The Combined Analysis of Transcriptome and Antioxidant Enzymes Revealed the Mechanism of EBL and ZnO NPs Enhancing Styrax tonkinensis Seed Abiotic Stress Resistance
Abstract
1. Introduction
2. Material and Methods
2.1. Site Information and Experimental Design
2.2. Determination of Physiological Indexes
2.3. RNA Extraction
2.4. Library Preparation and Sequencing
2.5. De Novo Assembly and Annotation
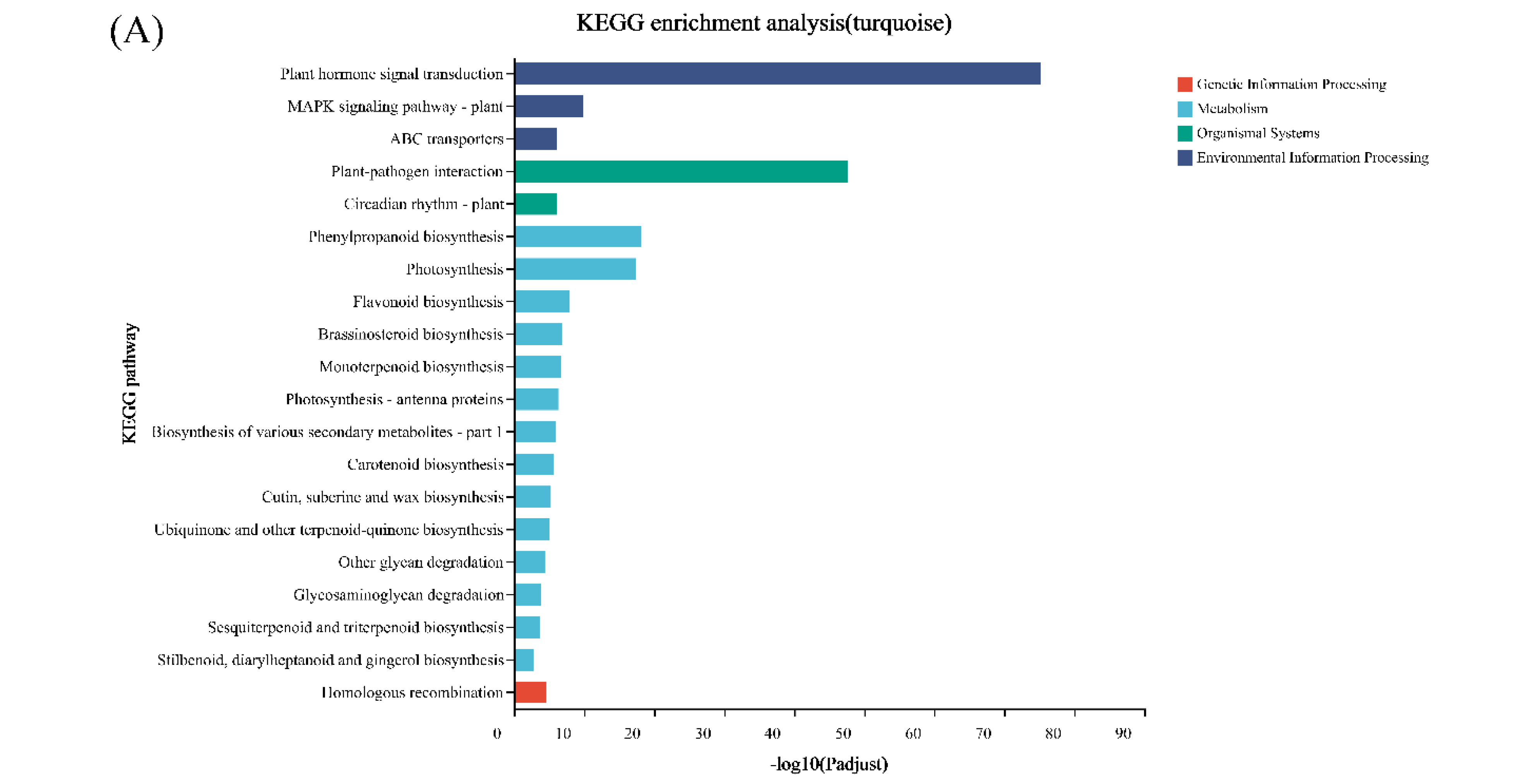
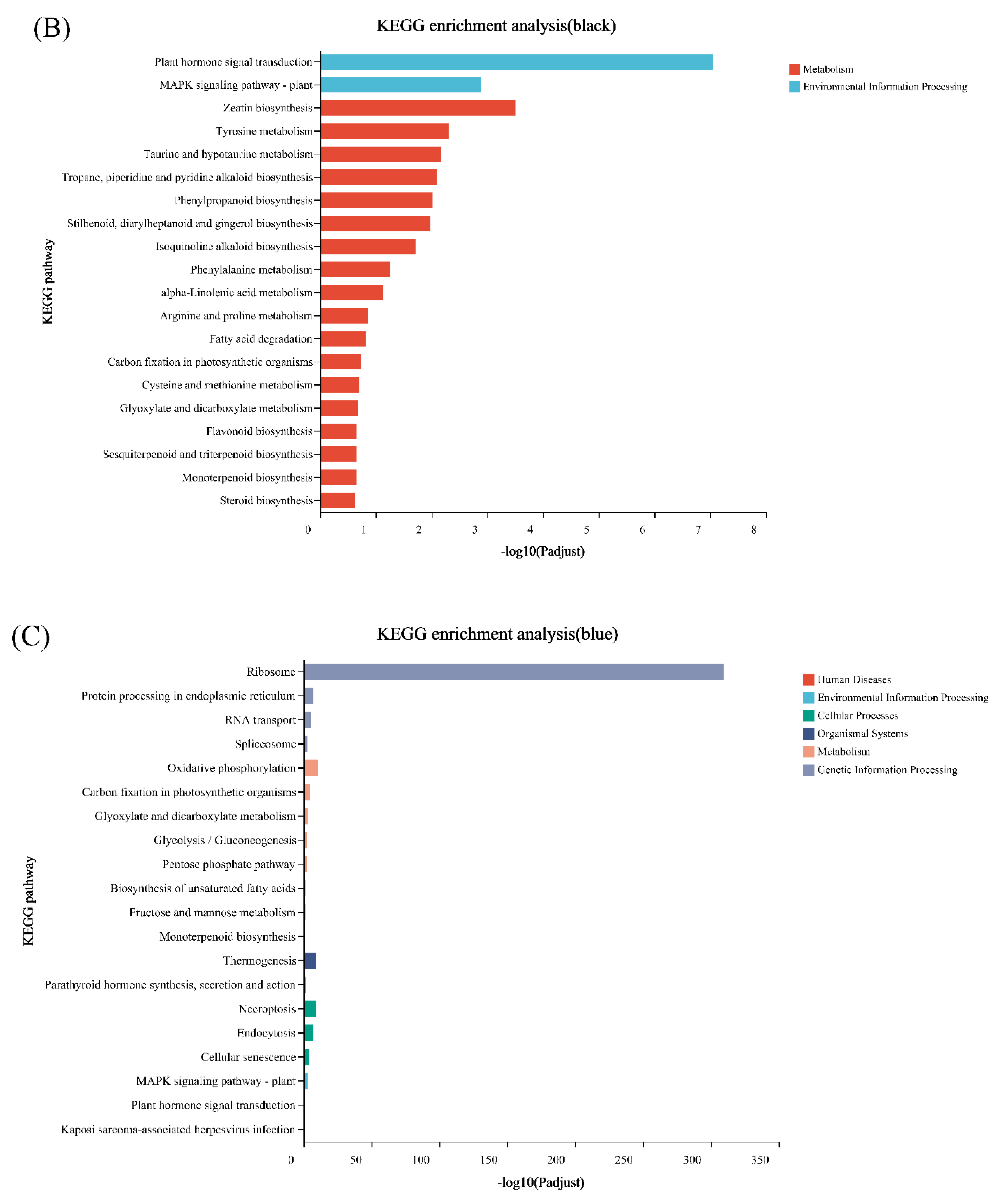
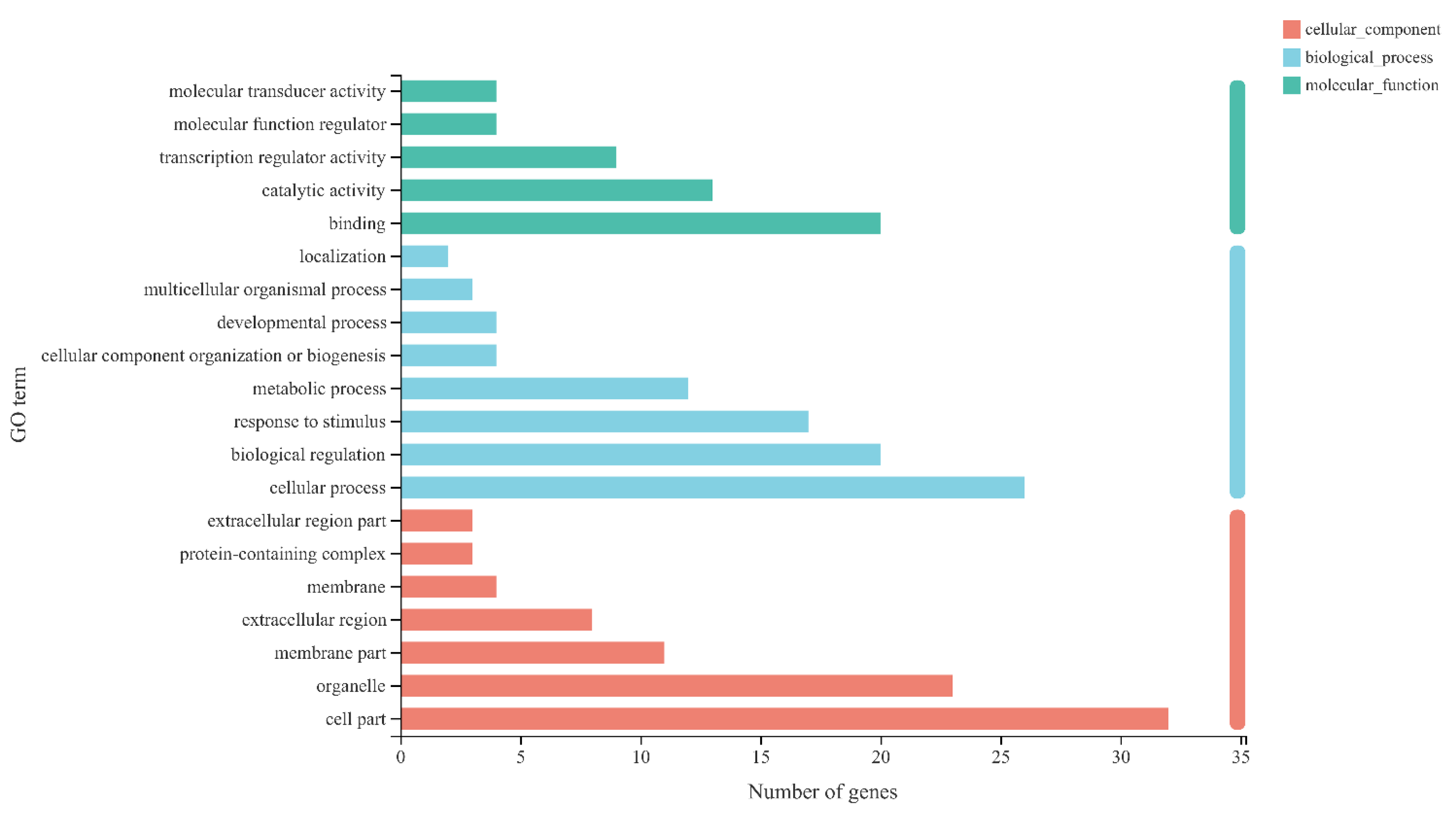
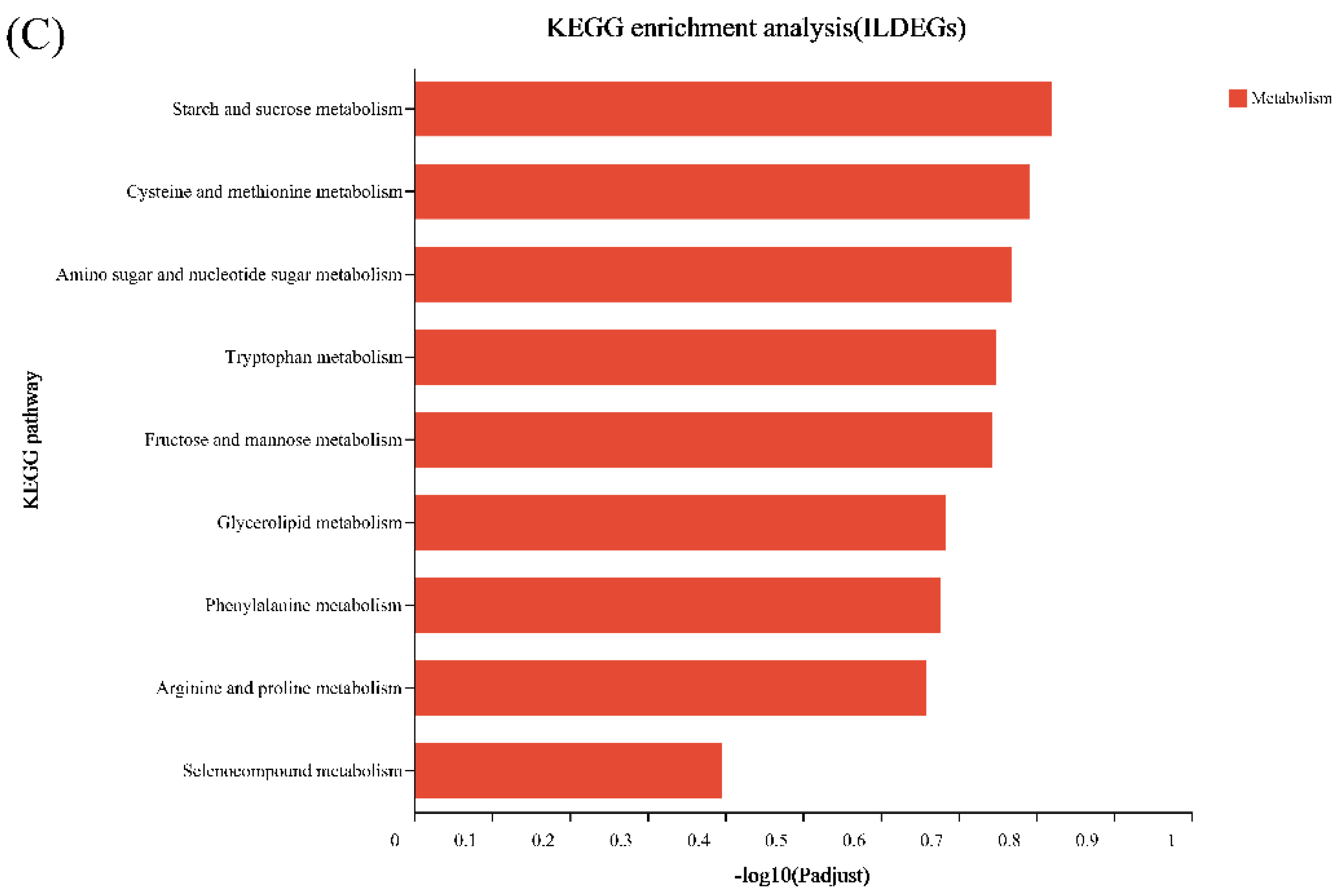
2.6. Differential Expression Analysis and Functional Enrichment
2.7. WGCNA Analysis
2.8. Statistics Analysis
3. Results
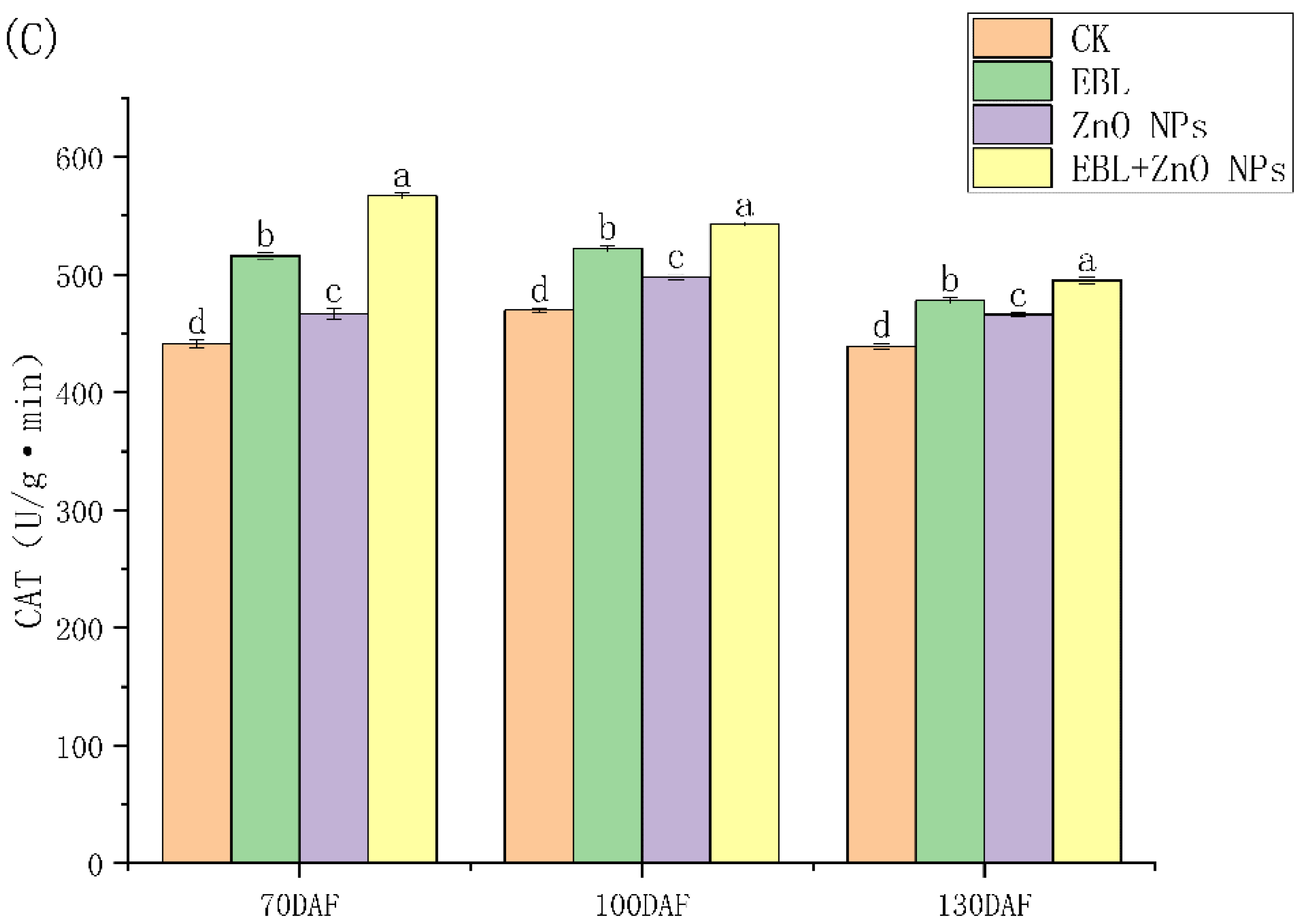
3.1. Response of the Antioxidant Enzyme to Treatments
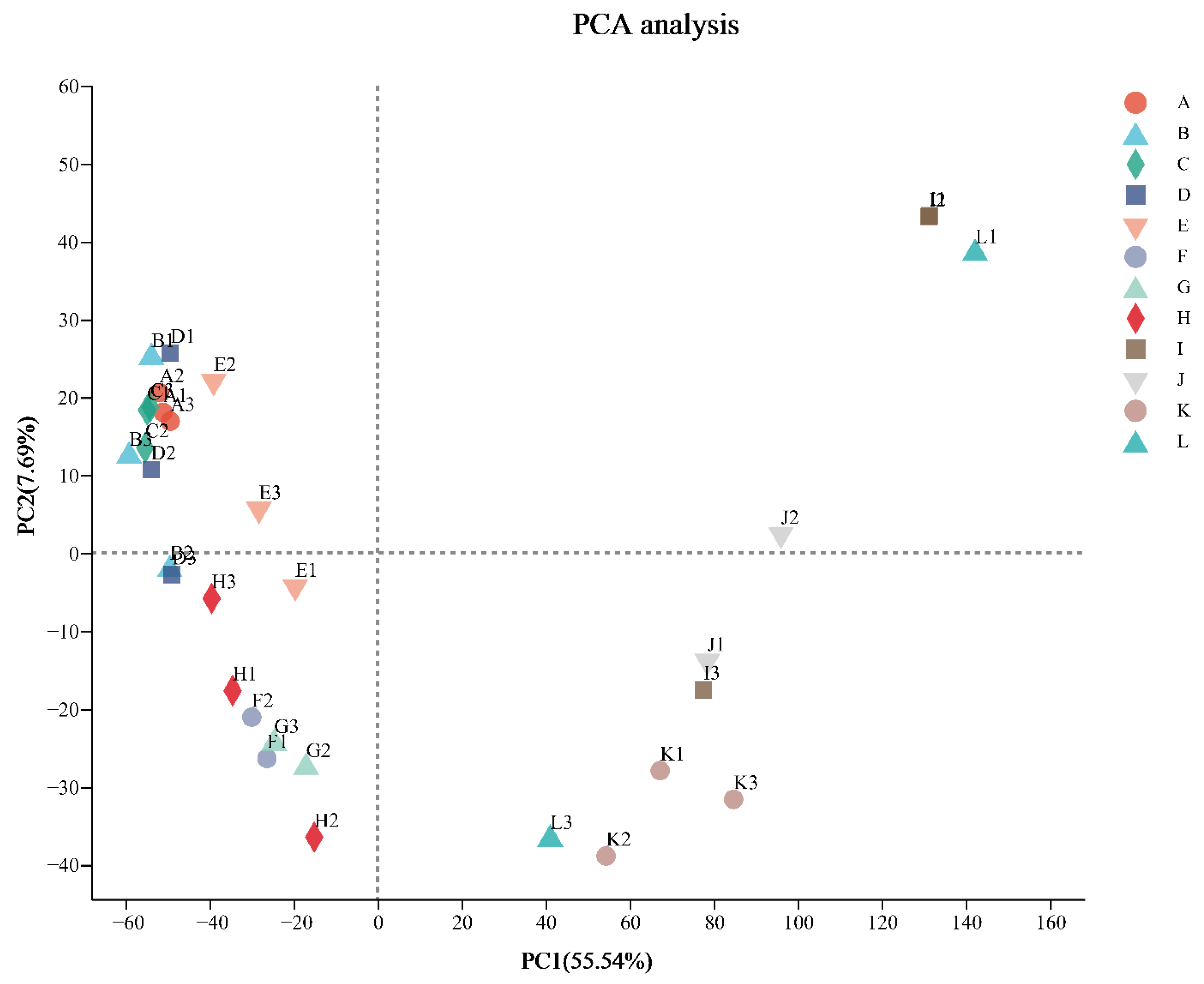
3.2. Sequencing, Assembly, and Sequence Analysis
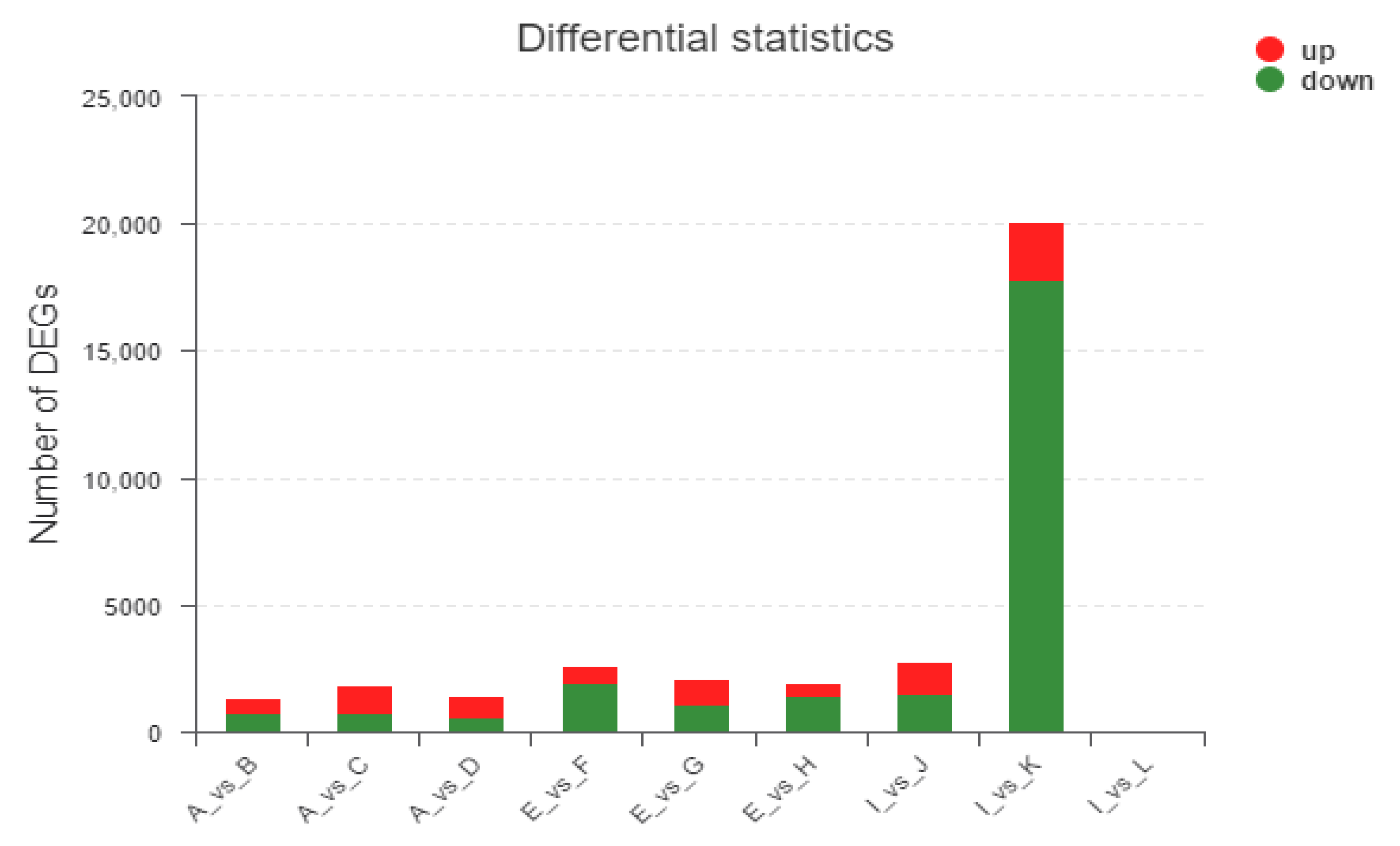
3.3. Differently Expressed Genes Obtained in Different Treatments
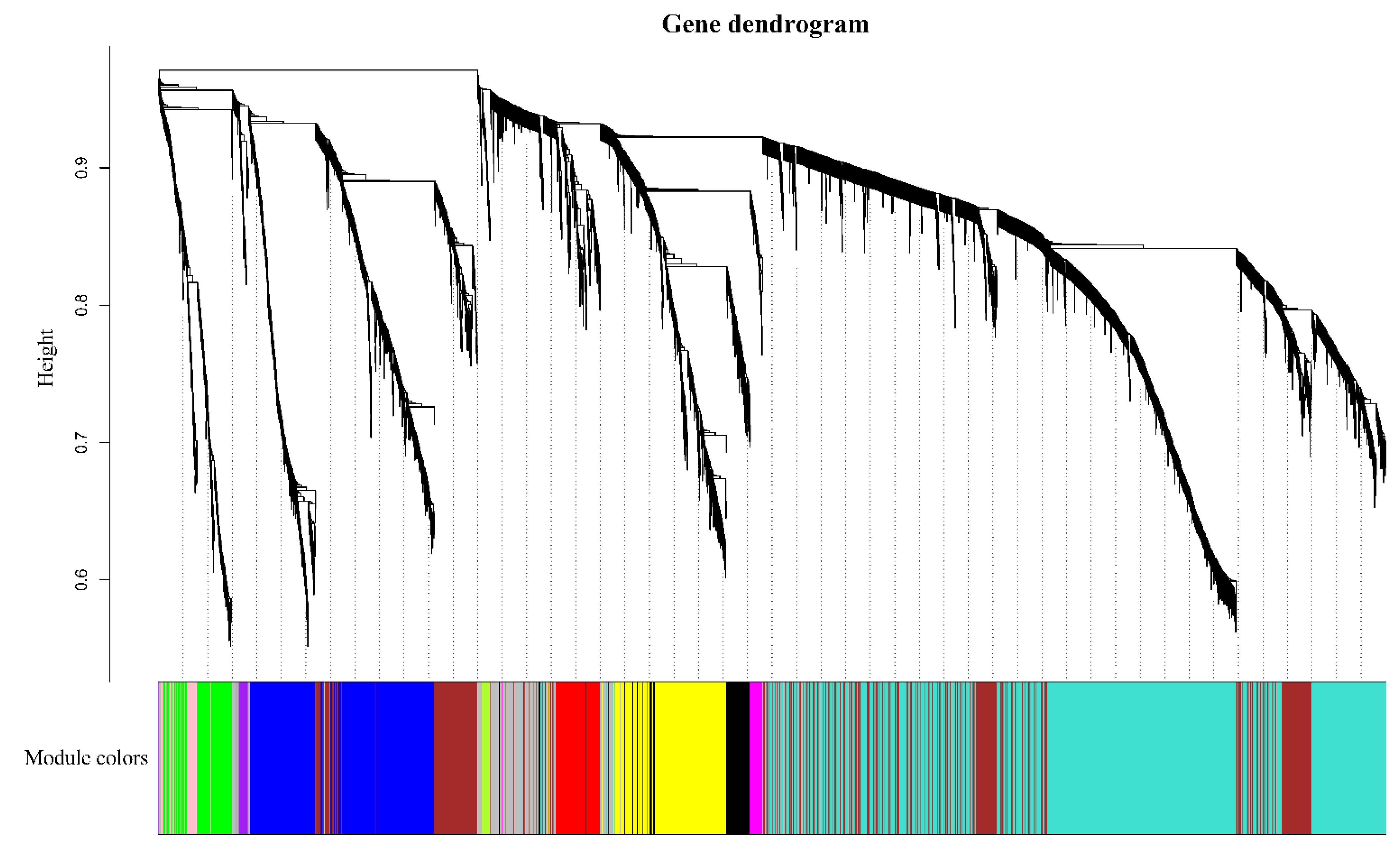
3.4. WGCNA Analysis
3.5. Search for Key Genes in Plant Hormone Signaling Pathway
3.6. About the Later Stages of Seed Development
4. Discussion
4.1. Changes in Antioxidant Enzyme Activities under Different Treatments
4.2. Key Genes in Hormone Signaling Pathways
4.3. Late Stage of Seed Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, C.-J.; Zhang, C.; Lu, Y.-N.; Jin, J.-Q.; Wang, X.-L. The Mechanisms of Brassinosteroids’ Action: From Signal Transduction to Plant Development. Mol. Plant 2011, 4, 588–600. [Google Scholar] [CrossRef]
- Vriet, C.; Russinova, E.; Reuzeau, C. From Squalene to Brassinolide: The Steroid Metabolic and Signaling Pathways across the Plant Kingdom. Mol. Plant 2013, 6, 1738–1757. [Google Scholar] [CrossRef] [PubMed]
- Fridman, Y.; Savaldi-Goldstein, S. Brassinosteroids in Growth Control: How, When and Where. Plant Sci. 2013, 209, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zhang, S.; Jiang, H.; Zhang, X.; Gao, H.; Yang, P.; Hu, T. Melatonin-Induced Cold and Drought Tolerance is Regulated by Brassinosteroids and Hydrogen Peroxide Signaling in Perennial Ryegrass. Environ. Exp. Bot. 2022, 196, 104815. [Google Scholar] [CrossRef]
