Migration/Differentiation-Associated LncRNA SENCR rs12420823*C/T: A Novel Gene Variant Can Predict Survival and Recurrence in Patients with Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. SENCR Mutation in Cancer Databases
2.3. Selection of SENCR Gene Variant
2.4. SENCR rs12420823*C/T Allelic Discrimination Analysis
2.5. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Study Population
3.2. Genotype and Allele Frequencies of SENCR rs12420823*C/T Polymorphism
3.3. Association of SENCR rs12420823*C/T Polymorphism with Breast Cancer Risk
3.4. Association of SENCR rs12420823*C/T Polymorphism and the Histopathological Types of Breast Cancer
3.5. Association of SENCR rs12420823*C/T with Polymorphism and Risk Factors
3.6. SENCR Polymorphism as a Prognostic Marker
3.7. SENCR rs12420823*C/T Polymorphism as a Predictive Marker
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Barrios, C.H. Global challenges in breast cancer detection and treatment. Breast 2022, 62 (Suppl. S1), S3–S6. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Castelli, G.; Pelosi, E. Breast Cancer: A Molecularly Heterogenous Disease Needing Subtype-Specific Treatments. Med. Sci. 2020, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Leitão, M.; Lopes, S.; Pereira, D.; Medeiros, R.; Vieira, C. Genetic Polymorphisms as Predictors of Survival in Breast Cancer: Future Lessons in Historical Data. Cureus 2022, 14, e21410. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Toraih, E.A.; Mohammed, E.A.; Farrag, S.; Ramsis, N.; Hosny, S. Pilot Study of Serum MicroRNA-21 as a Diagnostic and Prognostic Biomarker in Egyptian Breast Cancer Patients. Mol. Diagn. Ther. 2015, 19, 179–190. [Google Scholar] [CrossRef]
- Shi, T.; Gao, G.; Cao, Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis. Markers 2016, 2016, 9085195. [Google Scholar] [CrossRef]
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Toraih, E.A.; Alelwani, W.; Kattan, S.W.; Alnajeebi, A.M.; Hassan, R. The prognostic value of microRNA-biogenesis genes. Am. J. Transl. Res. 2020, 12, 1994–2006. [Google Scholar]
- Toraih, E.A.; El-Wazir, A.; Ageeli, E.A.; Hussein, M.H.; Eltoukhy, M.M.; Killackey, M.T.; Kandil, E.; Fawzy, M.S. Unleash multifunctional role of long noncoding RNAs biomarker panel in breast cancer: A predictor classification model. Epigenomics 2020, 12, 1215–1237. [Google Scholar] [CrossRef]
- El-Fattah, A.A.A.; Sadik, N.A.H.; Shaker, O.G.; Mohamed Kamal, A.; Shahin, N.N. Serum Long Non-Coding RNAs PVT1, HOTAIR, and NEAT1 as Potential Biomarkers in Egyptian Women with Breast Cancer. Biomolecules 2021, 11, 301. [Google Scholar] [CrossRef] [PubMed]
- Dvorská, D.; Braný, D.; Ňachajová, M.; Halašová, E.; Danková, Z. Breast Cancer and the Other Non-Coding RNAs. Int. J. Mol. Sci. 2021, 22, 3280. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Malih, S.; Saidijam, M.; Malih, N. A brief review on long noncoding RNAs: A new paradigm in breast cancer pathogenesis, diagnosis and therapy. Tumour Biol. 2016, 37, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Fu, F.; Chen, Y.; Qiu, W.; Lin, S.; Yang, P.; Huang, M.; Wang, C. Genetic variants in long noncoding RNA. Onco Targets Ther. 2017, 10, 4369–4378. [Google Scholar] [CrossRef]
