Genome-Wide Identification of Potential mRNAs in Drought Response in Wheat (Triticum aestivum L.)
Abstract
1. Introduction
2. Materials and Methods
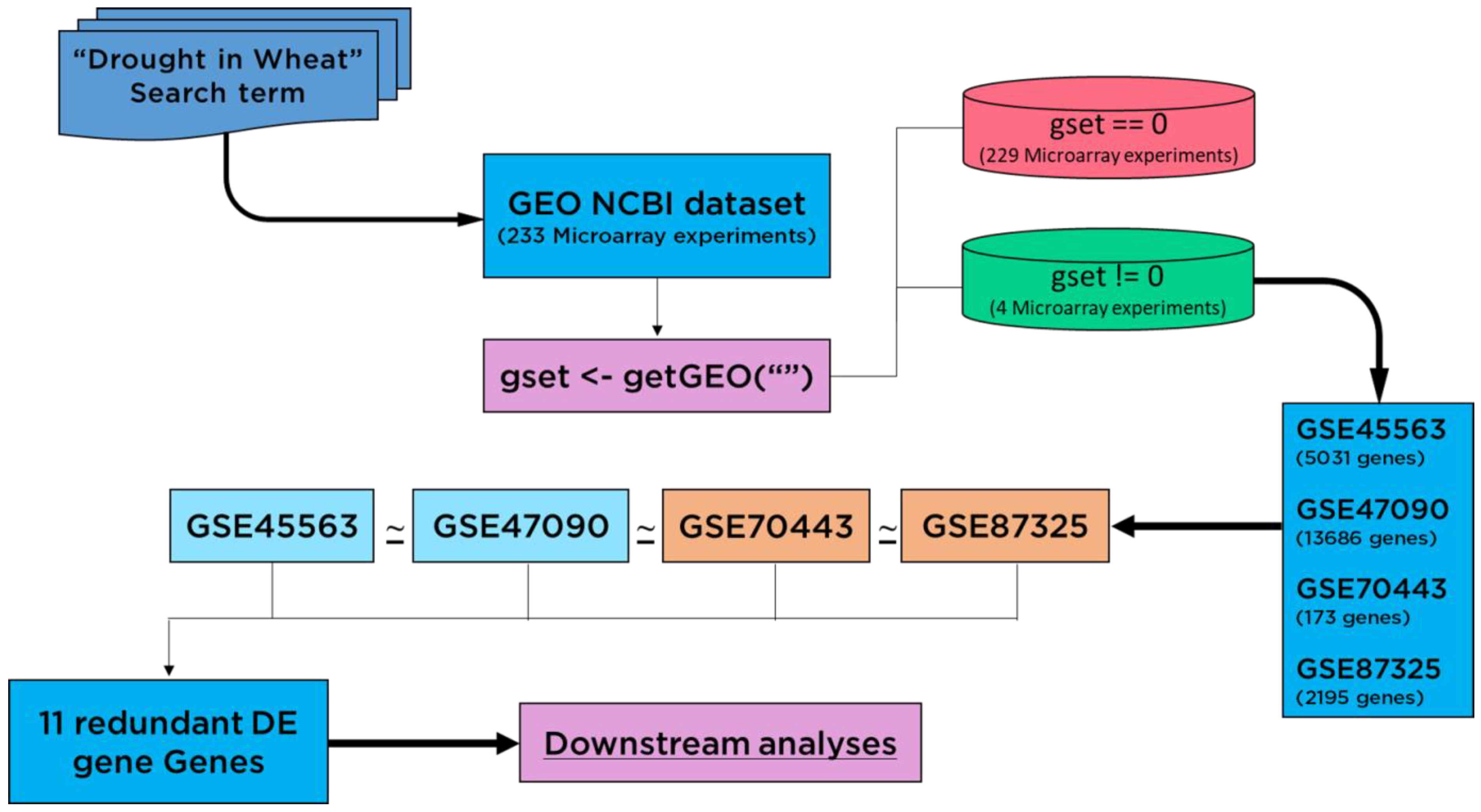
2.1. Meta-Analysis of Data from the Genome-Wide Transcriptome
2.2. Differential Expression Analyses
2.3. One-Sample Proportions Test for SDGs Screening
2.4. Bioinformatics Analyses
2.5. Functional and Pathway Enrichment Analyses of SDGs
2.6. Principal Component Analyses
2.7. Plant Ontology
3. Results
3.1. Meta-Analysis of Data from the Genome-Wide Transcriptome
3.2. Global Drought Genes Screening
3.3. IWGSC Gene IDs Identification
3.4. Identification of Analyzed Genes Transcripts
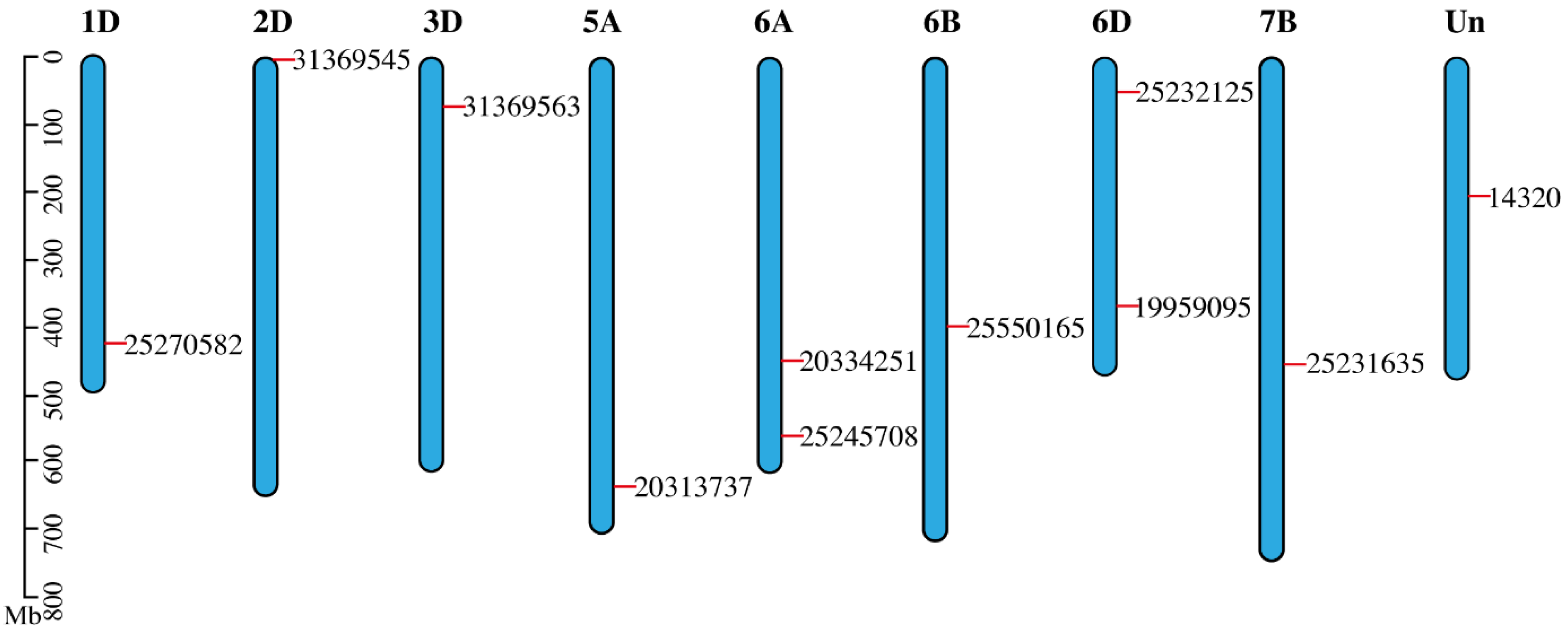
3.5. Mapping of SDGs on Wheat Genome
3.6. Expression Analyses
3.7. Statistical Analysis
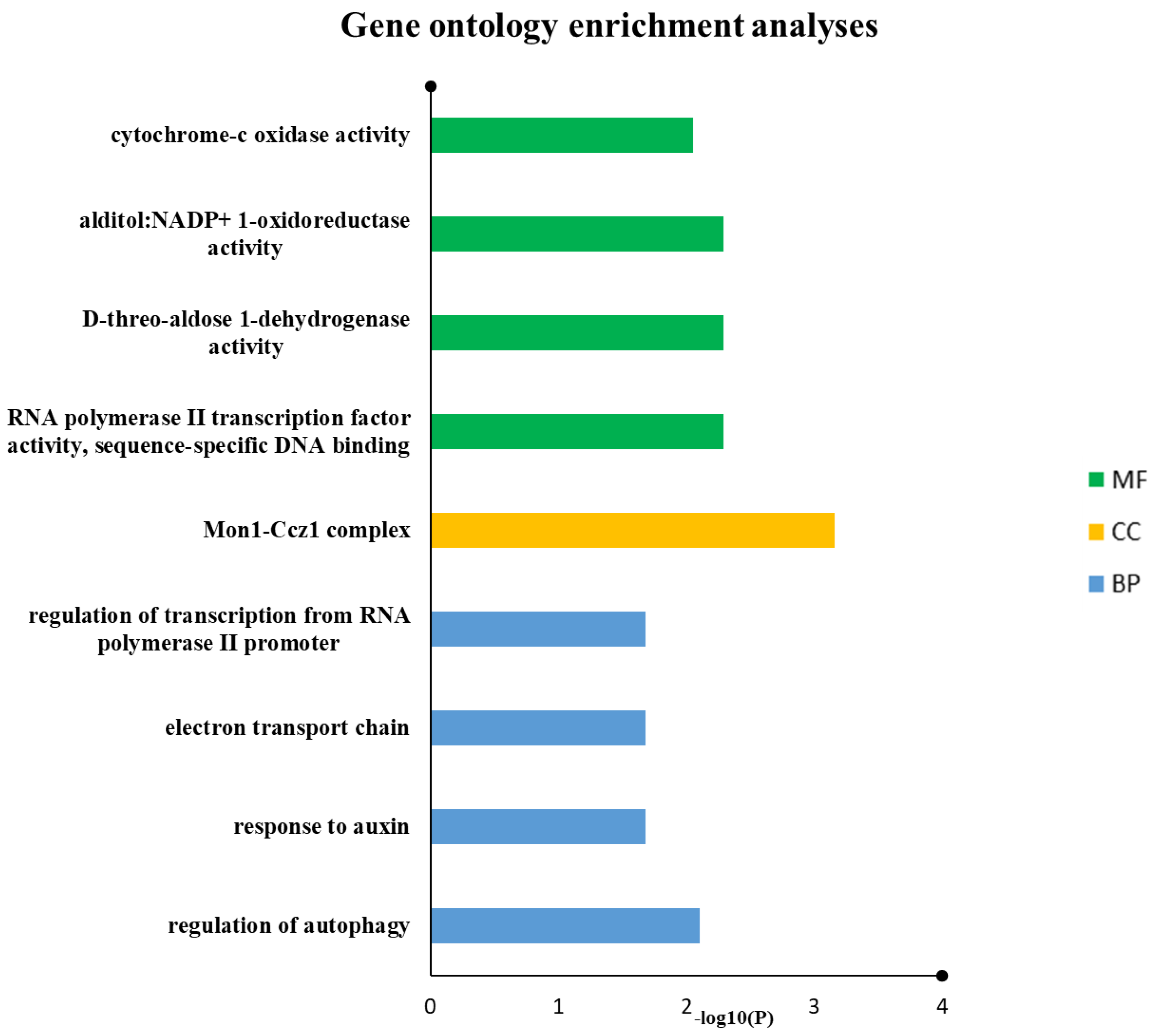
3.8. GO Categorization of SDGs
3.9. Gene Annotation from PlantRegMap Database
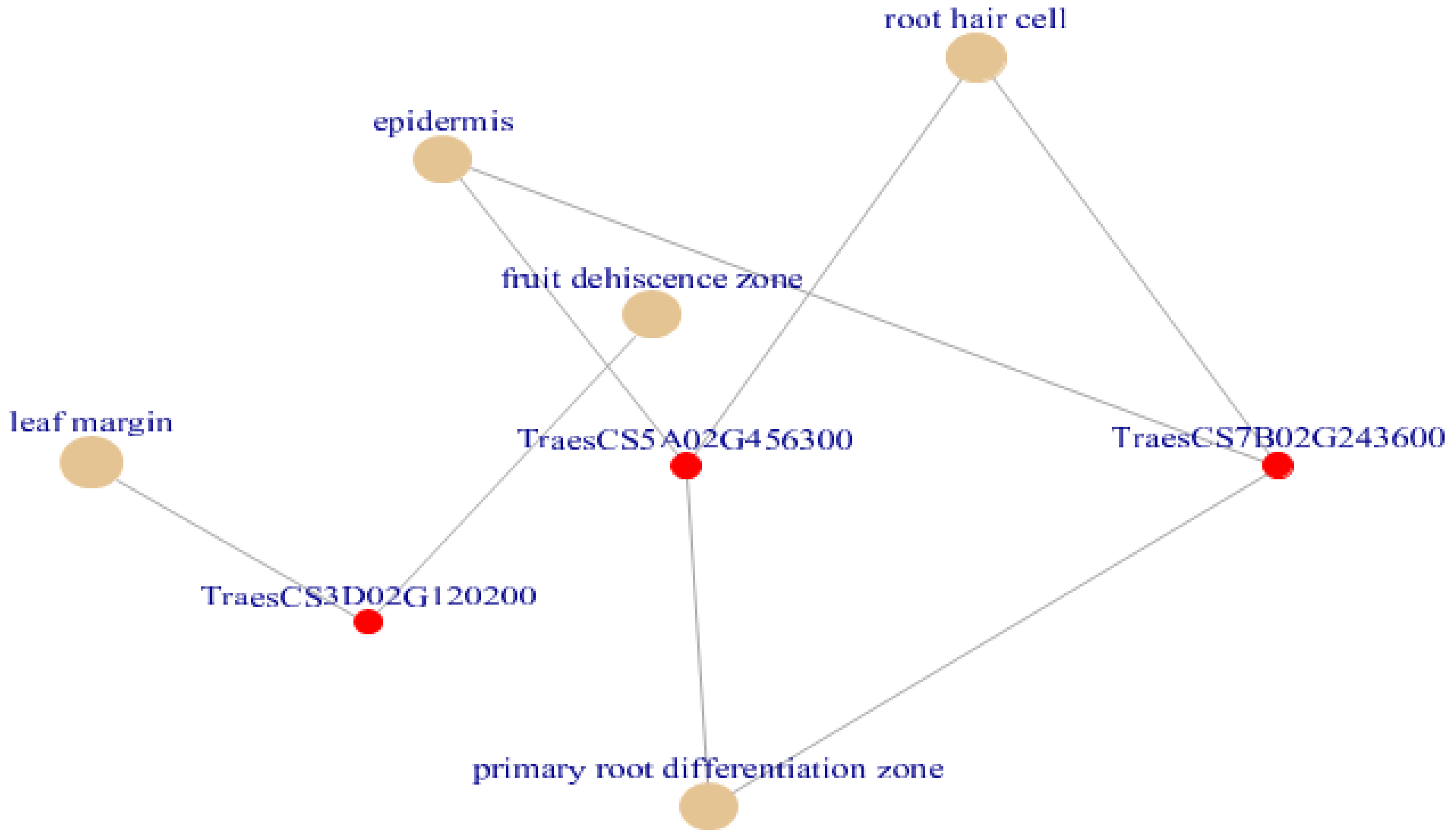
3.10. Plant Ontology Annotation Using Annotated Reference Genomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zulkiffal, M.; Ahsan, A.; Ahmed, J.; Musa, M.; Kanwal, A.; Saleem, M.; Anwar, J.; ur Rehman, A.; Ajmal, S.; Gulnaz, S.; et al. Heat and Drought Stresses in Wheat (Triticum aestivum L.): Substantial Yield Losses, Practical Achievements, Improvement Approaches, and Adaptive Mechanisms. Plant Stress Physiol. 2021, 3. [Google Scholar] [CrossRef]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef] [PubMed]
- De Flaviis, R.; Mutarutwa, D.; Sacchetti, G.; Mastrocola, D. Could environmental effect overcome genetic? A chemometric study on wheat volatiles fingerprint. Food Chem. 2022, 372, 131236. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, H.A.; Belgacem, A.O.; Behnassi, M.; Javed, K.; Baig, M.B. Impacts of Climate Change on Livestock and Related Food Security Implications—Overview of the Situation in Pakistan and Policy Recommendations. In Emerging Challenges to Food Production and Security in Asia, Middle East, and Africa; Springer: Cham, Switzherland, 2021; pp. 197–239. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, S.; Cheng, M.; Jiang, H.; Zhang, X.; Peng, C.; Lu, X.; Zhang, M.; Jin, J. Effect of drought on agronomic traits of rice and wheat: A meta-analysis. Int. J. Environ. Res. Public Health 2018, 15, 839. [Google Scholar] [CrossRef] [PubMed]
- Bapela, T.; Shimelis, H.; Tsilo, T.J.; Mathew, I. Genetic Improvement of Wheat for Drought Tolerance: Progress, Challenges and Opportunities. Plants 2022, 11, 1331. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Hao, Y.; Stebler, E.; Tanaka, N.; Zou, C.B. Impact of Plant Functional Types on Coherence Between Precipitation and Soil Moisture: A Wavelet Analysis. Geophys. Res. Lett. 2017, 44, 12,197–12,207. [Google Scholar] [CrossRef]