- Çoban, Ö.; Baydar, N.G. Brassinosteroid Effects on Some Physical and Biochemical Properties and Secondary Metabolite Accumulation in Peppermint (Mentha piperita L.) under Salt Stress. Ind. Crop. Prod. 2016, 86, 251–258. [Google Scholar] [CrossRef]
- Sharma, I.; Ching, E.; Saini, S.; Bhardwaj, R.; Pati, P.K. Exogenous Application of Brassinosteroid Offers Tolerance to Salinity by Altering Stress Responses in Rice Variety Pusa Basmati-1. Plant Physiol. Biochem. 2013, 69, 17–26. [Google Scholar] [CrossRef]
- Saini, S.; Kaur, N.; Pati, P.K. Phytohormones: Key Players in the Modulation of Heavy Metal Stress Tolerance in Plants. Ecotoxicol. Environ. Saf. 2021, 223, 112578. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Christie, P.; Zhang, S. Uptake, Translocation, and Transformation of Metal-Based Nanoparticles in Plants: Recent Advances and Methodological Challenges. Environ. Sci. Nano 2018, 6, 41–59. [Google Scholar] [CrossRef]
- Khalaki, M.A.; Moameri, M.; Ghorbani, A.; Alagoz, S.M.; Dolatabadi, N.; Lajayer, B.A.; van Hullebusch, E.D. Effects, Uptake and Translocation of Ag-Based Nanoparticles in Plants. In Toxicity of Nanoparticles in Plants; Academic Press: Cambridge, MA, USA, 2022; Volume 5, pp. 171–192. [Google Scholar] [CrossRef]
- Azarin, K.; Usatov, A.; Minkina, T.; Plotnikov, A.; Kasyanova, A.; Fedorenko, A.; Duplii, N.; Vechkanov, E.; Rajput, V.D.; Mandzhieva, S.; et al. Effects of ZnO Nanoparticles and Its Bulk Form on Growth, Antioxidant Defense System and Expression of Oxidative Stress Related Genes in Hordeum vulgare L. Chemosphere 2021, 287, 132167. [Google Scholar] [CrossRef]
- Yasmin, H.; Mazher, J.; Azmat, A.; Nosheen, A.; Naz, R.; Hassan, M.N.; Noureldeen, A.; Ahmad, P. Combined Application of Zinc Oxide Nanoparticles and Biofertilizer to Induce Salt Resistance in Safflower by Regulating Ion Homeostasis and Antioxidant Defence Responses. Ecotoxicol. Environ. Saf. 2021, 218, 112262. [Google Scholar] [CrossRef]