- Cui, P.; Zhao, Y.; Chu, X.; He, N.; Zheng, H.; Han, J.; Song, F.; Chen, K. SNP rs2071095 in LincRNA H19 is associated with breast cancer risk. Breast Cancer Res. Treat. 2018, 171, 161–171. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Heidari, R.; Akbariqomi, M.; Asgari, Y.; Ebrahimi, D.; Alinejad-Rokny, H. A systematic review of long non-coding RNAs with a potential role in breast cancer. Mutat. Res. Rev. Mutat. Res. 2021, 787, 108375. [Google Scholar] [CrossRef]
- Lyu, Q.; Xu, S.; Lyu, Y.; Choi, M.; Christie, C.K.; Slivano, O.J.; Rahman, A.; Jin, Z.G.; Long, X.; Xu, Y.; et al. stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc. Natl. Acad. Sci. USA 2019, 116, 546–555. [Google Scholar] [CrossRef]
- Bell, R.D.; Long, X.; Lin, M.; Bergmann, J.H.; Nanda, V.; Cowan, S.L.; Zhou, Q.; Han, Y.; Spector, D.L.; Zheng, D.; et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arter. Thromb. Vasc. Biol. 2014, 34, 1249–1259. [Google Scholar] [CrossRef]
- Mohammad, H.M.F.; Abdelghany, A.A.; Al Ageeli, E.; Kattan, S.W.; Hassan, R.; Toraih, E.A.; Fawzy, M.S.; Mokhtar, N. Long Non-Coding RNAs Gene Variants as Molecular Markers for Diabetic Retinopathy Risk and Response to Anti-VEGF Therapy. Pharmgenomics Pers. Med. 2021, 14, 997–1014. [Google Scholar] [CrossRef] [PubMed]
- Elwazir, M.Y.; Hussein, M.H.; Toraih, E.A.; Al Ageeli, E.; Esmaeel, S.E.; Fawzy, M.S.; Faisal, S. Association of Angio-LncRNAs MIAT rs1061540/MALAT1 rs3200401 Molecular Variants with Gensini Score in Coronary Artery Disease Patients Undergoing Angiography. Biomolecules 2022, 12, 137. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.S.; Alvarez, C.; Milikowski, C.; Olson, N.; Russo, I.; Russo, J.; Glass, A.; Zehnbauer, B.A.; Lister, K.; Parwaresch, R.; et al. Breast carcinoma malignancy grading by Bloom-Richardson system vs proliferation index: Reproducibility of grade and advantages of proliferation index. Mod. Pathol. 2005, 18, 1067–1078. [Google Scholar] [CrossRef]
- Singletary, S.E.; Connolly, J.L. Breast cancer staging: Working with the sixth edition of the AJCC Cancer Staging Manual. CA Cancer J. Clin. 2006, 56, 37–47, quiz 50–31. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.; Ali, I.; Silins, I.; Pyysalo, S.; Guo, Y.; Högberg, J.; Stenius, U.; Korhonen, A. Cancer Hallmarks Analytics Tool (CHAT): A text mining approach to organize and evaluate scientific literature on cancer. Bioinformatics 2017, 33, 3973–3981. [Google Scholar] [CrossRef]
- Nagy, Á.; Lánczky, A.; Menyhárt, O.; Győrffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar] [CrossRef]