- Panda, D.; Mishra, S.S.; Behera, P.K. Drought Tolerance in Rice: Focus on Recent Mechanisms and Approaches. Rice Sci. 2021, 28, 119–132. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, A.; Panda, D.; Modi, M.K.; Sen, P. Identification and characterization of drought responsive miRNAs from a drought tolerant rice genotype of Assam. Plant Gene 2020, 21, 100213. [Google Scholar] [CrossRef]
- Pandey, V.; Shukla, A. Acclimation and tolerance strategies of rice under drought stress. Rice Sci. 2015, 22, 147–161. [Google Scholar] [CrossRef]
- Naeem, M.; Waseem, M.; Zhu, Z.; Zhang, L. Downregulation of SlGRAS15 manipulates plant architecture in tomato (Solanum lycopersicum). Dev. Genes Evol. 2020, 230, 1–12. [Google Scholar] [CrossRef]
- Sunera; Amna; Saqib, S.; Uddin, S.; Zaman, W.; Ullah, F.; Ayaz, A.; Asghar, M.; ur Rehman, S.; Munis, M.F.H.; et al. Characterization and phytostimulatory activity of bacteria isolated from tomato (Lycopersicon esculentum Mill.) rhizosphere. Microb. Pathog. 2020, 140, 103966. [Google Scholar]
- Sadiqi, S.; Hamza, M.; Ali, F.; Alam, S.; Shakeela, Q.; Ahmed, S.; Ayaz, A.; Ali, S.; Saqib, S.; Ullah, F.; et al. Molecular Characterization of Bacterial Isolates from Soil Samples and Evaluation of their Antibacterial Potential against MDRS. Molecules 2022, 27, 6281. [Google Scholar] [PubMed]
- Scarano, A.; Murmura, G.; Vantaggiato, G.; Lauritano, D.; Silvestre-Rangil, J.; Di Cerbo, A.; Lorusso, F. Delayed expansion of atrophic mandible (deam): A case report. Oral Implantol. 2017, 10, 190. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Li, S.; Chen, B.; Jian, C.; Mei, F.; Zhang, Y.; Li, F.; Chen, N.; Li, T.; Du, L.; et al. Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant 2022, 15, 276–292. [Google Scholar] [CrossRef]
- Ma, J.; Tang, X.; Sun, B.; Wei, J.; Ma, L.; Yuan, M.; Zhang, D.; Shao, Y.; Li, C.; Chen, K.-M.; et al. A NAC transcription factor, TaNAC5D-2, acts as a positive regulator of drought tolerance through regulating water loss in wheat (Triticum aestivum L.). Environ. Exp. Bot. 2022, 196, 104805. [Google Scholar] [CrossRef]
- Wu, Z.; Shen, S.; Wang, Y.; Tao, W.; Zhao, Z.; Hu, X.; Yu, P. Genome-Wide Identification and Expression Analysis of the Zinc Finger Protein Gene Subfamilies under Drought Stress in Triticum aestivum. Plants 2022, 11, 2511. [Google Scholar] [CrossRef]
