Abstract
Azorhizobium caulinodans ORS571 contains an 87.6 kb integrative and conjugative element (ICEAc) that conjugatively transfers symbiosis genes to other rhizobia. Many hypothetical redundant gene fragments (rgfs) are abundant in ICEAc, but their potential function in horizontal gene transfer (HGT) is unknown. Molecular biological methods were employed to delete hypothetical rgfs, expecting to acquire a minimal ICEAc and consider non-functional rgfs as editable regions for inserting genes related to new symbiotic functions. We determined the significance of rgf4 in HGT and identified the physiological function of genes designated rihF1a (AZC_3879), rihF1b (AZC_RS26200), and rihR (AZC_3881). In-frame deletion and complementation assays revealed that rihF1a and rihF1b work as a unit (rihF1) that positively affects HGT frequency. The EMSA assay and lacZ-based reporter system showed that the XRE-family protein RihR is not a regulator of rihF1 but promotes the expression of the integrase (intC) that has been reported to be upregulated by the LysR-family protein, AhaR, through sensing host’s flavonoid. Overall, a conservative module containing rihF1 and rihR was characterized, eliminating the size of ICEAc by 18.5%. We propose the feasibility of constructing a minimal ICEAc element to facilitate the exchange of new genetic components essential for symbiosis or other metabolic functions between soil bacteria.
1. Introduction
Horizontal gene transfer (HGT) is a universal phenomenon that involves the transfer of extra genetic material from one cell to another through various mechanisms. Conjugation, transformation, and transduction are the main prevalent HGT methods [1,2,3], and other mechanisms are constantly being discovered, such as the membrane vesicle (MV) model [4,5]. HGT events exist extensively in prokaryotes and eukaryotes [6,7,8,9], thereby making numerous contributions to the ecosystem, especially adaptive evolution [10,11,12].
Rhizobia belong to various genera of α- and β-proteobacteria that interact with their legume hosts to facilitate nutrient (nitrogen) demanded by plants, and in return, plants provide energy, ultimately resulting in the formation of symbiotic organs named nodules [13,14,15]. The symbiosis genes in rhizobia that are associated with the process of nodule formation and nitrogen-fixing are generally found in extrachromosomal replicons or integrative and conjugative elements (ICEs) [16,17,18,19], which are spread mainly through conjugation, converting soil bacteria into symbionts and expanding the host range of other rhizobia [20,21,22,23]. The HGT of the symbiosis module is a significant player in bacterial evolution, biodiverse ecosystem network, material cycle, and sustainable production, promoting the diversity of rhizobia [24,25,26,27].
As gram-negative bacteria, the α-proteobacteria Azorhizobium caulinodans have a dual capacity to fix nitrogen both under free-living conditions and in symbiosis with the legume plant, Sesbania rostrata, forming both root and stem nodules [28]. Our previous work revealed that A. caulinodans ORS571 has an 87.6-kb integrative and conjugative element (ICEAc) containing nodulation genes, which can excise and form a circular DNA, then conjugatively transfer into various rhizobia species, allowing them to form nodules on S. rostrata. It interred that ICEAc may work as a synthetic biological element carrying special functional genes and endowing recipient cells’ new physiological phenotypes. The model strain Mesorrhizobium loti R7A contains an enormous symbiosis island (502-kb), limiting its modification and transformation to other rhizobia or soil bacteria [29]. The process of receiving and maintaining HGT elements may increase the genetic load cost in recipient cells [30,31]. Moreover, bacteria have evolved some tactics to prevent exogenous DNA fragment into the cells, such as TraS or CRISPR/Cas9 system [32]. A study indicated that only 63 kb (58 genes) on the 1.35 Mb plasmid pSymA of Sinorhizobium fredii NGR234 is required for symbiosis [33], and complementary 7 nodulation genes are sufficient for nodule formation after eliminating pSymA of NGR234 [34]. In contrast, ICEAc carries fewer native genes and provides a cleaner pedestal for gene insertion and modulation. The non-functional genes named redundant gene fragments (rgfs) are abundant in the ICEAc. They may work as potential modification sites to shorten the length of ICEAc, in order to increase the packing ratio. The smaller ICEAc can be regarded as a superior genetic tool, which may maximize the packing capacity and carry more properties into recipient cells. Additionally, several genes in ICEAc are required for ICEAc transfer, such as ahaR, intC, traB, and traG [35]. Modeling these genes may increase the transfer frequency of ICEAc or artificially control ICEAc transfer on special location. For instance, molecular biological methods were developed to modify and assemble specific genes such as mocB and nifA in A. caulinodans, and engineer a plant-controlled nitrogen-fixing bacterium only when in contact with RhiP barley roots [36]. A xenobiotic response element (XRE) family gene containing a DUF433 domain and a set of uncharacterized genes were identified as key HGT genes in Rhizobium etli CFN42 and Rhizobium leguminosarum strain VF39SM by deleting hypothetical genes [37,38,39]. The molecular biological methods offered the possibility of deleting potential rgfs to explore and modify crucial genes related to ICEAc HGT [30,31,32,33,34,40,41] in order to maintain a minimal ICEAc genetic tool that can efficiently pack large DNA fragments and transfer interesting functional genes into recipient cells under desired conditions.
In this study, we knocked out 4 hypothetical rgfs from ICEAc individually or together to shorten the length of ICEAc. We discovered 3 non-functional rgfs that can serve as editable regions fitting modifications. Interestingly, we found that rgf4 made an indispensable contribution to ICEAc transfer. Our preliminary analysis of genes in rgf4 revealed 3 genes that were directly correlated with the HGT process. Moreover, the mutual regulatory relationships have been elucidated and refined the regulation pathway of intC. In brief, we constructed a shorten transfer subset, which is conducive to the success of HGT and the exploration of crucial HGT genes and explicated a set of potential modeling proteins to promote HGT frequency. Our work will highlight synthetic remodeling thought to increase packing capacity and transferring frequency of ICEAc, carrying more interesting gene clusters into soil bacteria.
2. Materials and Methods
2.1. Bacterial Strains Growth Conditions
Bacterial strains and plasmids and their associated characteristics are listed in Table S1. Azorhizobium caulinodans ORS571 and its derivative strains were grown in TY medium at 28 °C [42]. Escherichia coli strains were routinely grown overnight in Luria-Bertani (LB) medium at 37 °C. Antibiotics were added to the medium as needed, with the final concentration as follows: ampicillin (Amp, 100 µg·mL−1), gentamicin (Gm, 20 µg·mL−1), kanamycin (Km, 100 µg·mL−1), tetracycline (Tet, 10 µg·mL−1), chloramphenicol (Chl, 30 µg·mL−1), streptomycin (Str, 20 µg·mL−1), and spectinomycin (Spe, 100 µg·mL−1). The bacteria were cultured for 72 h and optical density (OD600) measurements were taken at different times to compare the growth capability of the strains investigated. Meanwhile, 100 µL of bacterial suspension solution were extracted, gradient diluted, and placed on TY plates at 28 °C for 3 days to count visible colonies (colony-forming units, CFU). In addition, β-d-1-thiogalacto-pyranoside (IPTG) with a final concentration of 100 µg·mL−1 was added into the medium to induce gene expression when the strains containing recombinant plasmid pSRKGm.