- Toraih, E.A.; Fawz, M.S.; Elgazzaz, M.G.; Hussein, M.H.; Shehata, R.H.; Daoud, H.G. Combined Genotype Analyses of Precursor miRNA196a2 and 499a Variants with Hepatic and Renal Cancer Susceptibility a Preliminary Study. Asian Pac. J. Cancer Prev. 2016, 17, 3369–3375. [Google Scholar]
- Fawzy, M.S.; Toraih, E.A.; Hamed, E.O.; Hussein, M.H.; Ismail, H.M. Association of MIR-499a expression and seed region variant (rs3746444) with cardiovascular disease in Egyptian patients. Acta Cardiol. 2017, 73, 131–140. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Hussein, M.H.; Abdelaziz, E.Z.; Yamany, H.A.; Ismail, H.M.; Toraih, E.A. Association of MicroRNA-196a2 Variant with Response to Short-Acting β2-Agonist in COPD: An Egyptian Pilot Study. PLoS ONE 2016, 11, e0152834. [Google Scholar] [CrossRef]
- Toraih, E.A.; Fawzy, M.S.; Mohammed, E.A.; Hussein, M.H.; El-Labban, M.M. MicroRNA-196a2 Biomarker and Targetome Network Analysis in Solid Tumors. Mol. Diagn. Ther. 2016, 20, 559–577. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, H.; Yan, G.; Wu, T.; Liu, S.; Chen, W.; Ning, Y.; Lu, Z. Long Non-Coding RNA and Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819843889. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Du, W.; Huang, W.; Yan, J.; Tang, Q.; Chen, Y.; Zou, Z. lncRNA and breast cancer: Progress from identifying mechanisms to challenges and opportunities of clinical treatment. Mol. Ther. Nucleic Acids 2021, 25, 613–637. [Google Scholar] [CrossRef] [PubMed]
- Sideris, N.; Dama, P.; Bayraktar, S.; Stiff, T.; Castellano, L. LncRNAs in breast cancer: A link to future approaches. Cancer Gene Ther. 2022, 1–12. [Google Scholar] [CrossRef]
- Petkevicius, V.; Streleckiene, G.; Balciute, K.; Link, A.; Leja, M.; Malfertheiner, P.; Skieceviciene, J.; Kupcinskas, J. Association of Long Non-Coding RNA Polymorphisms with Gastric Cancer and Atrophic Gastritis. Genes 2020, 11, 1505. [Google Scholar] [CrossRef]
- Suvanto, M.; Beesley, J.; Blomqvist, C.; Chenevix-Trench, G.; Khan, S.; Nevanlinna, H. SNPs in lncRNA Regions and Breast Cancer Risk. Front. Genet. 2020, 11, 550. [Google Scholar] [CrossRef]
- Tong, G.; Tong, W.; He, R.; Cui, Z.; Li, S.; Zhou, B.; Yin, Z. MALAT1 Polymorphisms and Lung Cancer Susceptibility in a Chinese Northeast Han Population. Int. J. Med. Sci. 2022, 19, 1300–1306. [Google Scholar] [CrossRef]
- Bayram, S.; Sümbül, A.T.; Batmacı, C.Y.; Genç, A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumour Biol. 2015, 36, 3863–3870. [Google Scholar] [CrossRef]
- Peng, R.; Luo, C.; Guo, Q.; Cao, J.; Yang, Q.; Dong, K.; Wang, S.; Wang, K.; Song, C. Association analyses of genetic variants in long non-coding RNA MALAT1 with breast cancer susceptibility and mRNA expression of MALAT1 in Chinese Han population. Gene 2018, 642, 241–248. [Google Scholar] [CrossRef]