- Pan, Y.; Li, M.; Huang, J.; Pan, W.; Shi, T.; Guo, Q.; Yang, G.; Nie, X. Genome-Wide Identification and Characterization of RNA/DNA Differences Associated with Drought Response in Wheat. Int. J. Mol. Sci. 2022, 23, 1405. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, S.; Raina, M.; Kumar, D. Selenium transporters and their role in plant development and stress. In Cation Transporters in Plants; Elsevier: Amsterdam, The Netherlands, 2021; pp. 307–336. ISBN 9780323857901. [Google Scholar]
- Feng, X.; Ma, Q. Transcriptome and proteome profiling revealed molecular mechanism of selenium responses in bread wheat (Triticum aestivum L.). BMC Plant Biol. 2021, 21, 584. [Google Scholar] [CrossRef]
- Aprile, A.; Havlickova, L.; Panna, R.; Marè, C.; Borrelli, G.M.; Marone, D.; Perrotta, C.; Rampino, P.; De Bellis, L.; Curn, V.; et al. Different stress responsive strategies to drought and heat in two durum wheat cultivars with contrasting water use efficiency. BMC Genom. 2013, 14, 821. [Google Scholar] [CrossRef]
- Peremarti, A.; Marè, C.; Aprile, A.; Roncaglia, E.; Cattivelli, L.; Villegas, D.; Royo, C. Transcriptomic and proteomic analyses of a pale-green durum wheat mutant shows variations in photosystem components and metabolic deficiencies under drought stress. BMC Genom. 2014, 15, 125. [Google Scholar] [CrossRef]
- Begcy, K.; Walia, H. Drought stress delays endosperm development and misregulates genes associated with cytoskeleton organization and grain quality proteins in developing wheat seeds. Plant Sci. 2015, 240, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Gunapati, S.; Kianian, S.F.; Singh, S.P. Comparative analysis of transcriptome in two wheat genotypes with contrasting levels of drought tolerance. Protoplasma 2018, 255, 1487–1504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear models for microarray data BT. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Smyth, G.K.; Verbyla, A.P. A Conditional Likelihood Approach to Residual Maximum Likelihood Estimation in Generalized Linear Models. J. R. Stat. Soc. Ser. B 1996, 58, 565–572. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. others Factoextra: Extract and visualize the results of multivariate data analyses. R Package Version 2017, 1, 337–354. [Google Scholar]
- Baldoni, E.; Frugis, G.; Martinelli, F.; Benny, J.; Paffetti, D.; Buti, M. A comparative transcriptomic meta-analysis revealed conserved key genes and regulatory networks involved in drought tolerance in cereal crops. Int. J. Mol. Sci. 2021, 22, 13062. [Google Scholar] [CrossRef]
- Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 2015, 6, 84. [Google Scholar] [CrossRef]
- Muthuramalingam, P.; Krishnan, S.R.; Pothiraj, R.; Ramesh, M. Global transcriptome analysis of combined abiotic stress signaling genes unravels key players in Oryza sativa L.: An in silico approach. Front. Plant Sci. 2017, 8, 759. [Google Scholar] [CrossRef]
- Gallo, A.; Landi, R.; Rubino, V.; Di Cerbo, A.; Giovazzino, A.; Palatucci, A.T.; Centenaro, S.; Guidetti, G.; Canello, S.; Cortese, L.; et al. Oxytetracycline induces DNA damage and epigenetic changes: A possible risk for human and animal health? PeerJ 2017, 5, e3236. [Google Scholar] [CrossRef] [PubMed]
- Buti, M.; Baldoni, E.; Formentin, E.; Milc, J.; Frugis, G.; Schiavo, F.L.; Genga, A.; Francia, E. A meta-analysis of comparative transcriptomic data reveals a set of key genes involved in the tolerance to abiotic stresses in rice. Int. J. Mol. Sci. 2019, 20, 5662. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.K.; Bowles, D.J. A class of plant glycosyltransferases involved in cellular homeostatis. EMBO J. 2004, 23, 2915–2922. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.-J.; Zhang, F.-J.; Zhang, G.-Z.; Jiang, X.-Y.; Yu, H.-M.; Hou, B.-K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Li, Y.J.; Li, P.; Wang, T.; Zhang, F.J.; Huang, X.X.; Hou, B.K. The maize secondary metabolism glycosyltransferase UFGT2 modifies flavonols and contributes to plant acclimation to abiotic stresses. Ann. Bot. 2018, 122, 1203–1217. [Google Scholar] [CrossRef]
- Dong, N.Q.; Sun, Y.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C.; et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef]
- Pascuan, C.; Frare, R.; Alleva, K.; Ayub, N.D.; Soto, G. mRNA biogenesis-related helicase eIF4AIII from Arabidopsis thaliana is an important factor for abiotic stress adaptation. Plant Cell Rep. 2016, 35, 1205–1208. [Google Scholar] [CrossRef]
- Shivakumara, T.N.; Sreevathsa, R.; Dash, P.K.; Sheshshayee, M.S.; Papolu, P.K.; Rao, U.; Tuteja, N.; UdayaKumar, M. Overexpression of Pea DNA Helicase 45 (PDH45) imparts tolerance to multiple abiotic stresses in chili (Capsicum annuum L.). Sci. Rep. 2017, 7, 2760. [Google Scholar] [CrossRef]
- Harris, J.C.; Hrmova, M.; Lopato, S.; Langridge, P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011, 190, 823–837. [Google Scholar] [CrossRef]
- Cabello, J.V.; Chan, R.L. The homologous homeodomain-leucine zipper transcription factors HaHB1 and AtHB13 confer tolerance to drought and salinity stresses via the induction of proteins that stabilize membranes. Plant Biotechnol. J. 2012, 10, 815–825. [Google Scholar] [CrossRef]