2.2. Bioinformatics Analyses
Analysis was carried out of how relevant genes are organized in various genomes using an online website (http://www.microbesonline.org/, accessed on 1 July 2022). The online website (https://www.ncbi.nlm.nih.gov/genome/, accessed on 1 August 2022) provided the genomic information listed below: Azorhizobium caulinodans ORS571 (NC_009937.1), M. japonicum R7A (CP033366.1/CP051772.1), Mesorhizobium sp. BNC1 (NC_008254.1), Bradyrhizobium sp. BTAi1 (NC_009485), Bradyrhizobium japonicum USDA110 (NC_004463), Rhizobium sp. CIAT894 (CP020947.1), Paracoccus denitrificans PD1222 (NC_008687), Nitrobacter winogradskyi Nb-255 (NC_007406) and Xanthobacter autotrophicus Py2 (GCA_000017645.1).
The protein sequences of rgf4 were subjected to a multiple sequence alignment tool (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 1 August 2022) and protein homologues were identified by BLASTP analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 August 2022). Protein sequences were aligned and visualized in Geneious v. 2020.0 (Biomatters, Auckland, New Zealand) using the Clustal Omega algorithm.
Genes promoters were predicted by an online website (http://www.softberry.com/, accessed on 1 July 2022). We used the MEME Suite [43] to predict potential RihR binding motifs in the promoter regions of genes adjacent to rihR. The Tomtom program was employed to find similar motifs in published libraries.
2.3. Molecular Biology Techniques
Table S2 shows primers used to amplify corresponding DNA fragments. Figure S1 describes the construction of recombinant plasmids and the process of homologous recombination. Homologous fragments and Km fragment were cloned into a suicide vector pEX18Gm containing a sacB selectable marker [44,45], and the recombination plasmid was introduced into E. coli SM10 λpir as donor strain [46,47]. The plasmid was transferred from the donor into ORS571 by biparental conjugation. Donor and recipient cells were mixed in a 1:1 ratio and placed on a filter membrane, which was placed on TY solid medium to cultivate for 12 h at 28 °C. The cells mixture was evenly distributed on TY agar containing Km/Gm to screen conjugation colonies, which was the first homologous recombination. Subsequently, the colonies were streaked on TY plates to generate double-crossover events. TY-sucrose (10% sucrose) plates were used to exclude strains that stayed in the first homologous recombination state. The candidate strains were selected on TY supplemented with Km or Km/Gm at 28 °C for three days. The positive colonies that only grew on TY plates containing Km were verified by sequencing. In the same way, other in-frame deletion strains (Table S1) were constructed on the background of Azc1 [35] and Azc2, as follows: Δrgf1 (1.9 kb was deleted in Azc1), Δrgf2 (4.5 kb was deleted in Azc1), Δrgf3 (9.2 kb was deleted in Azc1), Δrgf4 (2.3 kb was deleted in Azc1), Δrgf12 (6.4 kb was deleted in Azc1), Δrgf123 (15.6 kb was deleted in Azc1), ΔrihR, ΔrihF1, ΔrihF2 and ΔrihR*. Because pVIK112 had a Km-resistant gene, the same as Azc1, we reconstructed a new tetracycline-resistant gene insertion strain, Azc2. In addition, we confirmed the deletion strains by amplification of the coding sequence of the genes (Figure S2). For complementation, the complemented genes were amplified by PCR. These fragments were inserted into the downstream of the pSRKGm promoter using the digestion sites of restriction enzymes. The recombinant plasmids were sequenced and transferred into corresponding mutants by electroporation. When β-d-1-thiogalacto-pyranoside (IPTG) was added, the complemented genes were induced through the activation of the pSRKGm promoter.
2.4. Measure HGT Frequency of ICEAc
Donor strains (Wild-type ORS571 [48] and its derivatives) and recipient strains (M. huakuii 93 [35]) were employed to determine the HGT frequency of ICEAc as previously reported [35]. Strains were grown overnight in a liquid medium, and 2 mL bacterial suspension were mixed in a 1:1 ratio, with starting optical density at 600 nm (OD600 = 1.0). Before mixing with the donor, recipient bacteria were graded and diluted on TY plates. The mixture was placed on a TY solid medium to cultivate for 12 h at 28 °C and then overlaid on TY plate containing Spe/Km or Spe/Tet. The colony-forming units (CFU) of transconjugants and recipient strains were counted to calculate HGT frequency. The experiment was repeated at least three times.
2.5. Measuring Transcriptional and Translational Expression of Genes
The construction of transcriptional fusion reporters and β-galactosidase activity assay followed the protocol as described previously [49,50]. The transcriptional regions and promoters of target genes were respectively cloned into expression plasmid pVIK112 and pRA302 [51,52], and the plasmids were integrated into the target genes locus by homologous recombination to indicate the intensity of gene expression in the chromosome. The 200 µL cultures were extracted to detect enzyme activity. The reaction time (Δt) was recorded, and the reading of OD420 was measured using ο-Nitrophenyl-β-d-galactopyranoside (ONPG) as the reaction substrate. The activity was normalized to the OD600 of the cultures and expressed in Miller units, One Miller unit of enzyme activity is defined as the amount of enzyme required to generate 1 mmol ο-Nitrophenyl (ONP) per minute. Three biological replicates were performed, and the experiment was repeated at least three times.
2.6. Bacterial One-Hybrid Assay
The bacterial one-hybrid (B1H) assay was an efficient method to detect the specific interaction between transcriptional regulators and target genes [53,54]. Candidate gene promoters were amplified and cloned into pBT-derived reporter vector pBXcmT. The coding region of rihR was cloned into pTRG to acquire recombination plasmid pTRG-rihR. The host strain for propagation was E. coli XL1-Blue MRF’ Kan [55], and rihR expressed in pTRG interacts with bait DNA in pBXcmT. Positive cotransformants grow well on selective medium containing 10 mM 3-amino-1,2,4-triazole (3-AT), 10 µg·mL−1 streptomycin (Str), 10 µg·mL−1 tetracycline, 30 µg·mL−1 chloramphenicol, and 50 µg·mL−1 kanamycin. All medium plates were incubated at 28 °C for 3 days. Cotransformants containing vectors pTRG and pBXcmT served as negative controls, and the strain cotransformed with pTRG-R3133 and pBXcmT-R2031 was employed as a positive control [53]. The assay was performed three times, and three replicates of different strains were examined in each time.