- Zeeshan, M.; Hu, Y.X.; Iqbal, A.; Salam, A.; Liu, Y.X.; Muhammad, I.; Ahmad, S.; Khan, A.H.; Hale, B.; Wu, H.Y.; et al. Amelioration of AsV Toxicity by Concurrent Application of ZnO-NPs and Se-NPs is Associated with Differential Regulation of Photosynthetic Indexes, Antioxidant Pool and Osmolytes Content in Soybean Seedling. Ecotoxicol. Environ. Saf. 2021, 225, 112738. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Ali, B.; Adrees, M.; Arshad, M.; Hussain, A.; ur Rehman, M.Z.; Waris, A.A. Zinc and Iron Oxide Nanoparticles Improved the Plant Growth and Reduced the Oxidative Stress and Cadmium Concentration in Wheat. Chemosphere 2019, 214, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Ghani, M.I.; Saleem, S.; Rather, S.A.; Rehmani, M.S.; Alamri, S.; Rajput, V.D.; Kalaji, H.M.; Saleem, N.; Sial, T.A.; Liu, M. Foliar Application of Zinc Oxide Nanoparticles: An Effective Strategy to Mitigate Drought Stress in Cucumber Seedling by Modulating Antioxidant Defense System and Osmolytes Accumulation. Chemosphere 2021, 289, 133202. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Tiwari, S.; Pandey, J.; Lata, C.; Singh, I.K. Role of Nanoparticles in Crop Improvement and Abiotic Stress Management. J. Biotechnol. 2021, 337, 57–70. [Google Scholar] [CrossRef]
- Liu, L.; Nian, H.; Lian, T. Plants and Rhizospheric Environment: Affected by Zinc Oxide Nanoparticles (ZnO NPs). A Review. Plant Physiol. Biochem. 2022, 185, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, D.; Singh, M.; Pandey-Rai, S. Crosstalk of Nanoparticles and Phytohormones Regulate Plant Growth and Metabolism under Abiotic and Biotic Stress. Plant Stress 2022, 6, 100107. [Google Scholar] [CrossRef]
- Hu, W.-L.; Li, Z.-L.; Chen, Q.-J.; Sun, Y.-W.; Zhai, S.; Lu, F.; Li, F.; Zhang, C.-F. Triterpenes and Lignans from the Leaves of Styrax tonkinensis. Biochem. Syst. Ecol. 2019, 86, 103891. [Google Scholar] [CrossRef]