- Riaz, M.; Berns, E.M.; Sieuwerts, A.M.; Ruigrok-Ritstier, K.; de Weerd, V.; Groenewoud, A.; Uitterlinden, A.G.; Look, M.P.; Klijn, J.G.; Sleijfer, S.; et al. Correlation of breast cancer susceptibility loci with patient characteristics, metastasis-free survival, and mRNA expression of the nearest genes. Breast Cancer Res. Treat. 2012, 133, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Royds, J.A.; Pilbrow, A.P.; Ahn, A.; Morrin, H.R.; Frampton, C.; Russell, I.A.; Moravec, C.S.; Sweet, W.E.; Tang, W.H.; Currie, M.J.; et al. The rs11515 Polymorphism Is More Frequent and Associated With Aggressive Breast Tumors with Increased ANRIL and Decreased p16 (INK4a) Expression. Front. Oncol. 2015, 5, 306. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Xiang, W.; Liu, J.; Tang, J.; Wang, J.; Long, Z.; Wang, L.; Yin, G. The regulatory role of antisense lncRNAs in cancer. Cancer Cell Int. 2021, 21, 459. [Google Scholar] [CrossRef] [PubMed]
- Boulberdaa, M.; Scott, E.; Ballantyne, M.; Garcia, R.; Descamps, B.; Angelini, G.D.; Brittan, M.; Hunter, A.; McBride, M.; McClure, J.; et al. A Role for the Long Noncoding RNA SENCR in Commitment and Function of Endothelial Cells. Mol. Ther. 2016, 24, 978–990. [Google Scholar] [CrossRef]
- Jin, K.T.; Yao, J.Y.; Fang, X.L.; Di, H.; Ma, Y.Y. Roles of lncRNAs in cancer: Focusing on angiogenesis. Life Sci. 2020, 252, 117647. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhou, H.; Zhang, M.; Wu, R.; Wang, L.; Wang, Y.; Chen, J. Super enhancer-LncRNA SENCR promoted cisplatin resistance and growth of NSCLC through upregulating FLI1. J. Clin. Lab. Anal. 2022, 36, e24460. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Karagkouni, D.; Vlachos, I.S.; Tastsoglou, S.; Hatzigeorgiou, A.G. microCLIP super learning framework uncovers functional transcriptome-wide miRNA interactions. Nat. Commun. 2018, 9, 3601. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Tastsoglou, S.; Skoufos, G.; Karavangeli, A.; Pierros, V.; Zacharopoulou, E.; Hatzigeorgiou, A.G. DIANA-LncBase v3: Indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2020, 48, D101–D110. [Google Scholar] [CrossRef]
- Gottwein, E.; Corcoran, D.L.; Mukherjee, N.; Skalsky, R.L.; Hafner, M.; Nusbaum, J.D.; Shamulailatpam, P.; Love, C.L.; Dave, S.S.; Tuschl, T.; et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 2011, 10, 515–526. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Corcoran, D.L.; Gottwein, E.; Frank, C.L.; Kang, D.; Hafner, M.; Nusbaum, J.D.; Feederle, R.; Delecluse, H.J.; Luftig, M.A.; et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012, 8, e1002484. [Google Scholar] [CrossRef]
- Ergün, S.; Ulasli, M.; Igci, Y.Z.; Igci, M.; Kırkbes, S.; Borazan, E.; Balik, A.; Yumrutaş, Ö.; Camci, C.; Cakmak, E.A.; et al. The association of the expression of miR-122-5p and its target ADAM10 with human breast cancer. Mol. Biol. Rep. 2015, 42, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Pasculli, B.; Barbano, R.; Fontana, A.; Biagini, T.; Di Viesti, M.P.; Rendina, M.; Valori, V.M.; Morritti, M.; Bravaccini, S.; Ravaioli, S.; et al. Hsa-miR-155-5p Up-Regulation in Breast Cancer and Its Relevance for Treatment With Poly [ADP-Ribose] Polymerase 1 (PARP-1) Inhibitors. Front. Oncol. 2020, 10, 1415. [Google Scholar] [CrossRef] [PubMed]