2.7. Expression and Purification of His-Tagged RihR
The reading region of rihR was cloned into pET28a (Novagen, Madison, WI, USA) to express His-tagged RihR in E. coli BL21 (DE3) [46] following standard procedures [42,56,57]. E. coli BL21 carrying pET28a-rihR was grown in 500 mL LB medium with the appropriate antibiotic and then induced with the addition of 0.5 mM β-d-1-thiogalacto-pyranoside (IPTG) at OD600 ≈ 0.6. Cells were incubated at 16 °C for 12 h and harvested by centrifugation at 4 °C. The cells were suspended in a pH 8.0 lysis buffer (50 mM NaH2PO4 and 300 mM NaCl) and sonicated to obtain cell lysate, which was centrifuged at 4 °C for 30 min to exclude insoluble sedimentation. The soluble proteins were loaded onto the Ni-NTA column (GE Healthcare, Chicago, IL, USA). Firstly, the column was washed with several column volumes of purification buffer (lysis buffer plus 20 mM imidazole). Subsequently, N-terminally His-tagged RihR was eluted with elution buffer (lysis buffer plus 250 mM imidazole) and assessed by SDS-PAGE. The purification was desalted by HiTrap Desalting Column (GE Healthcare, Chicago, IL, USA), and protein concentration was determined by the NanoDrop 2000c (Thermo Fisher Scientific, Waltham, MA, USA).
2.8. Electrophoretic Mobility Gel Shift Assay (EMSA)
The detailed protocol of EMSA has been described in previous research [58,59]. Briefly, approximately 300 bp promoter fragment of the candidate gene was amplified from A. caulinodans ORS571 genome and purified as a DNA probe using the PCR purification kit (Sangon Biotech, Shanghai, China). The DNA probes (50 ng) were mixed in a binding buffer containing different concentrations of RihR protein in a final volume of 20 µL. The binding buffer contained 50 mM Tris-HCl (pH 8.3), 0.25 M KCl, 2.5 mM dithiothreitol (DTT), 5 mM MgCl2, 0.25 mg·mL−1 bovine serum albumin, 0.05 mg·mL−1 poly(dI-dC), 2.5 mM EDTA, 1% glycerol. The reaction mixtures were incubated for 20 min at room temperature and then electrophoresed on a 6% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA buffer at 150 V for 70 min. Gels were stained and photographed using GelRed (Sangon Biotech, Shanghai, China) and the molecular imager Gel Doc XR system (Bio Rad, Hercules, CA, USA).
2.9. Quantitative Real-Time PCR Analysis
qRT-PCR was used to measure the transcriptional level of genes rihF1, rihR, and ahaR. Total RNA was extracted using TRIzol method [60]. A cDNA Synthesis Kit (Vazyme Biotech, Nanjing, China) was used to convert RNA into cDNA using specific amplified primers (Table S2), following the manufacturer’s instructions. The qPCR experimental program was described below: 30 s at 95 °C, followed by 40 cycles of 10 s at 95 °C and 30 s at 60 °C, and finally, 72 °C (30 s), according to the manufacturer’s protocol for the SYBR green detection Kit (Vazyme Biotech, Nanjing, China). Three independent experiments were performed (each in triplicate), the transcript levels were normalized by endogenous control 16S rRNA [61], and the relative expression levels of target genes were calculated by the Comparative CT method [62].
2.10. Statistical Analysis
GraphPad Prism 8.0 software (GraphPad, La Jolla, CA, USA) was applied to analyze and draw the experimental data, which at least referred to three biological samples and three independent experiments. All data were presented as mean values with standard deviations (±SD) indicated by error bars. Student’s t-test was used to compare two groups of data and calculate the p values, and p values < 0.05 were considered significant. One-way ANOVA with a post hoc Tukey-Kramer test of honestly significant difference was also applied in data analysis.
3. Results
3.1. Identification of rgfs Involved in ICEAc Transfer
Following our previous data and bioinformatics analysis [35], 4 hypothetical rgf regions were marked on the A. caulinodans symbiosis island (Figure 1A). In order to confirm whether these rgfs were involved in ICEAc transfer, we performed single or multi rgfs knockout. We found Δrgf1, Δrgf2, Δrgf3, Δrgf12, and Δrgf123 were consistent with that of WT strain on HGT frequency (Figure 1B). Surprisingly, a 100-fold decrease in the HGT frequency of Δrgf4 was observed. Additionally, the growth capacities of all the mutants were comparable to that of the WT strain (Figure S3A). Collectively, these data suggest that genes within or adjacent to rgf4 were vital for ICEAc transfer. In conclusion, we successfully shortened the ICEAc length by approximately 18.5% without affecting the transfer frequency by deleting rgf123, and these regions can be used as sites to insert editable interesting genes to remodel ICEAc.
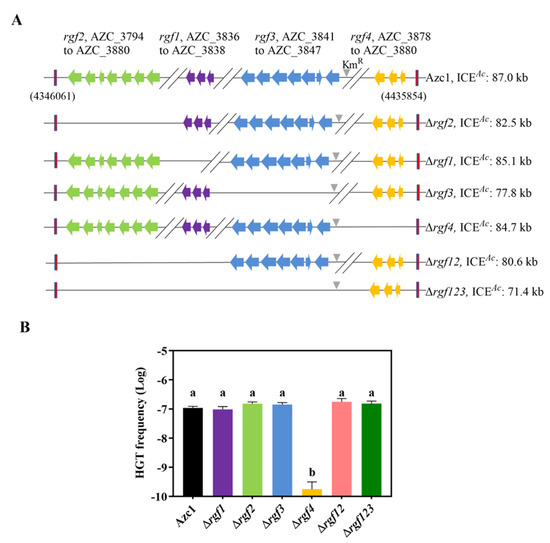
Figure 1.
Differential consequences of rgf mutants on ICEAc HGT frequency. (A) Distribution of 4 hypothetical rgf regions in ICEAc, with a deletion strategy starting from the second row down. Red rectangles represent the entire ICEAc region. The grey triangle represents the kanamycin-resistant gene, and other colors indicate different hypothetical rgfs. (B) Identification of rgf4 is required for ICEAc HGT frequency. Azc1 (wild-type) or mutants were mixed with M. huakuii 93 at 28 °C, respectively. The colony-forming units (CFUs) of transconjugants and the recipient were determined to calculate the HGT frequency. Data are mean ± SD of three independent experiments. Means not connected by the same letter are significantly different (p < 0.05).