- Wu, Q.; Cao, Y.; Chen, C.; Gao, Z.; Yu, F.; Guy, R.D. Transcriptome Analysis of Metabolic Pathways Associated with Oil Accumulation in Developing Seed Kernels of Styrax tonkinensis, a Woody Biodiesel Species. BMC Plant Biol. 2020, 20, 121. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, C.; Wang, X.; Zhang, Z.; Yu, F.; Guy, R.D. Proteomic Analysis of Metabolic Mechanisms Associated with Fatty Acid Biosynthesis during Styrax tonkinensis Kernel Development. J. Sci. Food Agric. 2021, 101, 6053–6063. [Google Scholar] [CrossRef]
- Courel, B.; Adam, P.; Schaeffer, P. The Potential of Triterpenoids as Chemotaxonomic Tools to Identify and Differentiate Genuine, Adulterated and Archaeological Balsams. Microchem. J. 2019, 147, 411–421. [Google Scholar] [CrossRef]
- Xie, Q.; Ma, R.; Guo, X.; Chen, H.; Wang, J. Benzoinum from Styrax tonkinensis (Pierre) Craib ex Hart Exerts a NVU Protective Effect by Inhibiting Cell Apoptosis in Cerebral Ischaemia Rats. J. Ethnopharmacol. 2020, 265, 113355. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Han, C.; Liu, Z.; Yu, F.; Wu, Q. 24-Epibrassinolide Promotes Fatty Acid Accumulation and the Expression of Related Genes in Styrax tonkinensis Seeds. Int. J. Mol. Sci. 2022, 23, 8897. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wu, Q.; Ni, M.; Chen, C.; Han, C.; Yu, F. Transcriptome Analysis of Endogenous Hormone Response Mechanism in Roots of Styrax tonkinensis under Waterlogging. Front. Plant Sci. 2022, 13, 896850. [Google Scholar] [CrossRef]
- Devireddy, A.R.; Zandalinas, S.I.; Fichman, Y.; Mittler, R. Integration of Reactive Oxygen Species and Hormone Signaling during Abiotic Stress. Plant J. 2021, 105, 459–476. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Dehghanian, Z.; Bandehagh, A.; Habibi, K.; Balilashaki, K.; Lajayer, B.A. Impact of Abiotic Stress on Plant Brassinosteroids. In Climate Change and the Microbiome; Choudhary, D.K., Mishra, A., Varma, A., Eds.; Springer: Cham, Switzerland, 2021; Volume 63, pp. 279–298. [Google Scholar] [CrossRef]
