, 1547. [Google Scholar] [CrossRef]
- Belotserkovskii, B.P.; Tornaletti, S.; D’Souza, A.D.; Hanawalt, P.C. R-loop generation during transcription: Formation, processing and cellular outcomes. DNA Repair 2018, 71, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Stork, C.T.; García-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef]
- Wilson-Sali, T.; Hsieh, T.S. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIbeta. Proc. Natl. Acad. Sci. USA 2002, 99, 7974–7979. [Google Scholar] [CrossRef]
- Bharati, A.P.; Singh, N.; Kumar, V.; Kashif, M.; Singh, A.K.; Singh, P.; Singh, S.K.; Siddiqi, M.I.; Tripathi, T.; Akhtar, M.S. The mRNA capping enzyme of Saccharomyces cerevisiae has dual specificity to interact with CTD of RNA Polymerase II. Sci. Rep. 2016, 6, 31294. [Google Scholar] [CrossRef]
- Teloni, F.; Michelena, J.; Lezaja, A.; Kilic, S.; Ambrosi, C.; Menon, S.; Dobrovolna, J.; Imhof, R.; Janscak, P.; Baubec, T.; et al. Efficient Pre-mRNA Cleavage Prevents Replication-Stress-Associated Genome Instability. Mol. Cell 2019, 73, 670–683.e12. [Google Scholar] [CrossRef] [PubMed]
- Mischo, H.E.; Gómez-González, B.; Grzechnik, P.; Rondón, A.G.; Wei, W.; Steinmetz, L.; Aguilera, A.; Proudfoot, N.J. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011, 41, 21–32. [Google Scholar] [CrossRef]
- Morales, J.C.; Richard, P.; Patidar, P.L.; Motea, E.A.; Dang, T.T.; Manley, J.L.; Boothman, D.A. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet. 2016, 12, e1006107. [Google Scholar] [CrossRef]
- Jimeno, S.; Aguilera, A. The THO complex as a key mRNP biogenesis factor in development and cell differentiation. J. Biol. 2010, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Gómez-González, B.; García-Rubio, M.; Bermejo, R.; Gaillard, H.; Shirahige, K.; Marín, A.; Foiani, M.; Aguilera, A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011, 30, 3106–3119. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Hou, Q.; Cheng, L.; Xu, W.; Hong, Y.; Li, S.; Sun, Q. RNase H1 Cooperates with DNA Gyrases to Restrict R-Loops and Maintain Genome Integrity in Arabidopsis Chloroplasts. Plant Cell 2017, 29, 2478–2497. [Google Scholar] [CrossRef] [PubMed]
- Alzu, A.; Bermejo, R.; Begnis, M.; Lucca, C.; Piccini, D.; Carotenuto, W.; Saponaro, M.; Brambati, A.; Cocito, A.; Foiani, M.; et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 2012, 151, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Sakasai, R.; Isono, M.; Wakasugi, M.; Hashimoto, M.; Sunatani, Y.; Matsui, T.; Shibata, A.; Matsunaga, T.; Iwabuchi, K. Aquarius is required for proper CtIP expression and homologous recombination repair. Sci. Rep. 2017, 7, 13808. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Grosse, F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair 2011, 10, 654–665. [Google Scholar] [CrossRef]
- Ribeiro de Almeida, C.; Dhir, S.; Dhir, A.; Moghaddam, A.E.; Sattentau, Q.; Meinhart, A.; Proudfoot, N.J. RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol. Cell 2018, 70, 650–662.e8. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 2002, 3, 430–440. [Google Scholar] [CrossRef]
- Zheng, X.; Peng, Q.; Wang, L.; Zhang, X.; Huang, L.; Wang, J.; Qin, Z. Serine/arginine-rich splicing factors: The bridge linking alternative splicing and cancer. Int. J. Biol. Sci. 2020, 16, 2442–2453. [Google Scholar] [CrossRef] [PubMed]
- Luna, R.; Gaillard, H.; González-Aguilera, C.; Aguilera, A. Biogenesis of mRNPs: Integrating different processes in the eukaryotic nucleus. Chromosoma 2008, 117, 319–331. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Crouch, R.J. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009, 276, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.A.; Gileadi, O. RecQ helicases in DNA repair and cancer targets. Essays Biochem. 2020, 64, 819–830. [Google Scholar] [CrossRef]