- Wang, P.; Chen, D.; Zheng, Y.; Jin, S.; Yang, J.; Ye, N. Identification and expression analyses of SBP-box genes reveal their involvement in abiotic stress and hormone response in tea plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 3404. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yan, C.; Sun, Q.; Wang, J.; Yuan, C.; Mou, Y.; Shan, S.; Zhao, X. The bHLH transcription factor AhbHLH112 improves the drought tolerance of peanut. BMC Plant Biol. 2021, 21, 540. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Fan, Z.; Qiu, L.; Che, Q.; Wang, T.; Li, Y.; Wang, Y. MdbHLH130, an Apple bHLH Transcription Factor, Confers Water Stress Resistance by Regulating Stomatal Closure and ROS Homeostasis in Transgenic Tobacco. Front. Plant Sci. 2020, 11, 543696. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Liu, H.; Ji, H.; Wang, Y.; Dong, B.; Qiao, Y.; Liu, M.; Li, X. The wheat GT factor TaGT2L1D negatively regulates drought tolerance and plant development. Sci. Rep. 2016, 6, 27042. [Google Scholar] [CrossRef]
- Xi, J.; Qiu, Y.; Du, L.; Poovaiah, B.W. Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci. 2012, 185, 274–280. [Google Scholar] [CrossRef]
- Amnan, M.A.M.; Aizat, W.M.; Khaidizar, F.D.; Tan, B.C. Drought Stress Induces Morpho-Physiological and Proteome Changes of Pandanus amaryllifolius. Plants 2022, 11, 221. [Google Scholar] [CrossRef]
- Dietz, K.J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Refay, Y.; Al-Suhaibani, N.; Al-Ashkar, I.; El-Hendawy, S.; Hafez, E.M. Integrative effects of rice-straw biochar and silicon on oil and seed quality, yield and physiological traits of Helianthus annuus L. grown under water deficit stress. Agronomy 2019, 9, 637. [Google Scholar] [CrossRef]
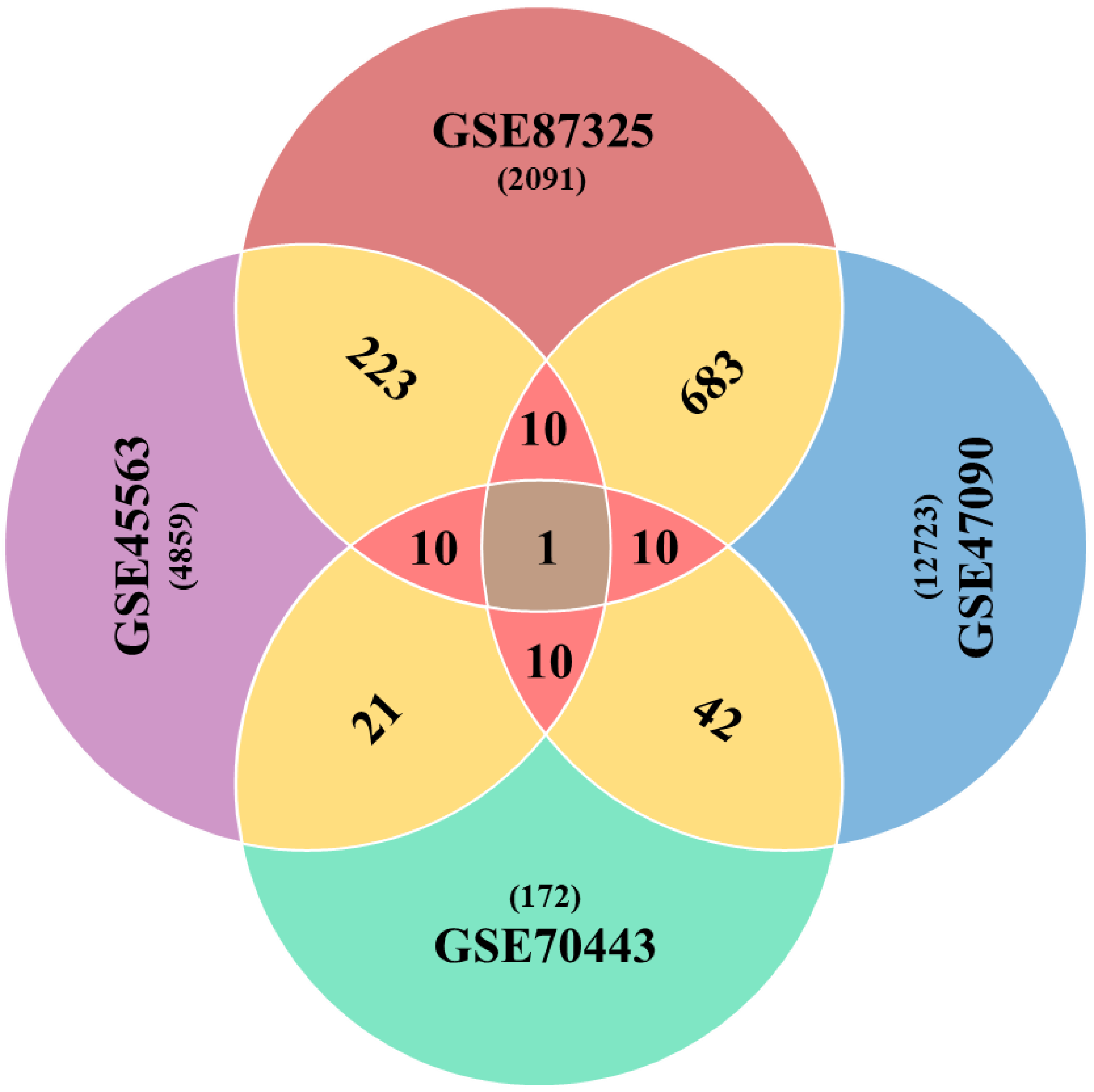
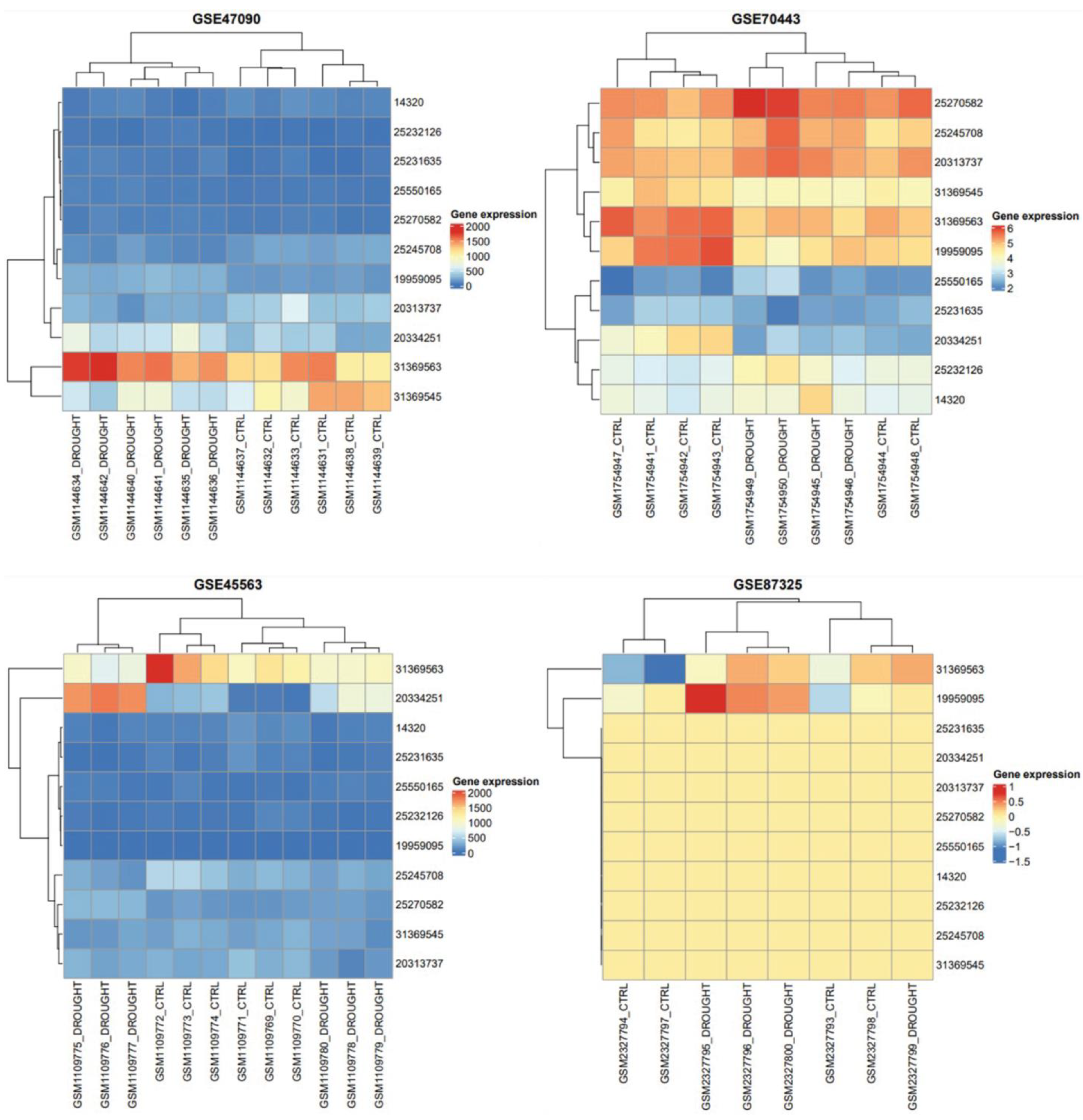
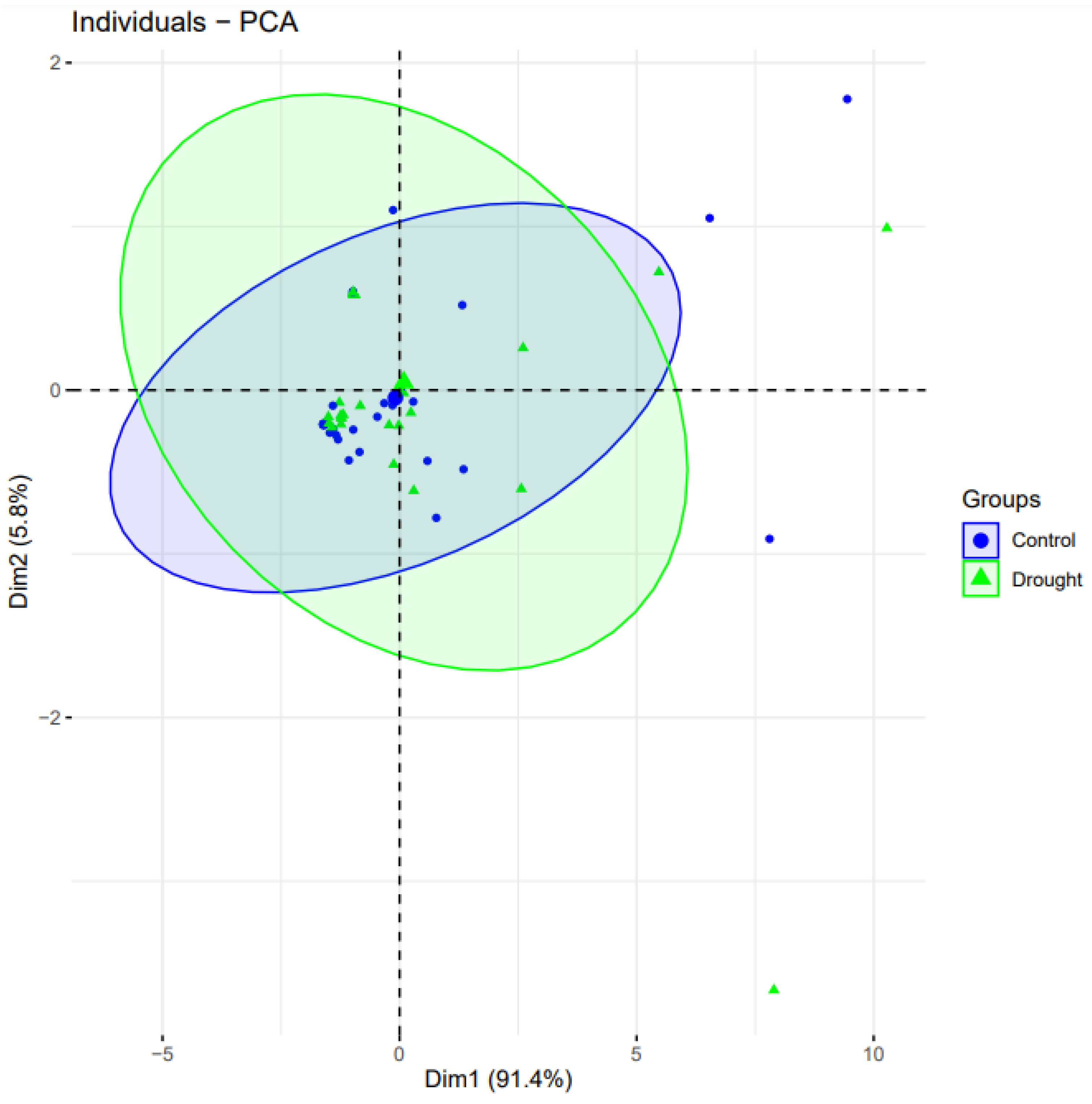
| Sr # | GSE45563 | GSE47090 | GSE70443 | GSE87325 | Coutif | p-Value |
|---|---|---|---|---|---|---|
| 1 | 31369563 | 31369563 | 31369563 | 31369563 | 4 | 4.16 × 10−23 |
| 2 | 25245708 | 25245708 | 25245708 | #N/A | 3 | 9.58 × 10−7 |
| 3 | 31369545 | 31369545 | 31369545 | #N/A | 3 | 9.58 × 10−7 |
| 4 | 25232126 | 25232126 | 25232126 | #N/A | 3 | 9.58 × 10−7 |
| 5 | #N/A | 19959095 | 19959095 | 19959095 | 3 | 9.58 × 10−7 |
| 6 | 14320 | 14320 | 14320 | #N/A | 3 | 9.58 × 10−7 |
| 7 | 25550165 | 25550165 | 25550165 | #N/A | 3 | 9.58 × 10−7 |
| 8 | 25270582 | 25270582 | 25270582 | #N/A | 3 | 9.58 × 10−7 |
| 9 | 20313737 | 20313737 | 20313737 | #N/A | 3 | 9.58 × 10−7 |
| 10 | 20334251 | 20334251 | 20334251 | #N/A | 3 | 9.58 × 10−7 |