- Faizan, M.; Bhat, J.A.; Chen, C.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P.; Yu, F. Zinc Oxide Nanoparticles (ZnO-NPs) Induce Salt Tolerance by Improving the Antioxidant System and Photosynthetic Machinery in Tomato. Plant Physiol. Biochem. 2021, 161, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhang, L.J.; Fan, J.J. Plant Physiology Experiment; China Agricultural University Press: Beijing, China, 2007. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; A Simão, F.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.M.; Kelly, S. TransRate: Reference-Free Quality Assessment of de Novo Transcriptome Assemblies. Genome Res. 2016, 26, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the Next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A One-Stop, Comprehensive Bioinformatic Platform for Multiomics Analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.-Q. Interplay between Reactive Oxygen Species and Hormones in the Control of Plant Development and Stress Tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef]
- Dvořák, P.; Krasylenko, Y.; Zeiner, A.; Šamaj, J.; Takáč, T. Signaling toward Reactive Oxygen Species-Scavenging Enzymes in Plants. Front. Plant Sci. 2021, 11, 618835. [Google Scholar] [CrossRef] [PubMed]
- Aliyari Rad, S.; Dehghanian, Z.; Asgari Lajayer, B.; Nobaharan, K.; Astatkie, T. Mitochondrial Respiration and Energy Production under Some Abiotic Stresses. J. Plant Growth Regul. 2021, 41, 3285–3299. [Google Scholar] [CrossRef]
- Jiang, G.; Hassan, M.A.; Muhammad, N.; Arshad, M.; Chen, X.; Xu, Y.; Xu, H.; Ni, Q.; Liu, B.; Yang, W.; et al. Comparative Physiology and Transcriptome Analysis of Young Spikes in Response to Late Spring Coldness in Wheat (Triticum aestivum L.). Front. Plant Sci. 2022, 13, 811884. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Ruan, Y.-P.; Zhou, J.; Xia, X.-J.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q. Brassinosteroid Alleviates Polychlorinated Biphenyls-Induced Oxidative Stress by Enhancing Antioxidant Enzymes Activity in Tomato. Chemosphere 2013, 90, 2645–2653. [Google Scholar] [CrossRef]
- Alam Khan, T.; Yusuf, M.; Ahmad, A.; Bashir, Z.; Saeed, T.; Fariduddin, Q.; Hayat, S.; Mock, H.-P.; Wu, T. Proteomic and Physiological Assessment of Stress Sensitive and Tolerant Variety of Tomato Treated with Brassinosteroids and Hydrogen Peroxide under Low-Temperature Stress. Food Chem. 2019, 289, 500–511. [Google Scholar] [CrossRef]