- Yang, Y.; McBride, K.M.; Hensley, S.; Lu, Y.; Chedin, F.; Bedford, M.T. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 2014, 53, 484–497. [Google Scholar] [CrossRef] [PubMed]
- Ogorodnikov, A.; Levin, M.; Tattikota, S.; Tokalov, S.; Hoque, M.; Scherzinger, D.; Marini, F.; Poetsch, A.; Binder, H.; Macher-Göppinger, S.; et al. Transcriptome 3′ end organization by PCF11 links alternative polyadenylation to formation and neuronal differentiation of neuroblastoma. Nat. Commun. 2018, 9, 5331. [Google Scholar] [CrossRef] [PubMed]
- Marini, F.; Scherzinger, D.; Danckwardt, S. TREND-DB-a transcriptome-wide atlas of the dynamic landscape of alternative polyadenylation. Nucleic Acids Res. 2021, 49, D243–D253. [Google Scholar] [CrossRef]
- Nourse, J.; Spada, S.; Danckwardt, S. Emerging Roles of RNA 3′-end Cleavage and Polyadenylation in Pathogenesis, Diagnosis and Therapy of Human Disorders. Biomolecules 2020, 10, 915. [Google Scholar] [CrossRef] [PubMed]
- Kilchert, C.; Wittmann, S.; Vasiljeva, L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016, 17, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Ogorodnikov, A.; Danckwardt, S. TRENDseq-A highly multiplexed high throughput RNA 3′ end sequencing for mapping alternative polyadenylation. Methods Enzymol. 2021, 655, 37–72. [Google Scholar] [CrossRef] [PubMed]
- Malik, I.; Kelley, C.P.; Wang, E.T.; Todd, P.K. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 2021, 22, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Paulson, H. Repeat expansion diseases. Handb. Clin. Neurol. 2018, 147, 105–123. [Google Scholar] [CrossRef] [PubMed]
- Ellerby, L.M. Repeat Expansion Disorders: Mechanisms and Therapeutics. Neurotherapeutics 2019, 16, 924–927. [Google Scholar] [CrossRef] [PubMed]
- Haeusler, A.R.; Donnelly, C.J.; Periz, G.; Simko, E.A.; Shaw, P.G.; Kim, M.S.; Maragakis, N.J.; Troncoso, J.C.; Pandey, A.; Sattler, R.; et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 2014, 507, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.; Schmidt, M.H.; Geist, J.M.; Thakkar, N.P.; Panigrahi, G.B.; Wang, Y.H.; Pearson, C.E. Processing of double-R-loops in (CAG)·(CTG) and C9orf72 (GGGGCC)·(GGCCCC) repeats causes instability. Nucleic Acids Res. 2014, 42, 10473–10487. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, C.S.; Li, D.; Wilton, S.D.; Aung-Htut, M.T. Polyglutamine Ataxias: Our Current Molecular Understanding and What the Future Holds for Antisense Therapies. Biomedicines 2021, 9, 1499. [Google Scholar] [CrossRef]
- Lin, Y.; Dent, S.Y.; Wilson, J.H.; Wells, R.D.; Napierala, M. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl. Acad. Sci. USA 2010, 107, 692–697. [Google Scholar] [CrossRef]
- Khristich, A.N.; Mirkin, S.M. On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability. J. Biol. Chem. 2020, 295, 4134–4170. [Google Scholar] [CrossRef] [PubMed]
- McMurray, C.T. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 2010, 11, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Paulson, H.L.; Fischbeck, K.H. Trinucleotide repeats in neurogenetic disorders. Annu. Rev. Neurosci. 1996, 19, 79–107. [Google Scholar] [CrossRef] [PubMed]
- Usdin, K.; House, N.C.; Freudenreich, C.H. Repeat instability during DNA repair: Insights from model systems. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 142–167. [Google Scholar] [CrossRef]
- Groh, M.; Lufino, M.M.; Wade-Martins, R.; Gromak, N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet. 2014, 10, e1004318. [Google Scholar] [CrossRef] [PubMed]
- Loomis, E.W.; Sanz, L.A.; Chédin, F.; Hagerman, P.J. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet. 2014, 10, e1004294. [Google Scholar] [CrossRef] [PubMed]
- James, P.A.; Talbot, K. The molecular genetics of non-ALS motor neuron diseases. Biochim. Biophys. Acta 2006, 1762, 986–1000. [Google Scholar] [CrossRef][Green Version]