3.2. Genomic Context of rgf4 Genes in Different Strains
In the rgf4 fragment (Figure 2A), a transposase gene, AZC_3878, was not associated with ICEAc transfer [35]. We hypothesized that other genes in rgf4 serve as relevant ICEAc HGT-related genes, respectively named them rihF1a (AZC_3879), rihF1b (AZC_RS26200), and rihF2 (AZC_3880). Bioinformatics analyses suggested that AZC_3879 and AZC_RS26200 shared the same promoter, which may be expressed as a unit named rihF1 (AZC_3879 and AZC_RS26200). Interestingly, the homologues of rihF1a, rihF1b, rihF2, and AZC_3881 are prevalent in different strains (Figure 2B,C), implying that these genes may serve as a conserved module in controlling the frequency of HGT. Moreover, XRE family proteins usually regulate their downstream genes to control various metabolic pathways [63,64,65]. Thus, we assumed that AZC_3881 might encode a relevant ICEAc HGT regulation protein named rihR. The RihF1a, RihF1b, RihF2, and RihR, respectively, share 35.35%, 33.80%, 46.05%, and 38.24% sequence homologies with the genes are required for ICEMlSymR7A transfer [66,67], which suggested the possibility that the module has significance for HGT. Furthermore, we statistically found that the module is distributed diversely in different ICEs, such as the symbiosis island ICEMlSymR7A (inside ICE) and the B. japonicum USDA110 symbiosis island (outside ICE) [68]. The uncertain distribution may be the result of gene rearrangement. On the other hand, HGT-related genes (marked with yellow) are conservative located in the upstream or downstream of the module and belong to different strains (Figure 2B), such as traG and traF. In previous work, we proved that AZC_3882 (intC) is critical for ICEAc HGT frequency [35]. We, therefore, hypothesized that the module could interact with adjacent intC to maintain ICEAc HGT.

Figure 2.
Composition and distribution of rgf4 genes in different strains. (A) The genetic structure of rgf4 in A. caulinodans. (B) The organization of rgf4 in different strains. Inside of the arrows showed gene function. HP, hypothetical protein. Protein sequence similarity in different strains compared with A. caulinodans is marked above the arrows. (C) Multiple sequence alignments of homologous rgf4 from various strains. Higher similarity is indicated by a darker gray backdrop for the amino acid letters.
3.3. rihF1 and rihR Contribute to the Conjugation Frequency of ICEAc
We individually deleted genes in the module in order to determine which gene contributes to ICEAc transfer. The growth curve experiment showed that mutant strains had no difference with WT stain Azc1 (Figure S3B). ΔrihF1 and ΔrihR stains showed approximately 1000- and 100-fold decreases in the HGT frequency, respectively (Figure 3A). In addition, these decreases were restored to WT levels following complementation with rihF1 and rihR (Figure 3B,C). These data supported that rihF1 and rihR are crucial genes relevant to ICEAc HGT frequency. However, the identical phenotype was not observed in ΔrihF2 (Figure 3A), indicating that rihF2 is a non-functional gene in the process of ICEAc HGT. We further investigated the individual contribution of rihF1a and rihF1b to ICEAc HGT. We introduced rihF1a and rihF1b into ΔrihF1 separately, and the results showed that the HGT frequency of each complementary stain remained the same ratio as that of ΔrihF1 strain, lower than that of the Azc1Vec strain (Figure 3D), verifying the hypothesis that rihF1a and rihF1b work as a unit (rihF1) to maintain ICEAc transfer. These results suggested that rihF1 and rihR are essentially required for ICEAc HGT.
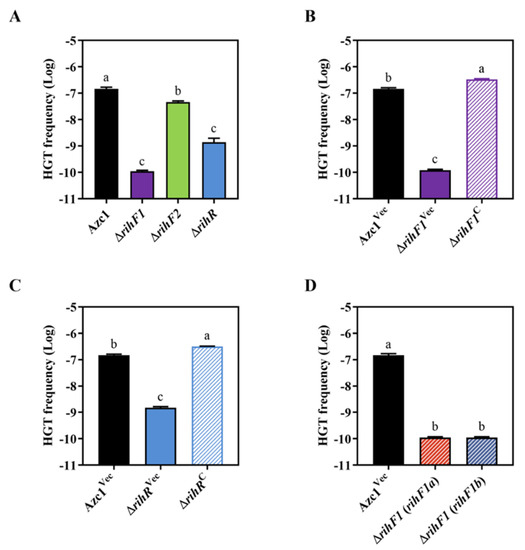
Figure 3.
Identification transfer capacities of ICEAc in different rgf4 gene deletion strains. (A) The transfer frequency of ICEAc in strains ΔrihF1, ΔrihF2, and ΔrihR. (B) The transfer frequency of ICEAc in ΔrihF1Vec and ΔrihF1C. (C) The transfer frequency of ICEAc in ΔrihRVec and ΔrihRC. (D) The transfer frequency of ICEAc in ΔrihF1 (rihF1a) and ΔrihF1 (rihF1b). The strains were mixed with M. huakuii 93, and the CFU of the transconjugants was determined. Data are mean ± SD of three independent experiments. Means not connected by the same letter are significantly different (p < 0.05). The same letter indicates no significant difference (p > 0.05).
3.4. ICEAc HGT Is Influenced by rihF1 and rihR as Two Indipendent Pathways
To further investigate how this module works, we first studied whether RihR regulates rihF1 or intC to affect conjugation frequency. We used the MEME motif discovery platform to identify shared sequence motifs in the promoter regions of rihF1a, rihF1b, rihF2, rihR, and intC, expecting to obtain a possible target sequence for RihR. The promoter sequences were submitted to MEME (Figure 4A) and showed a motif with high similarity to the HTH-type regulator RutR [69,70]. Moreover, the predicted motif was present in the intC and rihF1b promoter sequences (Figure 4B), suggesting that these two genes may be regulated by RihR. To determine the regulation relationship, the recombinant plasmids pRA302-intC::lacZ and pVIK112-rihF1::lacZ were introduced into ΔrihR* and Azc2 strains separately (Figure S3C). Subsequently, the expression of rihF1 and intC was measured through a lacZ-based reporter system, respectively (Figure 4C). As indicated, Azc2 showed significantly higher intC expression activity than that of the ΔrihR* strain(~2-fold), suggesting that RihR positively regulates intC but has no regulation effect on rihF1. Consequently, rihR and rihF1 maintain the conjugation frequency through different pathways, and the rihR performs its function by influencing intC expression.