- Tadaiesky, L.B.A.; da Silva, B.R.S.; Batista, B.L.; Lobato, A.K.D.S. Brassinosteroids Trigger Tolerance to Iron Toxicity in Rice. Physiol. Plant. 2020, 171, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.V.; Lobato, A.K.S. Brassinosteroids Improve Photosystem II Efficiency, Gas Exchange, Antioxidant Enzymes and Growth of Cowpea Plants Exposed to Water Deficit. Physiol. Mol. Biol. Plants 2017, 23, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.H.; Li, X.Q.; Wang, G.G.; Zhu, X.T. Brassinosteroids Alleviate High-Temperature Injury in Ficus Concinna Seedlings via Maintaining Higher Antioxidant Defence and Glyoxalase Systems. AoB Plants 2015, 7, plv009. [Google Scholar] [CrossRef] [PubMed]
- Ramzan, M.; Ayub, F.; Shah, A.A.; Naz, G.; Shah, A.N.; Malik, A.; Sardar, R.; Telesiński, A.; Kalaji, H.M.; Dessoky, E.S.; et al. Synergistic Effect of Zinc Oxide Nanoparticles and Moringa oleifera Leaf Extract Alleviates Cadmium Toxicity in Linum usitatissimum: Antioxidants and Physiochemical Studies. Front. Plant Sci. 2022, 13, 900347. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Ali, S.; Rizwan, M.; Zia ur Rehman, M.; Javed, M.R.; Imran, M.; Chatha, S.A.S.; Nazir, R. Zinc Oxide Nanoparticles Alter the Wheat Physiological Response and Reduce the Cadmium Uptake by Plants. Environ. Pollut. 2018, 242, 1518–1526. [Google Scholar] [CrossRef]
- Venkatachalam, P.; Jayaraj, M.; Manikandan, R.; Geetha, N.; Rene, E.R.; Sharma, N.; Sahi, S. Zinc Oxide Nanoparticles (ZnONPs) Alleviate Heavy Metal-Induced Toxicity in Leucaena leucocephala Seedlings: A Physiochemical Analysis. Plant Physiol. Biochem. 2017, 110, 59–69. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Han, Y.; Luthe, D. Identification and Evolution Analysis of the JAZ Gene Family in Maize. BMC Genom. 