- Pasculli, B.; Barbano, R.; Rendina, M.; Fontana, A.; Copetti, M.; Mazza, T.; Valori, V.M.; Morritti, M.; Maiello, E.; Graziano, P.; et al. Hsa-miR-210-3p expression in breast cancer and its putative association with worse outcome in patients treated with Docetaxel. Sci. Rep. 2019, 9, 14913. [Google Scholar] [CrossRef] [PubMed]
- Lindholm, E.M.; Leivonen, S.K.; Undlien, E.; Nebdal, D.; Git, A.; Caldas, C.; Børresen-Dale, A.L.; Kleivi, K. miR-342-5p as a Potential Regulator of HER2 Breast Cancer Cell Growth. Microrna 2019, 8, 155–165. [Google Scholar] [CrossRef]
- Uhr, K.; Prager-van der Smissen, W.J.C.; Heine, A.A.J.; Ozturk, B.; van Jaarsveld, M.T.M.; Boersma, A.W.M.; Jager, A.; Wiemer, E.A.C.; Smid, M.; Foekens, J.A.; et al. MicroRNAs as possible indicators of drug sensitivity in breast cancer cell lines. PLoS ONE 2019, 14, e0216400. [Google Scholar] [CrossRef]
- Jinghua, H.; Qinghua, Z.; Chenchen, C.; Lili, C.; Xiao, X.; Yunfei, W.; Zhengzhe, A.; Changxiu, L.; Hui, H. MicroRNA miR-92a-3p regulates breast cancer cell proliferation and metastasis via regulating B-cell translocation gene 2 (BTG2). Bioengineered 2021, 12, 2033–2044. [Google Scholar] [CrossRef]
- Lung, D.K.; Reese, R.M.; Alarid, E.T. Intrinsic and Extrinsic Factors Governing the Transcriptional Regulation of ESR1. Horm. Cancer 2020, 11, 129–147. [Google Scholar] [CrossRef]
- Vleugel, M.M.; Greijer, A.E.; Bos, R.; van der Wall, E.; van Diest, P.J. c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum. Pathol. 2006, 37, 668–674. [Google Scholar] [CrossRef]
- Brennan, A.; Leech, J.T.; Kad, N.M.; Mason, J.M. Selective antagonism of cJun for cancer therapy. J. Exp. Clin. Cancer Res. 2020, 39, 184. [Google Scholar] [CrossRef]
- Guo, H.; Ahmed, M.; Zhang, F.; Yao, C.Q.; Li, S.; Liang, Y.; Hua, J.; Soares, F.; Sun, Y.; Langstein, J.; et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 2016, 48, 1142–1150. [Google Scholar] [CrossRef]
| Characteristics | Levels | Controls (n = 93) | Patients (n = 110) | p-Value |
|---|---|---|---|---|
| Demographics | ||||
| Age | Adolescents (≤21 years) | 27 (29) | 22 (20) | 0.14 |
| Adults (>21 years) | 66 (71) | 88 (80) | ||
| Residency | Urban | 47 (50.5) | 67 (60.9) | 0.15 |
| Rural | 46 (49.5) | 43 (39.1) | ||
| Marital status | Divorced | 20 (21.5) | 19 (17.3) | 0.017 |
| Married | 60 (64.5) | 57 (51.8) | ||
| Single | 13 (14) | 34 (30.9) | ||
| Menopausal status | Pre-menopause | 69 (74.2) | 79 (71.8) | 0.75 |
| Post-menopause | 24 (25.8) | 31 (28.2) | ||
| Risk factors | ||||
| Family history of cancer | Negative | 73 (78.5) | 78 (70.9) | 0.25 |
| Positive | 20 (21.5) | 32 (29.1) | ||
| Prior breast disease | Negative | 93 (100) | 100 (90.9) | 0.002 |
| Positive | 0 (0) | 10 (9.1) | ||