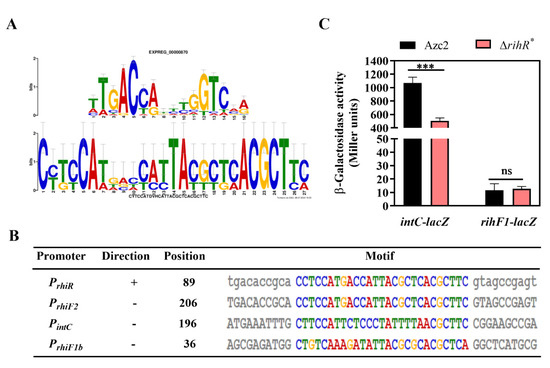
Figure 4.
RihR positively regulates intC expression in vivo. (A) The promoter motif was predicted by MEME in rgf4 genes and intC. Top: The motif results from a comparison with a known motif database by using the Tomtom program. Bottom: The motif is predicted based on the intC, rihR, rihF2, rihF1a, and rihF1b promoters. (B) The specific positions of the predicted motif in different promoters. +: The motif site was found in the sequence as it was supplied. −: The motif site is found in the reverse complement of the supplied sequence. (C) β-galactosidase activities of the transcriptional fusion rihF1-lacZ and the translational fusion intC-lacZ were measured in Azc2 and ΔrihR*. Data are mean ± SD of three independent experiments. ***: Student’s t-test p < 0.001, ns: no significant difference.
3.5. RihR Directly Binds to the Promoter Region of IntC
The EMSA and B1H assay were used to investigate whether RihR protein directly regulates intC. The primers used for promoter fragment amplification are listed in Table S2. RihR was expressed in BL21 (DE3) as a 6× His-tagged fusion protein (~14 kDa) and purified using a Ni-NTA resin column (Figure 5A). A protein-DNA complex was observed in different lanes, which generated different heights of shift with increasing concentrations of the purified RihR protein (Figure 5B). As controls, RihR did not bind to a 160 bp non-specific promoter (lanes 6 and 7). This result indicated that RihR specifically binds to the intC upstream sequence directly in vitro.
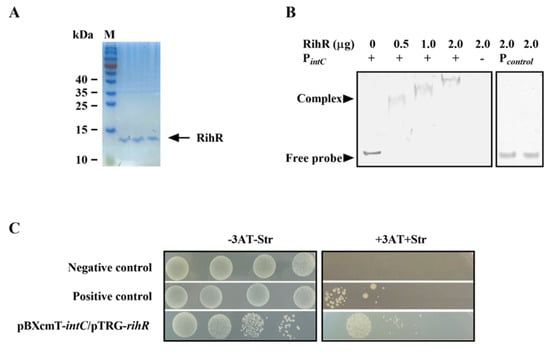
Figure 5.
RihR binds on the promoter region of intC with high affinity. (A) The purified RihR protein was analyzed through non-reducing SDS-PAGE. The soluble proteins in cells were washed down by the elution buffer and added to the three lanes. The arrow indicates the location of RihR protein. M: marker. (B) EMSA assay, RihR protein binds on the intC promoter. The DNA fragment containing the intC promoter was incubated with increasing concentrations of RihR protein at room temperature. The concentration of DNA fragments in the reaction system was 50 ng. The mixture was separated on polyacrylamide non-denaturing gel to observe the DNA-protein complex. (C) The B1H assay of the interaction between RihR and intC promoter. Co-transformant strains containing the plasmids pTRG-R3133, pBXcmT-R2031 and pTRG, pBXcmT served as positive control and negative control, respectively.
We employed the B1H assay to further verify the EMSA conclusion [53]. The cotransformant strain, containing pTRG-rihR and pBXcmT-intC, could be survived on the screening medium containing 3-amino-1,2,4-triazole (3AT) and streptomycin (Figure 5C), which suggests that regulator protein, RihR, interacts with the intC promoter on pBXcmT and initiates the expression of reporter genes. These results further support that RihR directly binds on the intC promoter to induce intC expression, which positively affects ICEAc transfer.
3.6. Regulation of rihR Is Independent of AhaR
Our previous results demonstrated that the transcription factor, AhaR, positively regulates intC gene expression by binding on the promoter region, and the regulation was dependent on the plant flavonoid naringenin (NAR) [35]. We then investigated whether there exists a relationship between ahaR and rihR when they regulate intC expression. The rihR expression in Azc1 and ΔahaR was measured, and the result showed that there had been no difference on rihR expression between WT and ΔahaR strain at different conditions, with or without the presence of NAR (Figure 6A). Consequently, AhaR had no regulatory impact on rihR. The qRT-PCR analysis also indicated that the expression of rhiF1 was not induced by AhaR (Figure S4A), revealing that rihF1 maintained ICEAc transfer independently of ahaR and rihR. These data showed that AhaR and rihR regulated intC separately, and AhaR did not regulate rihF1 to influence ICEAc HGT. In turn, we estimated whether RihR could regulate ahaR to influence ICEAc transfer. The result of ahaR-lacZ activity confirmed that ahaR was not regulated by RihR (Figure 6B); the qRT-PCR analysis also clarified the result (Figure S4B). Collectively, the aforementioned data supported the idea that rihR and ahaR respectively regulated intC expression to maintain and enhance ICEAc transfer. In order to maintain ICEAc transfer, ORS571 employs a complex regulation pathway and promotes ICEAc transferring into soil bacteria, broadening the host range of rhizobia.
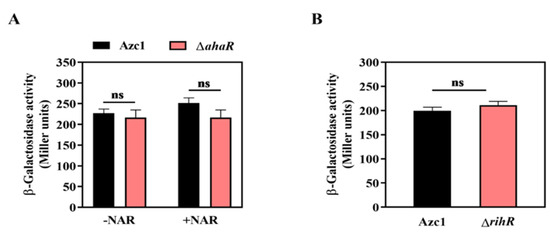
Figure 6.
The regulatory relationship between rihR and ahaR of A. caulinodans. (A) The β-galactosidase activities of the translational fusion rihR-lacZ was determined in Azc1 and ΔahaR. When indicated, 20 μM NAR was added to the medium. (B) The β-galactosidase activities of the translational fusion ahaR-lacZ were measured in Azc1 and ΔrihR. Data are mean ± SD of three independent experiments. ns: no significant difference.
4. Discussion
HGT plays an important role in bacteria evolution [71], sharing beneficial gene cluster among bacteria, such as symbiosis genes [22,23]. However, the transfer of macromolecular symbiotic islands may be an additional burden for the host cell [11], which makes the deletion of rgfs necessary.