| 11 | 25231635 | 25231635 | 25231635 | #N/A | 3 | 9.58 × 10−7 |
| Genes | Accession Numbers | IWGSC Genes | Score | E-Value | Identity (%) | Chromosome | Start | End |
|---|---|---|---|---|---|---|---|---|
| 31369563 | CD454935.1 | TraesCS3D02G120200 | 484 | 0 | 97.9 | 3D | 75946541 | 75948546 |
| 25245708 | CA667105.1 | TraesCS6A02G328700 | 146 | 1.50 × 10−75 | 99.3 | 6A | 562271850 | 562272738 |
| 31369545 | CD454917 | TraesCS2D02G000200 | 424 | 0 | 99.5 | 2D | 39478 | 40878 |
| 25232126 | CA653601 | TraesCS6D02G086600 | 248 | 2.50 × 10−136 | 100 | 6D | 52285210 | 52292603 |
| 19959095 | BJ220896 | TraesCS6D02G260700 | 696 | 0 | 99.9 | 6D | 368305109 | 368306633 |
| 14320 | X52867 | TraesCSU02G154600 | 507 | 0 | 98.3 | Un | 206782100 | 206784099 |
| 25550165 | CA734567 | TraesCS6B02G234100 | 105 | 9.40 × 10−51 | 100 | 6B | 393072749 | 393090605 |
| 25270582 | CA684029 | TraesCS1D02G333000 | 118 | 1.30 × 10−58 | 96.2 | 1D | 423261565 | 423269992 |
| 20313737 | BQ168410 | TraesCS5A02G456300 | 224 | 6.00 × 10−122 | 100 | 5A | 636429039 | 636430690 |
| 20334251 | BQ172428 | TraesCS6A02G240400 | 308 | 5.40 × 10−172 | 99.4 | 6A | 451659872 | 451662196 |
| 25231635 | CA653110 | TraesCS7B02G243600 | 235 | 3.20 × 10−128 | 99.6 | 7B | 452150743 | 452210823 |
| GO Type | Description | Negative Log10 (p Adjusted) | ID | Gene Ratio | Bg. Ratio | p Value | P Adjust | Q Value | Gene ID | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| BP | regulation of autophagy | 2.107492695 | GO:0010506 | ¼ | 3/7682 | 0.001561 | 0.007807 | 0.001644 | 25270582 | 1 |
| response to auxin | 1.683984604 | GO:0009733 | ¼ | 25/7682 | 0.012957 | 0.020702 | 0.004358 | 25245708 | 1 | |
| electron transport chain | 1.683984604 | GO:0022900 | ¼ | 31/7682 | 0.016047 | 0.020702 | 0.004358 | 14320 | 1 | |
| regulation of transcription from RNA polymerase II promoter | 1.683984604 | GO:0006357 | ¼ | 32/7682 | 0.016562 | 0.020702 | 0.004358 | 20334251 | 1 | |
| CC | Mon1-Ccz1 complex | 3.155214539 | GO:0035658 | 1/5 | 1/7148 | 0.000699 | 0.000699 | #N/A | 25270582 | 1 |
| MF | RNA polymerase II transcription factor activity, sequence-specific DNA binding | 2.290980192 | GO:0000981 | ¼ | 2/7814 | 0.001024 | 0.005117 | 0.001616 | 20334251 | 1 |
| D-threo-aldose 1-dehydrogenase activity | 2.290980192 | GO:0047834 | ¼ | 2/7814 | 0.001024 | 0.005117 | 0.001616 | 31369545 | 1 | |
| alditol:NADP+ 1-oxidoreductase activity | 2.290980192 | GO:0004032 | ¼ | 3/7814 | 0.001535 | 0.005117 | 0.001616 | 31369545 | 1 | |
| cytochrome-c oxidase activity | 2.048275717 | GO:0004129 | ¼ | 7/7814 | 0.003579 | 0.008948 | 0.002826 | 14320 | 1 |
