Inferring bona fide Differentially Expressed Genes and Their Variants Associated with Vitamin K Deficiency Using a Systems Genetics Approach
Abstract
1. Introduction
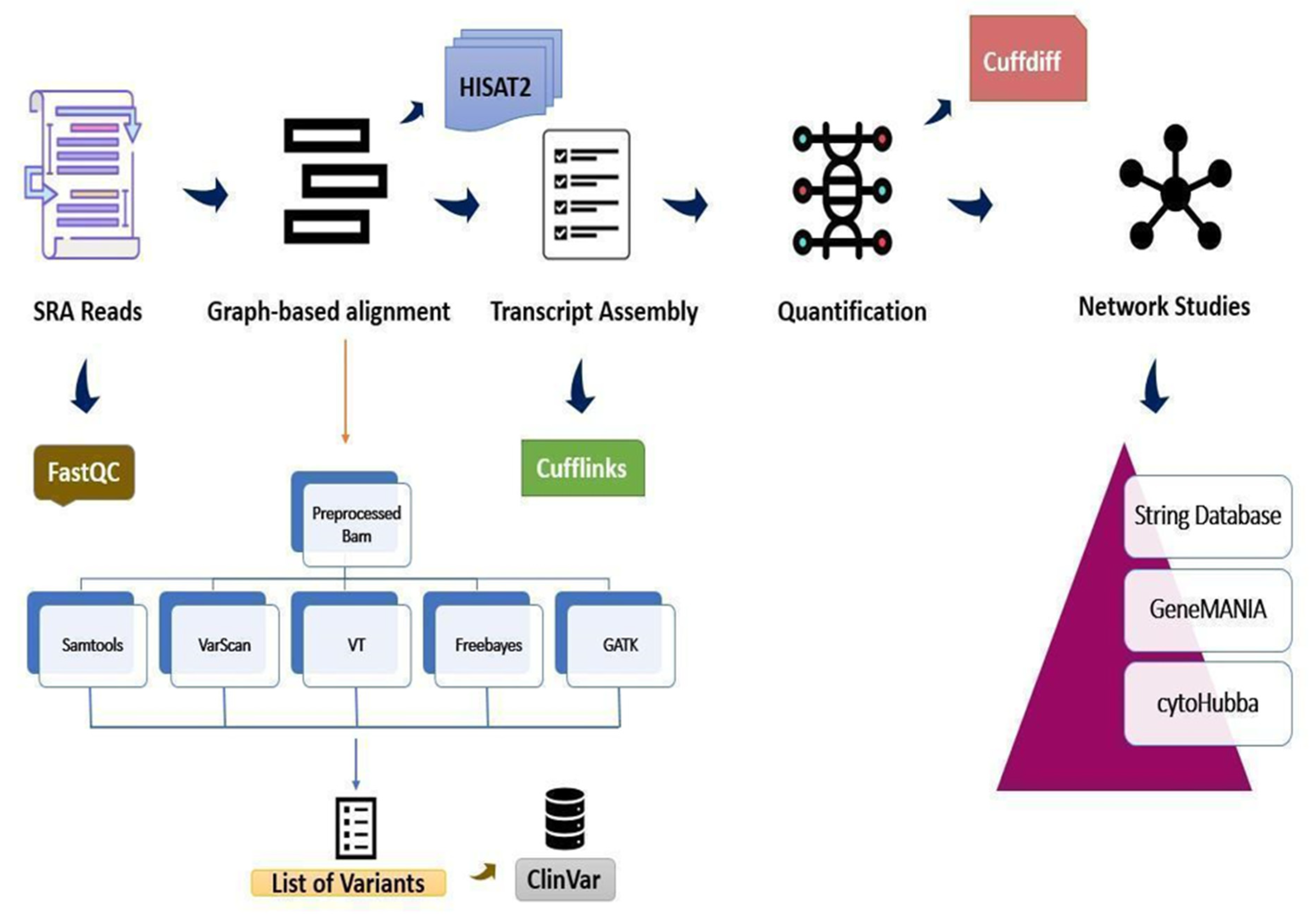
2. Materials and Methods
2.1. Datasets
2.2. RNA Sequencing Analysis, Statistics and Validation
2.3. Clustering Coefficient Network Analysis Using cytoHubba
3. Results
4. Discussion
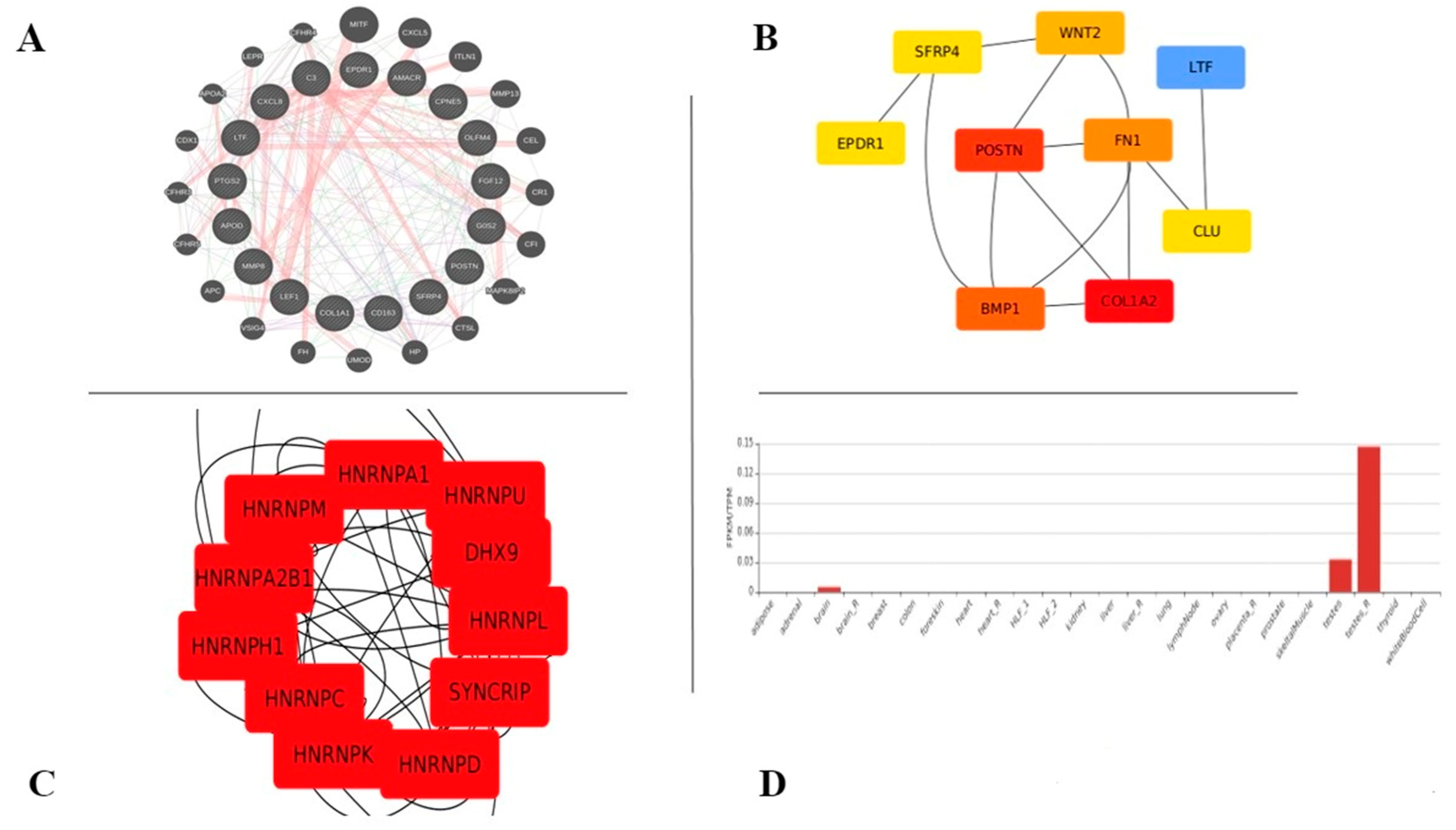
4.1. NONHSAT106693 among Significantly Enriched Genes
4.2. Heatmaps Showed Distinct Gene Expression Profiles with a Variance in Principal Components
4.3. CytoHubba Yielded Top Niche Ranks with Variant Analysis Showing No Mutations Attributed to DEGs
4.4. Variants of Unknown Significance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef]
- Buermans, H.P.J.; Den Dunnen, J.T. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2014, 1842, 1932–1941. [Google Scholar] [CrossRef]
- Luecken, M.D.; Theis, F.J. Current best practices in single-cell RNA-seq analysis: A tutorial. Mol. Syst. Biol. 2019, 15, e8746. [Google Scholar] [CrossRef]