- Palau, F.; Espinós, C. Autosomal recessive cerebellar ataxias. Orphanet. J. Rare Dis. 2006, 1, 47. [Google Scholar] [CrossRef]
- Sze, C.I.; Su, M.; Pugazhenthi, S.; Jambal, P.; Hsu, L.J.; Heath, J.; Schultz, L.; Chang, N.S. Down-regulation of WW domain-containing oxidoreductase induces Tau phosphorylation in vitro. A potential role in Alzheimer’s disease. J. Biol. Chem. 2004, 279, 30498–30506. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; Sanz, L.A.; Xu, X.; Hartono, S.R.; Chédin, F. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutières syndrome. Elife 2015, 4, e08007. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Noerenberg, M.; Whitehouse, A. A novel mechanism inducing genome instability in Kaposi’s sarcoma-associated herpesvirus infected cells. PLoS Pathog. 2014, 10, e1004098. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.S.; Rialdi, A.; Ho, J.S.; Tilove, M.; Martinez-Gil, L.; Moshkina, N.P.; Peralta, Z.; Noel, J.; Melegari, C.; Maestre, A.M.; et al. Senataxin suppresses the antiviral transcriptional response and controls viral biogenesis. Nat. Immunol. 2015, 16, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Stork, C.T.; Bocek, M.; Crossley, M.P.; Sollier, J.; Sanz, L.A.; Chédin, F.; Swigut, T.; Cimprich, K.A. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 2016, 5, e17548. [Google Scholar] [CrossRef]
- Hatchi, E.; Skourti-Stathaki, K.; Ventz, S.; Pinello, L.; Yen, A.; Kamieniarz-Gdula, K.; Dimitrov, S.; Pathania, S.; McKinney, K.M.; Eaton, M.L.; et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 2015, 57, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.J.; Rolland, T.; Adelmant, G.; Xia, X.; Owen, M.S.; Dricot, A.; Zack, T.I.; Sahni, N.; Jacob, Y.; Hao, T.; et al. Systematic screening reveals a role for BRCA1 in the response to transcription-associated DNA damage. Genes Dev. 2014, 28, 1957–1975. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, B.; Liu, X.; Gao, G.; Che, Z.; Fan, M.; Meng, S.; Zhao, X.; Sugimura, R.; Cao, H.; et al. ZFP281-BRCA2 prevents R-loop accumulation during DNA replication. Nat Commun. 2022, 13, 3493. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.; Barroso, S.I.; García-Rubio, M.L.; Tumini, E.; Herrera-Moyano, E.; Aguilera, A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 2014, 511, 362–365. [Google Scholar] [CrossRef]
- Chen, W.D.; Han, Z.J.; Skoletsky, J.; Olson, J.; Sah, J.; Myeroff, L.; Platzer, P.; Lu, S.; Dawson, D.; Willis, J.; et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J. Natl. Cancer Inst. 2005, 97, 1124–1132. [Google Scholar] [CrossRef]
- Boque-Sastre, R.; Soler, M.; Oliveira-Mateos, C.; Portela, A.; Moutinho, C.; Sayols, S.; Villanueva, A.; Esteller, M.; Guil, S. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. USA 2015, 112, 5785–5790. [Google Scholar] [CrossRef]
- Gorthi, A.; Romero, J.C.; Loranc, E.; Cao, L.; Lawrence, L.A.; Goodale, E.; Iniguez, A.B.; Bernard, X.; Masamsetti, V.P.; Roston, S.; et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 2018, 555, 387–391. [Google Scholar] [CrossRef]
- Becherel, O.J.; Yeo, A.J.; Stellati, A.; Heng, E.Y.; Luff, J.; Suraweera, A.M.; Woods, R.; Fleming, J.; Carrie, D.; McKinney, K.; et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 2013, 9, e1003435. [Google Scholar] [CrossRef]
- Bruno, T.; Corleone, G.; Catena, V.; Cortile, C.; de Nicola, F.; Fabretti, F.; Gumenyuk, S.; Pisani, F.; Mengarelli, A.; Passananti, C.; et al. AATF/Che-1 localizes to paraspeckles and suppresses R-loops accumulation and interferon activation in Multiple Myeloma. EMBO J. 2022, 41, e109711. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J.Y.; Huang, Y.J.; Gu, Y.; Qiu, J.; Qian, H.; Shao, C.; Zhang, X.; Hu, J.; Li, H.; et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol. Cell 2018, 69, 412–425.e6. [Google Scholar] [CrossRef] [PubMed]