| Oral contraceptive pills | Negative | 76 (81.7) | 89 (80.9) | 0.88 |
| Positive | 17 (18.3) | 21 (19.1) | ||
| Early menarche | Negative | 64 (68.8) | 38 (34.5) | <0.001 |
| Positive | 29 (31.2) | 72 (65.5) | ||
| Nullipara | Negative | 76 (81.7) | 94 (85.5) | 0.56 |
| Positive | 17 (18.3) | 16 (14.5) | ||
| Late first gravida | Negative | 88 (94.6) | 106 (96.4) | 0.73 |
| Positive | 5 (5.4) | 4 (3.6) | ||
| Late menopause | Negative | 83 (89.2) | 98 (89.1) | 0.97 |
| Positive | 10 (10.8) | 12 (10.9) | ||
| No breastfeeding | Negative | 81 (87.1) | 90 (81.8) | 0.33 |
| Positive | 12 (12.9) | 20 (18.2) | ||
| Night light exposure | Negative | 69 (74.2) | 98 (89.1) | 0.009 |
| Positive | 24 (25.8) | 12 (10.9) | ||
| Sedentary lifestyle | Negative | 24 (25.8) | 11 (10) | 0.005 |
| Positive | 69 (74.2) | 99 (90) | ||
| Smoking | Negative | 88 (94.6) | 98 (89.1) | 0.21 |
| Positive | 5 (5.4) | 12 (10.9) | ||
| Body weight | Underweight | 0 (0) | 15 (13.6) | 0.001 |
| Normal weight | 21 (22.6) | 27 (24.5) | ||
| Overweight | 37 (39.8) | 23 (20.9) | ||
| Obese | 29 (31.2) | 36 (32.7) | ||
| Morbid obesity | 6 (6.5) | 9 (8.2) |
| Variant | Total | Controls | Patients | p-Value | Crude OR (95%CI) |
|---|---|---|---|---|---|
| Total number | 203 | 93 | 110 | ||
| Allele frequency | |||||
| C | 194 (47.8%) | 105 (56%) | 89 (40.4%) | <0.001 | 1 |
| T | 212 (52.3%) | 81 (44%) | 131 (59.6%) | 1.91 (1.28–2.83) | |
| Genotype frequency | |||||
| C/C | 37 (18.2%) | 26 (28.0%) | 11 (10.0%) | 0.001 | 1 |
| C/T | 120 (59.1%) | 53 (57.0%) | 67 (60.9%) | 1.82 (0.88–3.70) | |
| T/T | 46 (22.7%) | 14 (15.0%) | 32 (29.1%) | 5.26 (2.08–14.3) |
| Model | Genotype | Control | Patients | Adjusted OR (95% CI) # | p-Value | AIC | Adjusted OR (95% CI) * | p-Value | AIC |
|---|---|---|---|---|---|---|---|---|---|
| Codominant | C/C | 26 (28%) | 11 (10%) | 1 | 0.001 | 272.6 | 1 | 0.001 | 233.7 |
| C/T | 53 (57%) | 67 (60.9%) | 1.61 (0.78–3.45) | 1.85 (0.72–4.76) | |||||
| T/T | 14 (15.1%) | 32 (29.1%) | 5.26 (2.04–14.29) | 8.33 (2.44–25.0) | |||||
| Dominant | C/C | 26 (28%) | 11 (10%) | 1 | 0.027 | 272.3 | 1 | 0.019 | 233.3 |
| T/T-C/T | 67 (72%) | 99 (90%) | 3.70 (1.72–8.33) | 5.56 (2.0–14.3) | |||||
| Recessive | C/T-C/C | 79 (85%) | 78 (70.9%) | 1 | <0.001 | 279.5 | 1 | <0.001 | 240 |
| T/T | 14 (15.1%) | 32 (29.1%) | 2.17 (1.08–4.55) | 2.86 (1.15–7.14) | |||||
| Over-dominant | C/C-T/T | 40 (43%) | 43 (39.1%) | 1 | 0.36 | 283.6 | 1 | 0.44 | 244.9 |
| C/T | 53 (57%) | 67 (60.9%) | 1.31 (0.73–2.53) | 1.34 (0.64–2.83) |
| Histopathological Type | C/C | C/T | T/T | p-Value |
|---|---|---|---|---|
| Duct carcinoma | 6 (54.5) | 24 (35.8) | 12 (37.5) | 0.68 |
| Lobular carcinoma | 2 (18.2) | 16 (23.9) | 10 (31.3) | |
| Invasive medullary carcinoma | 2 (18.2) | 10 (14.9) | 2 (6.3) | |
| Mucinous carcinoma | 0 (0) | 9 (13.4) | 2 (6.3) | |
| Tubular carcinoma | 0 (0) | 5 (7.5) | 3 (9.4) | |
| Metaplastic carcinoma | 1 (9.1) | 3 (4.5) | 3 (9.4) |