2021, 22, 256. [Google Scholar] [CrossRef]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 Family Member, OsGH3-2, Modulates Auxin and Abscisic Acid Levels and Differentially Affects Drought and Cold Tolerance in Rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The Master in Action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Li, L.; Yin, Y. Recent Advances in the Regulation of Brassinosteroid Signaling and Biosynthesis Pathways. J. Integr. Plant Biol. 2011, 53, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shen, J.; Liang, J. Genome-Wide Identification, Expression Profile, and Alternative Splicing Analysis of the Brassinosteroid-Signaling Kinase (BSK) Family Genes in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 1138. [Google Scholar] [CrossRef]
- de Figueiredo, M.R.A.; Strader, L.C. Intrinsic and Extrinsic Regulators of Aux/IAA Protein Degradation Dynamics. Trends Biochem. Sci. 2022, 47, 865–874. [Google Scholar] [CrossRef]
- Huang, X.; Bao, Y.; Wang, B.; Liu, L.; Chen, J.; Dai, L.; Baloch, S.U.; Peng, D. Identification of Small Auxin-up RNA (SAUR) Genes in Urticales Plants: Mulberry (Morus notabilis), Hemp (Cannabis sativa) and Ramie (Boehmeria nivea). J. Genet. 2016, 95, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Pontier, D.; Miao, Z.-H.; Lam, E. Trans-Dominant Suppression of Plant TGA Factors Reveals Their Negative and Positive Roles in Plant Defense Responses. Plant J. 2001, 27, 529–538. [Google Scholar] [CrossRef]
- Yang, G.; Yu, Z.; Gao, L.; Zheng, C. SnRK2s at the Crossroads of Growth and Stress Responses. Trends Plant Sci. 2019, 24, 672–676. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA Receptor PYL9 Promotes Drought Resistance and Leaf Senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant Hormone Regulation of Abiotic Stress Responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Jones, S.I.; Vodkin, L.O. Using RNA-Seq to Profile Soybean Seed Development from Fertilization to Maturity. PLoS ONE 2013, 8, e59270. [Google Scholar] [CrossRef]
- Rivera-Najera, L.Y.; Saab-Rincón, G.; Battaglia, M.; Amero, C.; Pulido, N.O.; García-Hernández, E.; Solórzano, R.M.; Reyes, J.L.; Covarrubias, A.A. A Group 6 Late Embryogenesis Abundant Protein from Common Bean Is a Disordered Protein with Extended Helical Structure and Oligomer-forming Properties. J. Biol. Chem. 2014, 289, 31995–32009. [Google Scholar] [CrossRef] [PubMed]