| Drought Genes | Hit ID | Organism | Description | Score | E-Value |
|---|---|---|---|---|---|
| TraesCS6D02G260700 | Zmw_sc01257.1. g00130.1 | Zea mays | Zoysia matrella SBP family protein | 110 | 7.00 × 10−6 |
| TraesCS6D02G260700 | AT1G53160.1 | Arabidopsis thaliana | Arabidopsis thaliana squamosa promoter-binding protein-like 4 | 278 | 3.00 × 10−29 |
| TraesCS6A02G240400 | AT2G46680.2 | Arabidopsis thaliana | Arabidopsis thaliana homeobox 7 | 264 | 6.00 × 10−27 |
| TraesCS2D02G000200 | 678321642 | Utricularia gibba | Utricularia gibba bHLH family protein | 138 | 3.00 × 10−8 |
| TraesCS2D02G000200 | AT3G59060.4 | Arabidopsis thaliana | phytochrome interacting factor 3-like 6 | - | 3.00 × 10−18 |
| TraesCS6D02G086600 | kfl00432_0070 | Klebsormidium flaccidum | Klebsormidium flaccidum C3H family protein | 709 | 2.00 × 10−78 |
| TraesCS6D02G086600 | AT1G29560.2 | Arabidopsis thaliana | Arabidopsis thaliana C3H family protein | 101 | 1.00 × 10−4 |
| TraesCS6B02G234100 | Medtr2g092960.1 | Medicago truncatula | Medicago truncatula Trihelix family protein | 104 | 0.001 |
| TraesCS6B02G234100 | AT3G58630.1 | Arabidopsis thaliana | sequence-specific DNA binding transcription factors | - | 2.00 × 10−24 |
| TraesCS6A02G328700 | AT1G70510.1 | Arabidopsis thaliana | Arabidopsis thaliana KNOTTED-like from Arabidopsis thaliana 2 | 64 | 0.04 |
| TraesCS1D02G333000 | No hits found | ||||
| TraesCS5A02G456300 | Zmw_sc03344.1. g00030.1 | Zoysia matrella | Zoysia matrella ERF family protein | 160 | 1.00 × 10−11 |
| TraesCS5A02G456300 | AT5G19790.1 | Arabidopsis thaliana | Arabidopsis thaliana related to AP2 11 | 229 | 4.00 × 10−22 |
| TraesCS3D02G120200 | Tp57577_TGAC_v2_mRNA33215 | Trifolium pratense | Trifolium pratense bHLH family protein | 325 | 5.00 × 10−31 |
| TraesCS3D02G120200 | AT2G22760.1 | Arabidopsis thaliana | bHLH family protein | - | 1.00 × 10−20 |
| TraesCS7B02G243600 | Do013987.1 | Dichanthelium oligosanthes | Dichanthelium oligosanthes ERF family protein | 244 | 1.00 × 10−20 |
| TraesCS7B02G243600 | AT5G19790.1 | Arabidopsis thaliana | related to AP2 11 | - | 2.00 × 10−18 |
| ID | Description | Gene Ratio | Bg Ratio | p Value | P Adjust | Q Value | Gene ID | Count |
|---|---|---|---|---|---|---|---|---|
| PO:0003015 | primary root differentiation zone | 2/9 | 42/24102 | 0.000106 | 0.001957 | 0.000309 | TraesCS5A02G456300/TraesCS7B02G243600 | 2 |
| PO:0005679 | Epidermis | 2/9 | 57/24102 | 0.000196 | 0.001957 | 0.000309 | TraesCS5A02G456300/TraesCS7B02G243600 | 2 |