| Characteristics | Levels | C/C-C/T | T/T | p-Value |
|---|---|---|---|---|
| Demographics | ||||
| Age | Adolescents (≤21 years) | 63 (80.8) | 25 (78.1) | 0.79 |
| Adults (>21 years) | 15 (19.2) | 7 (21.9) | ||
| Residency | Urban | 31 (39.7) | 12 (37.5) | 0.82 |
| Rural | 47 (60.3) | 20 (62.5) | ||
| Marital status | Divorced | 16 (20.5) | 3 (9.4) | 0.026 |
| Married | 34 (43.6) | 23 (71.9) | ||
| Single | 28 (35.9) | 6 (18.8) | ||
| Occupation | Housewife | 56 (71.8) | 24 (75) | 0.81 |
| Worker | 22 (28.2) | 8 (25) | ||
| Menopausal status | Pre-menopause | 58 (74.4) | 21 (65.6) | 0.31 |
| Post-menopause | 20 (25.6) | 11 (34.4) | ||
| Risk factors | ||||
| Family history of cancer | Negative | 55 (70.5) | 23 (71.9) | 0.88 |
| Positive | 23 (29.5) | 9 (28.1) | ||
| Oral contraceptive pills | Negative | 60 (76.9) | 29 (90.6) | 0.15 |
| Positive | 18 (23.1) | 3 (9.4) | ||
| Early menarche | Negative | 28 (35.9) | 10 (31.3) | 0.82 |
| Positive | 50 (64.1) | 22 (68.8) | ||
| Nullipara | Negative | 66 (84.6) | 28 (87.5) | 0.77 |
| Positive | 12 (15.4) | 4 (12.5) | ||
| Late first gravida | Negative | 75 (96.2) | 31 (96.9) | 0.85 |
| Positive | 3 (3.8) | 1 (3.1) | ||
| Late menopause | Negative | 69 (88.5) | 29 (90.6) | 0.74 |
| Positive | 9 (11.5) | 3 (9.4) | ||
| No breastfeeding | Negative | 63 (80.8) | 27 (84.4) | 0.78 |
| Positive | 15 (19.2) | 5 (15.6) | ||
| Night light exposure | Negative | 70 (89.7) | 28 (87.5) | 0.74 |
| Positive | 8 (10.3) | 4 (12.5) | ||
| Sedentary lifestyle | Negative | 9 (11.5) | 2 (6.3) | 0.50 |
| Positive | 69 (88.5) | 30 (93.8) | ||
| Smoking | Negative | 69 (88.5) | 29 (90.6) | 0.71 |
| Positive | 9 (11.5) | 3 (9.4) | ||
| Obesity | Negative | 30 (38.5) | 12 (37.5) | 0.92 |
| Positive | 48 (61.5) | 20 (62.5) |
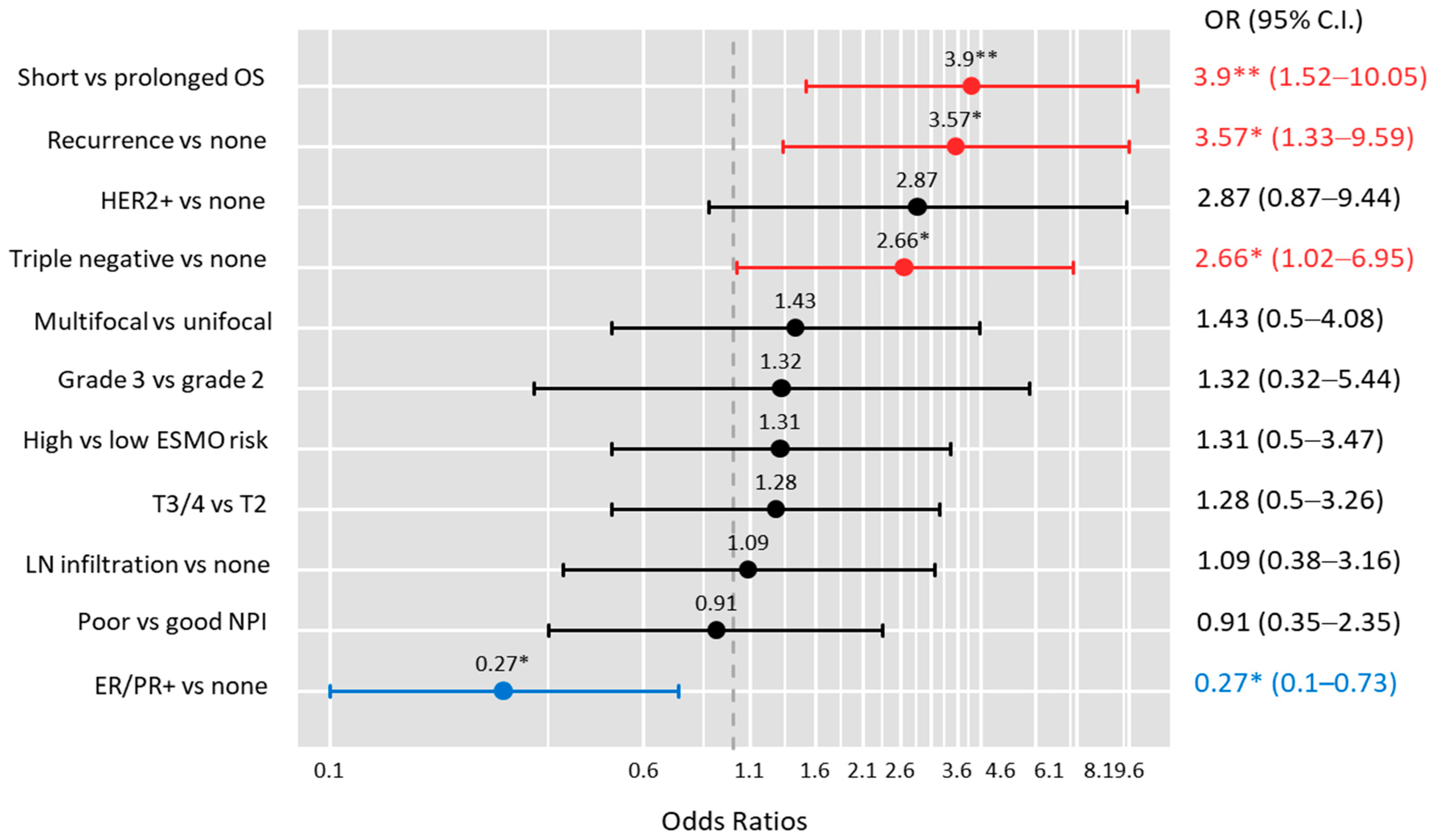
| Characteristics | Levels | C/C-C/T | T/T | p-Value | OR (95%CI) |
|---|---|---|---|---|---|
| Clinical presentation | |||||
| Mastalgia | Positive | 27 (34.6) | 8 (25) | 0.37 | 0.63 (0.25–1.59) |
| Breast mass | Positive | 64 (82.1) | 28 (87.5) | 0.58 | 1.53 (0.46–5.07) |
| Skin changes | Positive | 12 (15.4) | 5 (15.6) | 0.97 | 1.02 (0.33–3.17) |
| Nipple changes | Positive | 16 (20.5) | 2 (6.3) | 0.08 | 0.26 (0.06–1.2) |
| Axillary pain | Positive | 5 (6.4) | 2 (6.3) | 0.97 | 0.97 (0.18–5.3) |
| Axillary mass | Positive | 5 (6.4) | 2 (6.3) | 0.97 | 0.97 (0.18–5.3) |
| Pathological data | |||||
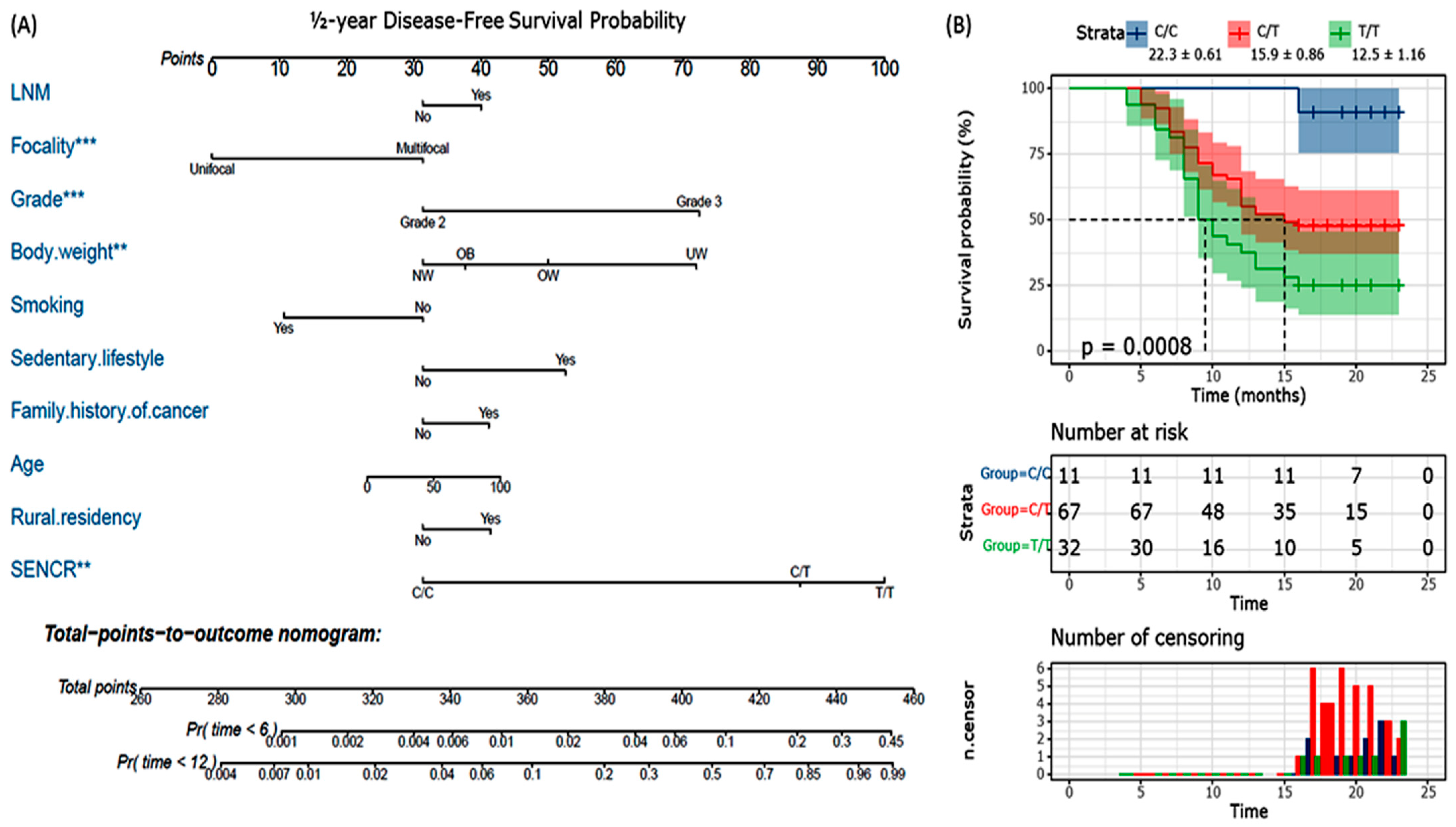
| Focality | Unifocal | 62 (79.5) | 22 (68.8) | 0.32 | Reference |
| Multifocal | 16 (20.5) | 10 (31.3) | 1.76 (0.7–4.45) | ||
| Pathological grade | Grade 2 | 62 (79.5) | 26 (81.3) | 0.83 | Reference |
| Grade 3 | 16 (20.5) | 6 (18.8) | 0.89 (0.31–2.54) | ||
| Tumor stage | T2 stage | 38 (48.7) | 13 (40.6) | 0.52 | Reference |
| T3/4 stages | 40 (51.3) | 19 (59.4) | 1.39 (0.6–3.2) | ||
| Nodal stage | Negative infiltration | 21 (26.9) | 8 (25) | 0.83 | Reference |