- Sagie, S.; Toubiana, S.; Hartono, S.R.; Katzir, H.; Tzur-Gilat, A.; Havazelet, S.; Francastel, C.; Velasco, G.; Chédin, F.; Selig, S. Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA:RNA hybrids. Nat. Commun. 2017, 8, 14015. [Google Scholar] [CrossRef] [PubMed]
- Thrasher, A.J. New insights into the biology of Wiskott-Aldrich syndrome (WAS). Hematol. Am. Soc. Hematol. Educ. Program. 2009, 2009, 132–138. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chandra, S.; Bronicki, L.; Nagaraj, C.B.; Zhang, K. WAS-Related Disorders. In Gene Reviews; Adam, M.P., Ardinger, H.H., Pagon, R.A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Muralimanoharan, S.; Shamby, R.; Stansbury, N.; Schenken, R.; de la Pena Avalos, B.; Javanmardi, S.; Dray, E.; Sung, P.; Boyer, T.G. Aberrant R-loop-induced replication stress in MED12-mutant uterine fibroids. Sci. Rep. 2022, 12, 6169. [Google Scholar] [CrossRef] [PubMed]
- Struve, N.; Hoffer, K.; Weik, A.S.; Riepen, B.; Krug, L.; Cetin, M.H.; Burmester, J.; Ott, L.; Liebing, J.; Gatzemeier, F.; et al. Increased replication stress and R-loop accumulation in EGFRvIII-expressing glioblastoma present new therapeutic opportunities. Neurooncol. Adv. 2021, 4, vdab180. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Qian, T.; Huang, Z.; Liu, Y.; Cui, L.; Zhu, P.; Zhong, Q.; Zeng, T.; Fu, L.; Si, C.; et al. Increased expression of IFI16 predicts adverse prognosis in multiple myeloma. Pharm. J. 2021, 21, 520–532. [Google Scholar] [CrossRef]
- Giannini, M.; Bayona-Feliu, A.; Sproviero, D.; Barroso, S.I.; Cereda, C.; Aguilera, A. TDP-43 mutations link Amyotrophic Lateral Sclerosis with R-loop homeostasis and R loop-mediated DNA damage. PLoS Genet. 2020, 16, e1009260. [Google Scholar] [CrossRef] [PubMed]
- Boguslawski, S.J.; Smith, D.E.; Michalak, M.A.; Mickelson, K.E.; Yehle, C.O.; Patterson, W.L.; Carrico, R.J. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 1986, 89, 123–130. [Google Scholar] [CrossRef]
- Phillips, D.D.; Garboczi, D.N.; Singh, K.; Hu, Z.; Leppla, S.H.; Leysath, C.E. The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit. 2013, 26, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhang, A.; Storz, G.; Gottesman, S.; Leppla, S.H. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res. 2006, 34, e52. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yeo, A.J.; Becherel, O.J.; Luff, J.E.; Cullen, J.K.; Wongsurawat, T.; Jenjaroenpun, P.; Kuznetsov, V.A.; McKinnon, P.J.; Lavin, M.F. R-loops in proliferating cells but not in the brain: Implications for AOA2 and other autosomal recessive ataxias. PLoS ONE 2014, 9, e90219. [Google Scholar] [CrossRef] [PubMed]
- Wahba, L.; Costantino, L.; Tan, F.J.; Zimmer, A.; Koshland, D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 2016, 30, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Sanz, L.A.; Chédin, F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2019, 14, 1734–1755. [Google Scholar] [CrossRef] [PubMed]
- Nadel, J.; Athanasiadou, R.; Lemetre, C.; Wijetunga, N.A.; Broin, P.Ó.; Sato, H.; Zhang, Z.; Jeddeloh, J.; Montagna, C.; Golden, A.; et al. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin 2015, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Dumelie, J.G.; Jaffrey, S.R. Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. Elife 2017, 6, e28306. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.J.; Hamperl, S.; Swigut, T.; Cimprich, K.A. qDRIP: A method to quantitatively assess RNA-DNA hybrid formation genome-wide. Nucleic Acids Res. 2020, 48, e84. [Google Scholar] [CrossRef]
- Malig, M.; Hartono, S.R.; Giafaglione, J.M.; Sanz, L.A.; Chedin, F. Ultra-deep Coverage Single-molecule R-loop Footprinting Reveals Principles of R-loop Formation. J. Mol. Biol. 2020, 432, 2271–2288. [Google Scholar] [CrossRef]