- Englert, J.A.; Cho, M.H.; Lamb, A.E.; Shumyatcher, M.; Barragan-Bradford, D.; Basil, M.C.; Higuera, A.; Isabelle, C.; Vera, M.P.; Dieffenbach, P.B.; et al. Whole blood RNA sequencing reveals a unique transcriptomic profile in patients with ARDS following hematopoietic stem cell transplantation. Respir. Res. 2019, 20, 15. [Google Scholar] [CrossRef]
- Zheng, T.; Ellinghaus, D.; Juzenas, S.; Cossais, F.; Burmeister, G.; Mayr, G.; Jørgensen, I.F.; Teder-Laving, M.; Skogholt, A.H.; Chen, S.; et al. Genome-wide analysis of 944 133 individuals provides insights into the etiology of haemorrhoidal disease. Gut 2021, 70, 1538–1549. [Google Scholar] [CrossRef]
- Li, F.; Yan, K.; Wu, L.; Zheng, Z.; Du, Y.; Liu, Z.; Zhao, L.; Li, W.; Sheng, Y.; Ren, L.; et al. Single-cell RNA-seq reveals cellular heterogeneity of mouse carotid artery under disturbed flow. Cell Death Discov. 2021, 7, 180. [Google Scholar] [CrossRef]
- Ng, C.J.; Liu, A.; Venkataraman, S.; Ashworth, K.J.; Baker, C.D.; O’Rourke, R.; Vibhakar, R.; Jones, K.L.; Di Paola, J. Single-cell transcriptional analysis of human endothelial colony-forming cells from patients with low VWF levels. Blood 2022, 139, 2240–2251. [Google Scholar] [CrossRef]
- Wang, Y.; Han, S.; Ran, R.; Li, A.; Liu, H.; Liu, M.; Duan, Y.; Zhang, X.; Zhao, Z.; Song, S.; et al. A longitudinal sampling study of transcriptomic and epigenetic profiles in patients with thrombocytopenia syndrome. Nat. Commun. 2021, 12, 5629. [Google Scholar] [CrossRef]
- Liu, X.; Ma, Y.; Yin, K.; Li, W.; Chen, W.; Zhang, Y.; Zhu, C.; Li, T.; Han, B.; Liu, X.; et al. Long non-coding and coding RNA profiling using strand-specific RNA-seq in human hypertrophic cardiomyopathy. Sci. Data 2019, 6, 90. [Google Scholar] [CrossRef]
- Ren, S.; Peng, Z.; Mao, J.-H.; Yu, Y.; Yin, C.; Gao, X.; Cui, Z.; Zhang, J.; Yi, K.; Xu, W.; et al. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012, 22, 806–821. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 13 October 2022).