| PO:0000256 | root hair cell | 2/9 | 208/24102 | 0.002564 | 0.015719 | 0.002482 | TraesCS5A02G456300/TraesCS7B02G243600 | 2 |
| PO:0004707 | fruit dehiscence zone | 1/9 | 11/24102 | 0.004101 | 0.015719 | 0.002482 | TraesCS3D02G120200 | 1 |
| PO:0020128 | leaf margin | 1/9 | 16/24102 | 0.00596 | 0.015719 | 0.002482 | TraesCS3D02G120200 | 1 |
| PO:0009064 | receptacle | 1/9 | 17/24102 | 0.006331 | 0.015719 | 0.002482 | TraesCS6A02G328700 | 1 |
| PO:0000112 | shoot axis epidermis | 1/9 | 18/24102 | 0.006702 | 0.015719 | 0.002482 | TraesCS3D02G120200 | 1 |
| PO:0000033 | fruit valve | 1/9 | 19/24102 | 0.007074 | 0.015719 | 0.002482 | TraesCS3D02G120200 | 1 |
| PO:0004006 | mesophyll cell | 1/9 | 19/24102 | 0.007074 | 0.015719 | 0.002482 | TraesCS3D02G120200 | 1 |
| PO:0006016 | leaf epidermis | 1/9 | 23/24102 | 0.008557 | 0.017114 | 0.002702 | TraesCS3D02G120200 | 1 |
| PO:0009053 | Peduncle | 1/9 | 31/24102 | 0.011518 | 0.019813 | 0.003128 | TraesCS3D02G120200 | 1 |
| PO:0020127 | primary root | 1/9 | 32/24102 | 0.011888 | 0.019813 | 0.003128 | TraesCS3D02G120200 | 1 |
| PO:0006036 | root epidermis | 1/9 | 62/24102 | 0.022919 | 0.035259 | 0.005567 | TraesCS3D02G120200 | 1 |
| PO:0006504 | leaf trichome | 1/9 | 81/24102 | 0.029848 | 0.04264 | 0.006733 | TraesCS3D02G120200 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aqeel, M.; Ajmal, W.; Mujahid, Q.; Murtaza, M.; Almuqbil, M.; Ghazanfar, S.; Uzair, M.; Wadood, A.; Asdaq, S.M.B.; Abid, R.; et al. Genome-Wide Identification of Potential mRNAs in Drought Response in Wheat (Triticum aestivum L.). Genes 2022, 13, 1906. https://doi.org/10.3390/genes13101906
Aqeel M, Ajmal W, Mujahid Q, Murtaza M, Almuqbil M, Ghazanfar S, Uzair M, Wadood A, Asdaq SMB, Abid R, et al. Genome-Wide Identification of Potential mRNAs in Drought Response in Wheat (Triticum aestivum L.). Genes. 2022; 13(10):1906. https://doi.org/10.3390/genes13101906
Chicago/Turabian StyleAqeel, Muhammad, Wajya Ajmal, Quratulain Mujahid, Maryam Murtaza, Mansour Almuqbil, Shakira Ghazanfar, Muhammad Uzair, Ayesha Wadood, Syed Mohammed Basheeruddin Asdaq, Rameesha Abid, and et al. 2022. "Genome-Wide Identification of Potential mRNAs in Drought Response in Wheat (Triticum aestivum L.)" Genes 13, no. 10: 1906. https://doi.org/10.3390/genes13101906
APA StyleAqeel, M., Ajmal, W., Mujahid, Q., Murtaza, M., Almuqbil, M., Ghazanfar, S., Uzair, M., Wadood, A., Asdaq, S. M. B., Abid, R., Ali, G. M., & Khan, M. R. (2022). Genome-Wide Identification of Potential mRNAs in Drought Response in Wheat (Triticum aestivum L.). Genes, 13(10), 1906. https://doi.org/10.3390/genes13101906