- El Hage, A.; Webb, S.; Kerr, A.; Tollervey, D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014, 10, e1004716. [Google Scholar] [CrossRef]
- Yan, Q.; Shields, E.J.; Bonasio, R.; Sarma, K. Mapping Native R-Loops Genome-wide Using a Targeted Nuclease Approach. Cell Rep. 2019, 29, 1369–1380.e5. [Google Scholar] [CrossRef]
- Wang, K.; Wang, H.; Li, C.; Yin, Z.; Xiao, R.; Li, Q.; Xiang, Y.; Wang, W.; Huang, J.; Chen, L.; et al. Genomic profiling of native R loops with a DNA-RNA hybrid recognition sensor. Sci. Adv. 2021, 7, eabe3516. [Google Scholar] [CrossRef]
- Yan, Q.; Wulfridge, P.; Doherty, J.; Fernandez-Luna, J.L.; Real, P.J.; Tang, H.Y.; Sarma, K. Proximity labeling identifies a repertoire of site-specific R-loop modulators. Nat. Commun. 2022, 13, 53. [Google Scholar] [CrossRef]
- Jenjaroenpun, P.; Wongsurawat, T.; Yenamandra, S.P.; Kuznetsov, V.A. QmRLFS-finder: A model, web server and stand-alone tool for prediction and analysis of R-loop forming sequences. Nucleic Acids Res. 2015, 43, W527–W534. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Havlík, J.; Kolomazník, J.; Trenz, O.; Šťastný, J. R-Loop Tracker: Web Access-Based Tool for R-Loop Detection and Analysis in Genomic DNA Sequences. Int. J. Mol. Sci. 2021, 22, 2857. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, K.; Li, S.; Hou, Q.; Zhang, Y.; Liu, K.; Sun, Q. The R-Loop Atlas of Arabidopsis Development and Responses to Environmental Stimuli. Plant Cell 2020, 32, 888–903. [Google Scholar] [CrossRef] [PubMed]
- Jenjaroenpun, P.; Wongsurawat, T.; Sutheeworapong, S.; Kuznetsov, V.A. R-loopDB: A database for R-loop forming sequences (RLFS) and R-loops. Nucleic Acids Res. 2017, 45, D119–D127. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J.Y.; Zhang, X.; Gu, Y.; Xiao, R.; Shao, C.; Tang, P.; Qian, H.; Luo, D.; Li, H.; et al. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol. Cell 2017, 68, 745–757.e5. [Google Scholar] [CrossRef]
- Shaw, N.N.; Xi, H.; Arya, D.P. Molecular recognition of a DNA:RNA hybrid: Sub-nanomolar binding by a neomycin-methidium conjugate. Bioorg. Med. Chem. Lett. 2008, 18, 4142–4145. [Google Scholar] [CrossRef]
- Lin, R.; Zhong, X.; Zhou, Y.; Geng, H.; Hu, Q.; Huang, Z.; Hu, J.; Fu, X.D.; Chen, L.; Chen, J.Y. R-loopBase: A knowledgebase for genome-wide R-loop formation and regulation. Nucleic Acids Res. 2022, 50, D303–D315. [Google Scholar] [CrossRef]
| R-Loop Binding Proteins | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AQR | BUB3 | EWSR1 | RNASEH1 | RNASEH2A | RNASEH2C | THOC5 | TREX1 | DDX5 | EXOSC3 | |||
| Transcription | YBX1 | |||||||||||
| CTCF | ||||||||||||
| LEO1 | ||||||||||||
| XRN2 | ||||||||||||
| SSU72 | ||||||||||||
| POLR2B | ||||||||||||
| POLR2C | ||||||||||||
| Translation | CIRBP | |||||||||||
| Cleavage and Polyadenylation | CPSF | CPSF1 | ||||||||||
| CPSF2 | ||||||||||||
| CPSF3 | ||||||||||||
| CPSF4 | ||||||||||||
| FIP1L1 | ||||||||||||
| CSTF | CSTF1 | |||||||||||
| CSTF2 | ||||||||||||
| CSTF3 | ||||||||||||
| CFIm | CPSF6 | |||||||||||
| CPSF7 | ||||||||||||
| NUDT21 | ||||||||||||
| CFIIm | PCF11 | |||||||||||
| CLP1 | ||||||||||||
| Integrator complex | CPSF3L | |||||||||||
| SYMPK | ||||||||||||
| CSTF21 | ||||||||||||
| WDR33 | ||||||||||||
| RBBP6 | ||||||||||||
| CPEB1 | ||||||||||||
| PAP | PAPOLA | |||||||||||
| PAPOLG | ||||||||||||
| NC PAP | PAPD4 | |||||||||||
| PAPD5 | ||||||||||||
| PAPD7 | ||||||||||||
| PABP | PABPC4 | |||||||||||
| PABPN1 | ||||||||||||
| PABPC1 | ||||||||||||
| RNA processing | RBM5 | |||||||||||
| PTBP1 | ||||||||||||
| DDX39B | ||||||||||||
| DDX23 | ||||||||||||
| U2AF1 | ||||||||||||
| SCAF1 | ||||||||||||
| SRSF1 | ||||||||||||
| SRSF3 | ||||||||||||
| SRSF4 | ||||||||||||
| RNPS1 | ||||||||||||
| SCAF1 | ||||||||||||
| XRN2 | ||||||||||||
| DIS3L | ||||||||||||
| SKIV2L2 | ||||||||||||
| DCP2 | ||||||||||||
| ZFP36 | ||||||||||||
| KHSRP | ||||||||||||
| TARBP2 | ||||||||||||