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Gupta, A.; Rajagopal, S.; Gupta, S.; Mishra, A.K.; Suravajhala, P. A Bioinformatics Pipeline for Processing and Analysis of Whole Transcriptome Sequence Data. 2021. Available online: https://dx.doi.org/10.17504/protocols.io.brz8m79w (accessed on 13 October 2022).
- Dilworth, R.P. A Decomposition Theorem for Partially Ordered Sets. Ann. Math. 1950, 51, 161. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Chen, K.; Wylie, T.; Larson, D.E.; McLellan, M.D.; Mardis, E.R.; Weinstock, G.M.; Wilson, R.K.; Ding, L. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009, 25, 2283–2285. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar] [CrossRef]
- Tan, A.; Abecasis, G.R.; Kang, H.M. Unified representation of genetic variants. Bioinformatics 2015, 31, 2202–2204. [Google Scholar] [CrossRef]
- RNAseq-Short-Variant-Discovery-SNPs-INDELS. (n.d.). Available online: https://gatk.broadinstitute.org/hc/en-us/articles/360035531192-RNAseq-short-variant-discovery-SNPs-Indels- (accessed on 13 October 2022).
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2018, 18, 623–632. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Drayna, D.T.; McLEAN, J.W.; Wion, K.L.; Trent, J.M.; Drabkin, H.A.; Lawn, R.M. Human Apolipoprotein D Gene: Gene Sequence, Chromosome Localization, and Homology to the α2u-Globulin Superfamily. DNA 1987, 6, 199–204. [Google Scholar] [CrossRef]
- Ritter, M.; Buechler, C.; Langmann, T.; Schmitz, G. Genomic Organization and Chromosomal Localization of the Human CD163 (M130) Gene: A Member of the Scavenger Receptor Cysteine-Rich Superfamily. Biochem. Biophys. Res. Commun. 1999, 260, 466–474. [Google Scholar] [CrossRef]
- Mansour, R.G.; Stamper, L.; Jaeger, F.; McGuire, E.; Fouda, G.; Amos, J.; Barbas, K.; Ohashi, T.; Alam, S.M.; Erickson, H.; et al. The Presence and Anti-HIV-1 Function of Tenascin C in Breast Milk and Genital Fluids. PLoS ONE 2016, 11, e0155261. [Google Scholar] [CrossRef]
- Ashizawa, Y.; Kuboki, S.; Nojima, H.; Yoshitomi, H.; Furukawa, K.; Takayashiki, T.; Takano, S.; Miyazaki, M.; Ohtsuka, M. OLFM4 Enhances STAT3 Activation and Promotes Tumor Progression by Inhibiting GRIM19 Expression in Human Hepatocellular Carcinoma. Hepatol. Commun. 2019, 3, 954–970. [Google Scholar] [CrossRef]
- Siena, Á.D.D.; Plaça, J.R.; Araújo, L.F.; de Barros, I.I.; Peronni, K.; Molfetta, G.; de Biagi, C.A.O.; Espreafico, E.M.; Sousa, J.F.; Silva, W.A. Whole transcriptome analysis reveals correlation of long noncoding RNA ZEB1-AS1 with invasive profile in melanoma. Sci. Rep. 2019, 9, 11350. [Google Scholar] [CrossRef]
- Herbert, A. Complement controls the immune synapse and tumors control complement. J. Immunother. Cancer 2020, 8, e001712. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Gillan, L.; Matei, D.; Fishman, D.A.; Gerbin, C.S.; Karlan, B.Y.; Chang, D.D. Periostin secreted by epithelial ovarian carcinoma is a ligand for α(V)β(3) and α(V)β(5) integrins and promotes cell motility. Cancer Res. 2002, 62, 5358–5364. [Google Scholar]