In a successful instance of synthetic biology, the nif operons from Klebsiella pneumoniae are integrated into E. coli after deleting redundant genes, giving it equal nitrogen-fixing activity [72]. ICEAc also contains a large number of redundant genes. Gene knockout offers an established approach for the study of unknown function rgfs [73]. We deleted the hypothetical rgfs and identified 3 genes located on rgf4 required for ICEAc transfer through the gene deleting method. Excision of the rgf123 fragment reduced the ICEAc size by 18.5%, discovering a 15.6-kb gene editable region (Figure 1A,B). Meanwhile, the construction of minimal ICEAc can excavate more crucial genes relevant to HGT. In a classic case, the nitrogenase activity of A. caulinodans shows no ammonium repression through the modification of nifA gene [74]. Thus, these crucial HGT genes can be further modified to enhance the conjugation frequency of ICEAc. The bacteria, such as Ralstonia solanacearum, transferred with nodulation and nitrogen-fixation genes can provide both themselves and hosts with competitive advantages [75,76,77]. These findings show that the transfer of functional genes is vital for bacteria in nature. For the subsequent study, modifiable regions in ICEAc could be substituted by nitrogen fixation and other functional genes, conferring more advantage phenotypes to recipient cells. The regions can also be replaced by reporter genes such as green fluorescence protein, which can assist in the observation of ICEAc transfer into diverse soil bacteria in different environments [78]. We expect to develop a remodeling ICEAc genetic tool that can pack more functional gene clusters into soil bacteria.
The protein sequence alignment suggests that the module located on rgf4 and rihR may be widespread in bacteria (Figure 2A–C), participating in HGT regulation. We have shown that rihR and rihF1 in A. caulinodans are engaged in the HGT process (Figure 3), but the module in other strains requires further experimental investigation. Only one of the strains, M. loti R7A, with a homologous module, has been reported to promote ICEMlSymR7A HGT [66,67]. The +1 programmed ribosomal frameshift (PRF) fuses two TraR-activated genes, msi172 and msi171, producing an activator FseA to promote HGT. The HGT process is mediated by the regulation of quorum-sensing (QS) gene traR. Conversely, the homologous groups of msi172 and msi171 in A. caulinodans, rihF1b, and rihF1a contain individual start and end codons themselves. Thus, the PRF phenomenon and traR are absent in A. caulinodans [79,80,81]. These data indicate that rihF1 contains a different regulation mechanism from M. loti strain R7A to maintain ICEAc transfer. rihF1 and rihR may work as potential remodeling sites to promote ICEAc transfer frequency.
In this study, we improved a new regulation feature of intC. In brief, intC can be expressed at a basal level regulated by a new regulon RihR; it also is positively regulated by AhaR sensing host signal, NAR. A previous study has proven that the XRE-family genes located among or nearby HGT-related genes are required to conjugative HGT [37,38]. In A. caulinodans, the XRE-family RihR plays a significant role in maintaining ICEAc transfer by promoting intC expression (Figure 4). The response to plant-exudation of soil bacteria is a key parameter through which bacteria control HGT in rhizosphere [82,83]. AhaR responds to the plant inducer NAR and promotes intC expression to enhance HGT [35], but no regulatory relationship exists between ahaR and rihR (Figure 6). Thus, we enrich our previous study results by finding that there exists a complex regulation pathway of intC expression. In Xanthomonas oryzae MAFF311018, GamR employed two independent regulators, HrpG and HrpX, to regulate hrp expression [84]. We found that A. caulinodans has developed multiple pathways to regulate ICEAc transfer through a long period of evolution. In the absence of NAR, rihR and rihF1 serve as two key components to maintain the general level of HGT. In the presence of NAR, the HGT level is enhanced by ahaR (Figure 7). This study offers strong evidence that A. caulinodans has evolved multiple strategies to maintain or enhance HGT under different environmental conditions, and the complex mechanism remains to be further investigated. Our work contributes to the preliminary elucidation of the vital HGT regulation pathways in A. caulinodans, which may be applied in ICEAc remodeling. Meanwhile, our works highlight the thought that ICEAc may work as a new bacterial remodeling element through increasing package ratio and transferring frequency by deleting rgfs and embellishing special genes.
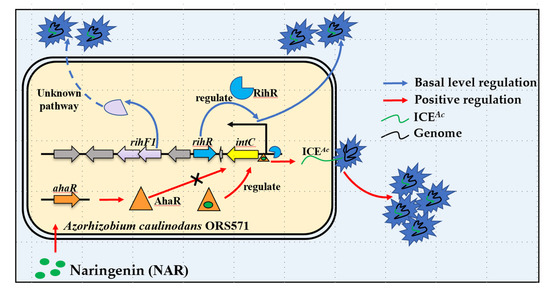
Figure 7.
Model of ICEAc transfer. Efficient transfer pathway: The host plant (S. rostrata) produces a flavonoid signal (NAR) that is recognized by AhaR. The AhaR protein is activated, then binds on the intC promoter to promote HGT frequency (~10−4). General transfer pathway: The RihR protein binds on the intC promoter and maintains the general level of HGT frequency (~10−7), and RihF1 maintains the same HGT frequency through an unknown process.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13101895/s1, Figure S1: Schematic for the construction of gene insertion and knockout; Figure S2: Confirmation of mutant strains; Figure S3: Bacterial growth curves of A. caulinodans and its derivative strains; Figure S4: qRT-PCR analysis of the transcriptional levels of the rihF1, rihR, and ahaR; Table S1: Strains and plasmids used in this study; Table S2: PCR primers used in this study. References [35,45,46,47,48,51,52,53,55,57] are cited in the supplementary materials.