| HNRNPH1 | ||||||||||||
| Epigenetics | CHD1 | |||||||||||
| PARN | ||||||||||||
| Others | MAPK9 | |||||||||||
| Detection Method | Method Name | Processing Method | Advantages | Disadvantages | |
|---|---|---|---|---|---|
| Wet lab techniques | |||||
| Techniques for detecting R loop | S9.6 antibody staining DNA/RNA hybrid | Immunoprecipitation (IP)/Immunohistochemistry (IHC) [128]. | Immunostaining of DNA–RNA hybrid | Good signal, likely useful for the analysis of samples from tissue banks | Limited to the microscopic examination of R- loops |
| DRIP [56,70,129,130,131] | Restriction digestion (RE) of genome followed by IP | Robust signal | Better resolution than IP/IHC but is still low | ||
| DRIP-seq [31] | RE of genome followed by IP and dsDNA sequencing | Robust signal is widely adopted, and is easy to set up | Low resolution, no strand specificity, and cannot be used in situ | ||
| S1-DRIP-seq [132] | Sonication of samples followed by IP and dsDNA sequencing | Higher resolution than DRIP-seq | No strand specificity and cannot be used in situ. S1 nuclease is delicate, and it is difficult to control the reaction, which may make it challenging to reproduce the data in clinical setting | ||
| DRIPc-seq [133] | RE of genome followed by RNA sequencing | Strand-specific, high resolution | Not in situ, requires longer sample preparation, S9.6 may recognize dsRNA | ||
| RDIP-seq [134] | Sonication of genome followed by RNA sequencing | Not in situ, tedious preparation | |||
| Bis-DRIP-seq [135] | RE of genome followed by sequencing dsDNA with bisulfite conversions | Strand-specific, provides additional control to ensure S9.6 signal arises from an R-loop in situ | Requires many replicates and shows R-loop enrichment in promoter regions only | ||
| qDRIP [136] | RE of genome followed by IP of DNA–RNA hybrid and synthetic DNA/RNA hybrid used as internal standards followed by dsDNA sequencing and quantification using internal standards as a reference. | Internal standards help with high-resolution, strand-specific sequencing | Spikes in hybrids shorter than 150 bp are unlikely to be useful for normalization. Additional spike-in may be required | ||
| SMRF-seq [137] | RE of genome followed by IP of DNA/RNA hybrid and sequencing of dsDNA with bisulfite conversions at single molecule level | Strand-specific, single-molecule resolution, avoids biases inherent to read-count normalization by accurately profiling signals in regions unaffected by transcription inhibition thus providing accurate differential peak calling between conditions | As with any foot-printing method, SMF is agnostic to the distinguishing of DNA-binding proteincreating the footprints. | ||
| Catalytically inactive RNase H | DRIVE-seq [31] | RE of genome followed by targeting catalytically inactive RNaseHs and dsDNA sequencing | Provides independent verification of some DRIP-seq results | Low enrichment, low resolution, reagent not commercially available, no strand specificity, not in situ | |
| R-ChIP-seq [138] | Sonication followed by targeting catalytically inactive RNaseHs and ssDNA sequencing | Strand specific, in situ capture | Cell line must be engineered to express catalytically inactive RNase H construct, inactive RNase H may alter hybrid dynamics | ||
| RNase H to guide micrococcal nuclease to R-loops | MapR [139] | Antibody-independent R-loop-profiling technique that utilizes RNase H to guide micrococcal nuclease to R-loops, which are subsequently cleaved, released, and identified by sequencing | Heavily based on CUT&RUN, a new and fast method to identify transcription factor binding sites genome-wide | Does not discriminate between the template and non-template strands and, therefore, cannot identify which DNA strand is involved in DNA–RNA hybrid formation. | |