- Gupte, A.A.; Sabek, O.M.; Fraga, D.; Minze, L.J.; Nishimoto, S.K.; Liu, J.Z.; Afshar, S.; Gaber, L.; Lyon, C.J.; Gaber, A.O.; et al. Osteocalcin Protects Against Nonalcoholic Steatohepatitis in a Mouse Model of Metabolic Syndrome. Endocrinology 2014, 155, 4697–4705. [Google Scholar] [CrossRef]
- Delbeck, M.; Nickel, K.F.; Perzborn, E.; Ellinghaus, P.; Strassburger, J.; Kast, R.; Laux, V.; Schäfer, S.; Schermuly, R.; Von Degenfeld, G. A role for coagulation factor Xa in experimental pulmonary arterial hypertension. Cardiovasc. Res. 2011, 92, 159–168. [Google Scholar] [CrossRef]
- Verma, D.; Kumar, R.; Pereira, R.S.; Karantanou, C.; Zanetti, C.; Minciacchi, V.R.; Fulzele, K.; Kunz, K.; Hoelper, S.; Zia-Chahabi, S.; et al. Vitamin K antagonism impairs the bone marrow microenvironment and hematopoiesis. Blood 2019, 134, 227–238. [Google Scholar] [CrossRef]
- Xiao, H.; Chen, J.; Duan, L.; Li, S. Role of emerging vitamin K-dependent proteins: Growth arrest-specific protein 6, Gla-rich protein and periostin (Review). Int. J. Mol. Med. 2020, 47, 4835. [Google Scholar] [CrossRef]
- Boudin, E.; Fijalkowski, I.; Piters, E.; Van Hul, W. The role of extracellular modulators of canonical Wnt signaling in bone metabolism and diseases. Semin. Arthritis Rheum. 2013, 43, 220–240. [Google Scholar] [CrossRef]
- Shen, E.H.; Overly, C.C.; Jones, A.R. The Allen Human Brain Atlas. Trends Neurosci. 2012, 35, 711–714. [Google Scholar] [CrossRef]
- Fine, D.H. Lactoferrin. J. Dent. Res. 2015, 94, 768–776. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajagopal, S.; Sharma, A.; Simlot, A.; Mathur, P.; Mehta, S.; Mehta, S.; Naravula, J.; Medicherla, K.M.; Kumar, A.; Kanga, U.; et al. Inferring bona fide Differentially Expressed Genes and Their Variants Associated with Vitamin K Deficiency Using a Systems Genetics Approach. Genes 2022, 13, 2078. https://doi.org/10.3390/genes13112078
Rajagopal S, Sharma A, Simlot A, Mathur P, Mehta S, Mehta S, Naravula J, Medicherla KM, Kumar A, Kanga U, et al. Inferring bona fide Differentially Expressed Genes and Their Variants Associated with Vitamin K Deficiency Using a Systems Genetics Approach. Genes. 2022; 13(11):2078. https://doi.org/10.3390/genes13112078
Chicago/Turabian StyleRajagopal, Shalini, Akanksha Sharma, Anita Simlot, Praveen Mathur, Sudhir Mehta, Sumita Mehta, Jalaja Naravula, Krishna Mohan Medicherla, Anil Kumar, Uma Kanga, and et al. 2022. "Inferring bona fide Differentially Expressed Genes and Their Variants Associated with Vitamin K Deficiency Using a Systems Genetics Approach" Genes 13, no. 11: 2078. https://doi.org/10.3390/genes13112078
APA StyleRajagopal, S., Sharma, A., Simlot, A., Mathur, P., Mehta, S., Mehta, S., Naravula, J., Medicherla, K. M., Kumar, A., Kanga, U., Suravajhala, R., Bhandari, R. K., Nair, B. G., Kishor, P. B. K., & Suravajhala, P. (2022). Inferring bona fide Differentially Expressed Genes and Their Variants Associated with Vitamin K Deficiency Using a Systems Genetics Approach. Genes, 13(11), 2078. https://doi.org/10.3390/genes13112078