| Positive infiltration | 57 (73.1) | 24 (75) | 1.11 (0.43–2.84) | ||
| NPI | Good | 37 (47.4) | 17 (53.1) | 0.67 | 0.8 (0.35–1.82) |
| Poor | 41 (52.6) | 15 (46.9) | |||
| ESMO | Low risk | 29 (37.2) | 11 (34.4) | 0.83 | 1.13 (0.48–2.68) |
| High risk | 49 (62.8) | 21 (65.6) | |||
| Receptor status | |||||
| ER/PR | Positive | 47 (60.3) | 12 (37.5) | 0.036 | 0.4 (0.17–0.92) |
| HER2+ | Positive | 9 (11.5) | 8 (25) | 0.08 | 2.56 (0.89–7.37) |
| TNBC | Positive | 28 (35.9) | 17 (53.1) | 0.13 | 2.02 (0.88–4.66) |
| IHPI | Good | 47 (60.3) | 12 (37.5) | 0.08 | Reference |
| Moderate | 28 (35.9) | 17 (53.1) | 0.84 (0.35–2.02) | ||
| Poor | 3 (3.8) | 3 (9.4) | 2.33 (0.41–13.2) | ||
| Clinical outcomes | |||||
| Recurrence | Negative | 42 (53.8) | 8 (25) | 0.006 | 3.5 (1.4–8.74) |
| Positive | 36 (46.2) | 24 (75) | |||
| Survival | Prolonged > 12 months | 55 (70.5) | 13 (40.6) | 0.005 | 3.5 (1.49–8.33) |
| Short ≤ 12 months | 23 (29.5) | 19 (59.4) |
| Position (hg38) | LD (r²) | LD (D′) | Variant | Ref | Alt | AFR Freq | AMR Freq | ASN Freq | EUR Freq | Promoter Histone Marks | Enhancer Histone Marks | Proteins Bound | Motifs Changed | dbSNP Func Annot |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 128693497 | 1 | 1 | rs12420823 | C | T | 0.51 | 0.40 | 0.20 | 0.47 | 21 tissues | 4 tissues | CTCF, CJUN | intronic | |
| 128693518 | 1 | 1 | rs12420835 | C | A | 0.39 | 0.38 | 0.19 | 0.47 | 22 tissues | 4 tissues | CTCF, CJUN | 9 altered motifs | intronic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ageeli, E.A.; Attallah, S.M.; Mohamed, M.H.; Almars, A.I.; Kattan, S.W.; Toraih, E.A.; Fawzy, M.S.; Darwish, M.K. Migration/Differentiation-Associated LncRNA SENCR rs12420823*C/T: A Novel Gene Variant Can Predict Survival and Recurrence in Patients with Breast Cancer. Genes 2022, 13, 1996. https://doi.org/10.3390/genes13111996
Ageeli EA, Attallah SM, Mohamed MH, Almars AI, Kattan SW, Toraih EA, Fawzy MS, Darwish MK. Migration/Differentiation-Associated LncRNA SENCR rs12420823*C/T: A Novel Gene Variant Can Predict Survival and Recurrence in Patients with Breast Cancer. Genes. 2022; 13(11):1996. https://doi.org/10.3390/genes13111996
Chicago/Turabian StyleAgeeli, Essam Al, Samy M. Attallah, Marwa Hussein Mohamed, Amany I. Almars, Shahad W. Kattan, Eman A. Toraih, Manal S. Fawzy, and Marwa K. Darwish. 2022. "Migration/Differentiation-Associated LncRNA SENCR rs12420823*C/T: A Novel Gene Variant Can Predict Survival and Recurrence in Patients with Breast Cancer" Genes 13, no. 11: 1996. https://doi.org/10.3390/genes13111996
APA StyleAgeeli, E. A., Attallah, S. M., Mohamed, M. H., Almars, A. I., Kattan, S. W., Toraih, E. A., Fawzy, M. S., & Darwish, M. K. (2022). Migration/Differentiation-Associated LncRNA SENCR rs12420823*C/T: A Novel Gene Variant Can Predict Survival and Recurrence in Patients with Breast Cancer. Genes, 13(11), 1996. https://doi.org/10.3390/genes13111996