| Sensor that binds to R loop | R loop CUT&Tag [140] | Combines CUT&Tag and GST-His6-2×HBD (glutathione S-transferase–hexahistidine–2× hybrid-binding domain) tags as an artificial R loop hybrid sensor to specifically recognize the DNA–RNA hybrids. | Sensitive, reproducible and generates good resolution to sense the R-loop instead of capture strategies that largely contribute to disparities in the previous techniques including R-loop Mapping | Current form of R-loop CUT&Tag does not provide strand information about R loops | |
| Techniques for detecting R loop and R loop-binding proteins | Fusion protein that binds to hybrid-binding domain (HBD) of RNaseH1 and an engineered variant of ascorbate peroxidase | RDProx (RNA–DNA Proximity Proteomics) [141] | Provides a snapshot of the R-loop-proximal proteome | In vivo labelling of R-loop-proximal proteins is performed, difficult to solubilize proteins that are amenable to the analysis can be identified, even transient spatiotemporal interactions with low affinity and transient interactions are detected. | Unable to distinguish between direct protein-binding or indirect proteins associated with RNA |
| Staining of both DNA–RNA hybrid (S9.6 antibody)+ R-loop-binding proteins (Antibody specific to the desired protein) | IP/IHC (R-loops+ R-Loop-binding proteins) [124] | Immunostaining of DNA–RNA hybrid and their binding proteins | Fast analysis of pathology specimens | Low resolution and limited to routine microscopic analysis | |
| Bioinformatics tools and databases | |||||
| Techniques for detecting R loop | Structure-based detection and prediction based on existing wet-lab data | QmRLFS-finder [142] | Identifies three structural features of R loop including a short G-cluster-rich region (R-loop initiation zone or RIZ), a structurally non-specified linker (linker), and long downstream region that has high G-density R-loop elongation zone (or REZ) based on experimental data | User-friendly web server and stand-alone tool for rapid and accurate prediction of RLFSs in DNA or RNA sequences shows strong agreement with existing genes and genome-scale experimentally determined R-loops | Information is limited and an updated version needs to be integrated with the growing experimental data |
| R loop tracker [143] | |||||
| R-loop atlas [144] | About 63 million peaks called from 254 plant species by ssDRIP-seq and deepR-loopPre are available | User-friendly web server for plants species based on experimental data | Limited to plant species only | ||
| Techniques for R loops and their binding proteins | R-loop DB [145] | Consists of computationally predicted R-loop-forming sequences (RLFSs) in human genic regions. Using the QmRLFS, the updated version of this database now has an increased number of RLFSs predicted in the human genes and in the genomes of other organisms | Provides comprehensive annotation of Ensembl RLFS-positive genes to study comparative evolution and genome-scale analyses, also R loop-binding proteins | Limited information and an updated version needs to be integrated with the growing experimental data | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, E.S.; Danckwardt, S. Pathophysiological Role and Diagnostic Potential of R-Loops in Cancer and Beyond. Genes 2022, 13, 2181. https://doi.org/10.3390/genes13122181
Khan ES, Danckwardt S. Pathophysiological Role and Diagnostic Potential of R-Loops in Cancer and Beyond. Genes. 2022; 13(12):2181. https://doi.org/10.3390/genes13122181
Chicago/Turabian StyleKhan, Essak S., and Sven Danckwardt. 2022. "Pathophysiological Role and Diagnostic Potential of R-Loops in Cancer and Beyond" Genes 13, no. 12: 2181. https://doi.org/10.3390/genes13122181
APA StyleKhan, E. S., & Danckwardt, S. (2022). Pathophysiological Role and Diagnostic Potential of R-Loops in Cancer and Beyond. Genes, 13(12), 2181. https://doi.org/10.3390/genes13122181