Author Contributions
Conceptualization, Z.Z., C.C., Z.H. and M.L.; methodology, M.L., C.C. and Z.H.; software, M.L. and Q.C.; validation, M.L., C.W., Q.C., Y.L. (Yiyang Li) and S.W.; formal analysis, X.C., M.L., C.W., Q.C. and B.Q.; investigation, M.L., C.W., Q.C., Y.L. (Yiyang Li), S.W., Y.L. (Yuxin Li), D.M. and H.L.; resources, Y.C. and D.Y.; data curation, X.C., M.L., C.W. and Q.C.; writing—original draft preparation, M.L.; writing—review and editing, Z.Z., C.C. and Z.H.; visualization, M.L., C.W. and Q.C.; supervision, Z.Z.; project administration, C.C. and Z.H.; funding acquisition, Z.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (2019YFA0904700), National Natural Science Foundation of China (32270254, 31970266, 31770096) and “Yazhou Bay” Elite Talent Project of Hainan Province.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Hui Wang helpful discussion.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Heinemann, J.A.; Sprague, G.F., Jr. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 1989, 340, 205–209. [Google Scholar] [CrossRef]
- Chen, I.; Dubnau, D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004, 2, 241–249. [Google Scholar] [CrossRef]
- Goh, S. Phage Transduction. Methods Mol. Biol. 2016, 1476, 177–185. [Google Scholar]
- Abe, K.; Nomura, N.; Suzuki, S. Biofilms: Hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 2020, 96, fiaa031. [Google Scholar] [CrossRef]
- Dubey, G.; Ben-Yehuda, S. Intercellular nanotubes mediate bacterial communication. Cell 2011, 144, 590–600. [Google Scholar] [CrossRef]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef]
- Dagan, T.; Artzy-Randrup, Y.; Martin, W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 10039–10044. [Google Scholar] [CrossRef]
- Bansal, A.K.; Meyer, T.E. Evolutionary analysis by whole-genome comparisons. J. Bacteriol. 2002, 184, 2260–2272. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Ren, X.; Mason, A.S.; Liu, H.; Xiao, M.; Li, J.; Fu, D. Horizontal gene transfer in plants. Funct. Integr. Genomics 2014, 14, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Palmer, J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008, 9, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Soucy, S.M.; Huang, J.; Gogarten, J. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Arnold, B.J.; Huang, I.T.; Hanage, W. Horizontal gene transfer and adaptive evolution in bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef]
- Moulin, L.; Munive, A.; Dreyfus, B.; Boivin-Masson, C. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 2001, 411, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Martinezromero, E. Recent Developments in Rhizobium Taxonomy. Plant Soil 1994, 161, 11–20. [Google Scholar] [CrossRef]
- Martínez, E.; Palacios, R.; Sánchez, F. Nitrogen-fixing nodules induced by Agrobacterium tumefaciens harboring Rhizobium phaseoli plasmids. J. Bacteriol. 1987, 169, 2828–2834. [Google Scholar] [CrossRef] [PubMed]
- Geddes, B.A.; Kearsley, J.; Morton, R.; Finan, T.M. The Genomes of Rhizobia, in Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2020; pp. 213–249. [Google Scholar]
- Kelly, S.; Sullivan, J.; Ronson, C.; Tian, R.; Bräu, L.; Munk, C.; Goodwin, L.; Han, C.; Woyke, T.; Reddy, T.; et al. Genome sequence of the Lotu ssp. microsymbiont Mesorhizobium loti strain R7A. Stand. Genomic Sci. 2014, 9, 6. [Google Scholar] [CrossRef]
- Johnson, C.M.; Grossman, A.D. Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu. Rev. Genet. 2015, 49, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Remigi, P.; Zhu, J.; Young, J.P.W.; Masson-Boivin, C. Symbiosis within Symbiosis: Evolving Nitrogen-Fixing Legume Symbionts. Trends Microbiol. 2016, 24, 63–75. [Google Scholar] [CrossRef]
- Sullivan, J.T.; Ronson, C.W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 1998, 95, 5145–5149. [Google Scholar] [CrossRef]
- Sullivan, J.T.; Patrick, H.N.; Lowther, W.L.; Scott, D.B.; Ronson, C.W. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. USA 1995, 92, 8985–8989. [Google Scholar] [CrossRef]
- Marchetti, M.; Capela, D.; Glew, M.; Cruveiller, S.; Chane-Woon-Ming, B.; Gris, C.; Masson-Boivin, C. Experimental evolution of a plant pathogen into a legume symbiont. PLoS Biol. 2010, 8, e1000280. [Google Scholar] [CrossRef]
- Graham, P.H.; Vance, C. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef]
- Smil, V. Nitrogen in crop production: An account of global flows. Glob. Biogeochem. Cycles 1999, 13, 647–662. [Google Scholar] [CrossRef]
- Jackson, L.E.; Burger, M.; Cavagnaro, T.R. Roots, nitrogen transformations, and ecosystem services. Annu. Rev. Plant Biol. 2008, 59, 341–363. [Google Scholar] [CrossRef]
- Dreyfus, B.L.; Elmerich, C.; Dommergues, Y.R. Free-living Rhizobium strain able to grow on n(2) as the sole nitrogen source. Appl. Environ. Microbiol. 1983, 45, 711–713. [Google Scholar] [CrossRef][Green Version]
- Sullivan, J.T.; Trzebiatowski, J.R.; Cruickshank, R.W.; Gouzy, J.; Brown, S.D.; Elliot, R.M.; Ronson, C.W. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 2002, 184, 3086–3095. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.N.T.; Woods, L.C.; Gorrell, R.; Ramanan, S.; Kwok, T.; McDonald, M.J. Recombination resolves the cost of horizontal gene transfer in experimental populations of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 2022, 119, e2119010119. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.; Serbus, L.R. Gene Transfer Agents in Symbiotic Microbes. Results Probl. Cell Differ. 2020, 69, 25–76. [Google Scholar] [PubMed]
- Audette, G.F.; Manchak, J.; Beatty, P.; Klimke, W.A.; Frost, L.S. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 2007, 153 Pt 2, 442–451. [Google Scholar] [CrossRef]
- Geddes, B.A.; Kearsley, J.V.; Huang, J.; Zamani, M.; Muhammed, Z.; Sather, L.; Finan, T.M. Minimal gene set from Sinorhizobium (Ensifer) meliloti pSymA required for efficient symbiosis with Medicago. Proc. Natl. Acad. Sci. USA 2021, 118, e2018015118. [Google Scholar] [CrossRef] [PubMed]