- Wise, M.J.; Tunnacliffe, A. POPP the Question: What Do LEA Proteins Do? Trends Plant Sci. 2004, 9, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Covarrubias, A.A. Late Embryogenesis Abundant (LEA) Proteins in Legumes. Front. Plant Sci. 2013, 4, 190. [Google Scholar] [CrossRef] [PubMed]



| Treatments | The Specific Composition |
|---|---|
| CK | clean water |
| T1 | 5 mL/L EBL |
| T2 | 50 mL/L ZnO NPs |
| T3 | 5 mL/L EBL + 50 mL/L ZnO NPs |
| Type | Unigene | Transcript |
|---|---|---|
| total number | 213,566 | 329,559 |
| total base | 238,692,947 | 405,460,296 |
| largest length (bp) | 16,047 | 16,047 |
| smallest length (bp) | 201 | 201 |
| average length (bp) | 1117.65 | 1230.31 |
| N50 length (bp) | 1586 | 1807 |
| GC percent (%) | 47.46 | 46.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.-M.; Faizan, M.; Chen, C.; Zheng, L.-H.; Yu, F.-Y. The Combined Analysis of Transcriptome and Antioxidant Enzymes Revealed the Mechanism of EBL and ZnO NPs Enhancing Styrax tonkinensis Seed Abiotic Stress Resistance. Genes 2022, 13, 2170. https://doi.org/10.3390/genes13112170
Liu Z-M, Faizan M, Chen C, Zheng L-H, Yu F-Y. The Combined Analysis of Transcriptome and Antioxidant Enzymes Revealed the Mechanism of EBL and ZnO NPs Enhancing Styrax tonkinensis Seed Abiotic Stress Resistance. Genes. 2022; 13(11):2170. https://doi.org/10.3390/genes13112170
Chicago/Turabian StyleLiu, Ze-Mao, Mohammad Faizan, Chen Chen, Li-Hong Zheng, and Fang-Yuan Yu. 2022. "The Combined Analysis of Transcriptome and Antioxidant Enzymes Revealed the Mechanism of EBL and ZnO NPs Enhancing Styrax tonkinensis Seed Abiotic Stress Resistance" Genes 13, no. 11: 2170. https://doi.org/10.3390/genes13112170
APA StyleLiu, Z.-M., Faizan, M., Chen, C., Zheng, L.-H., & Yu, F.-Y. (2022). The Combined Analysis of Transcriptome and Antioxidant Enzymes Revealed the Mechanism of EBL and ZnO NPs Enhancing Styrax tonkinensis Seed Abiotic Stress Resistance. Genes, 13(11), 2170. https://doi.org/10.3390/genes13112170