- Unay, J.; Perret, X. A Minimal Genetic Passkey to Unlock Many Legume Doors to Root Nodulation by Rhizobia. Genes 2020, 11, 521. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Wang, H.; Wu, P.; Li, T.; Tang, Y.; Naseer, N.; Zhu, J. Plant nodulation inducers enhance horizontal gene transfer of Azorhizobium caulinodans symbiosis island. Proc. Natl. Acad. Sci. USA 2016, 113, 13875–13880. [Google Scholar] [CrossRef] [PubMed]
- Haskett, T.L.; Paramasivan, P.; Mendes, M.D.; Green, P.; Geddes, B.A.; Knights, H.E.; Jorrin, B.; Ryu, M.; Brett, P.; Voigt, C.A.; et al. Engineered plant control of associative nitrogen fixation. Proc. Natl. Acad. Sci. USA 2022, 119, e2117465119. [Google Scholar] [CrossRef]
- Wathugala, N.D.; Hemananda, K.M.; Yip, C.B.; Hynes, M.F. Defining the requirements for the conjugative transfer of Rhizobium leguminosarum plasmid pRleVF39b. Microbiology 2020, 166, 318–331. [Google Scholar] [CrossRef]
- López-Fuentes, E.; Torres-Tejerizo, G.; Cervantes, L.; Brom, S. Genes encoding conserved hypothetical proteins localized in the conjugative transfer region of plasmid pRet42a from Rhizobium etli CFN42 participate in modulating transfer and affect conjugation from different donors. Front. Microbiol. 2014, 5, 793. [Google Scholar]
- Ding, H.; Yip, C.B.; Hynes, M.F. Genetic characterization of a novel rhizobial plasmid conjugation system in Rhizobium leguminosarum bv. viciae strain VF39SM. J. Bacteriol. 2013, 195, 328–339. [Google Scholar] [CrossRef][Green Version]
- Wang, X.; Lv, S.; Liu, T.; Wei, J.; Qu, S.; Lu, Y.; Liu, H. CRISPR/Cas9 genome editing shows the important role of AZC_2928 gene in nitrogen-fixing bacteria of plants. Funct. Integr. Genomics 2020, 20, 657–668. [Google Scholar] [CrossRef]
- Pistorio, M.; Torres Tejerizo, G.A.; Del Papa, M.F.; de los Angeles Giusti, M.; Lozano, M.; Lagares, A. rptA, a novel gene from Ensifer (Sinorhizobium) meliloti involved in conjugal transfer. FEMS Microbiol. Lett. 2013, 345, 22–30. [Google Scholar] [CrossRef]
- Si, Y.; Guo, D.; Deng, S.; Lu, X.; Zhu, J.; Rao, B.; Zhu, J. Ohr and OhrR Are Critical for Organic Peroxide Resistance and Symbiosis in Azorhizobium caulinodans ORS571. Genes 2020, 11, 335. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids. Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Metcalf, W.W.; Jiang, W.; Daniels, L.L.; Kim, S.K.; Haldimann, A.; Wanner, B.L. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 1996, 35, 1–13. [Google Scholar] [CrossRef]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef]
- Chart, H.; Smith, H.; La Ragione, R.; Woodward, M. An investigation into the pathogenic properties of Escherichia coli strains BLR, BL21, DH5alpha and EQ1. J. Appl. Microbiol. 2000, 89, 1048–1058. [Google Scholar] [CrossRef]
- Ferrières, L.; Hémery, G.; Nham, T.; Gueérout, A.-M.; Mazel, D.; Beloin, C.; Ghigo, J.-M. Silent Mischief: Bacteriophage Mu Insertions Contaminate Products of Escherichia coli Random Mutagenesis Performed Using Suicidal Transposon Delivery Plasmids Mobilized by Broad-Host-Range RP4 Conjugative Machinery. J. Bacteriol. 2010, 192, 6418–6427. [Google Scholar] [CrossRef] [PubMed]
- Goethals, K.; Van Montagu, M.; Holsters, M. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc. Natl. Acad. Sci. USA 1992, 89, 1646–1650. [Google Scholar] [CrossRef]
- Zhao, Y.; Nickels, L.M.; Wang, H.; Ling, J.; Zhong, Z.; Zhu, J. OxyR-regulated catalase activity is critical for oxidative stress resistance, nodulation and nitrogen fixation in Azorhizobium caulinodans. FEMS Microbiol. Lett. 2016, 363, fnw130. [Google Scholar] [CrossRef]
- Miller, J. Experiments in Molecular Genetics 1972 Cold Spring Harbor; Cold Spring Harbor Laboratory: Laurel Hollow, NY, USA, 1972; pp. 431–433. [Google Scholar]
- Kalogeraki, V.S.; Winans, S.C. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 1997, 188, 69–75. [Google Scholar] [CrossRef]
- Jiang, G.; Yang, J.; Li, X.; Cao, Y.; Liu, X.; Ling, J.; Wang, H.; Zhong, Z.; Zhu, J. Alkyl hydroperoxide reductase is important for oxidative stress resistance and symbiosis in Azorhizobium caulinodans. FEMS Microbiol. Lett. 2019, 366, fnz014. [Google Scholar] [CrossRef]
- Guo, M.; Feng, H.; Zhang, J.; Wang, W.; Wang, Y.; Li, Y.; Gao, C.; Chen, H.; Feng, Y.; He, Z.-G. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009, 19, 1301–1308. [Google Scholar] [CrossRef]
- Wang, P.; Chen, H.; Qian, G.; Liu, F. LetR is a TetR family transcription factor from Lysobacter controlling antifungal antibiotic biosynthesis. Appl. Microbiol. Biotechnol. 2017, 101, 3273–3282. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Shen, D.; Yang, N.; Chou, S.H.; Gomelsky, M.; Qian, G. Coordinated control of the type IV pili and c-di-GMP-dependent antifungal antibiotic production in Lysobacter by the response regulator PilR. Mol. Plant Pathol. 2021, 22, 602–617. [Google Scholar] [CrossRef]
- Allemann, M.N.; Allen, E.E. Genetic Regulation of the Bacterial Omega-3 Polyunsaturated Fatty Acid Biosynthesis Pathway. J. Bacteriol. 2020, 202, e00050-20. [Google Scholar] [CrossRef] [PubMed]
- Guan, G.; Dai, P.-H.; Osborne, T.F.; Kim, J.B.; Shechter, I. Multiple Sequence Elements are Involved in the Transcriptional Regulation of the Human Squalene Synthase Gene. J. Biol. Chem. 1997, 272, 10295–10302. [Google Scholar] [CrossRef]
- Liu, M.; Hao, G.; Li, Z.; Zhou, Y.; Garcia-Sillas, R.; Li, J.; Wang, H.; Kan, B.; Zhu, J. CitAB Two-Component System-Regulated Citrate Utilization Contributes to Vibrio cholerae Competitiveness with the Gut Microbiota. Infect. Immun. 2019, 87, e00746-18. [Google Scholar] [CrossRef]
- Shao, X.; Zhang, X.; Zhang, Y.; Zhu, M.; Yang, P.; Yuan, J.; Zhou, Y.X.T.; Wang, W.; Chen, S.; Liang, H.; et al. RpoN-Dependent Direct Regulation of Quorum Sensing and the Type VI Secretion System in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2018, 200, e00205-18. [Google Scholar] [CrossRef]
- Jahn, C.E.; Charkowski, A.O.; Willis, D.K. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J. Microbiol. Methods 2008, 75, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ゔゔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.L.; Wood, T.K.; Peti, W. Rebecca Page Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J. Biol. Chem. 2011, 286, 2285–2296. [Google Scholar] [CrossRef] [PubMed]
- Talavera, A.; Tamman, H.; Ainelo, A.; Konijnenberg, A.; Hadži, S.; Sobott, F.; Garcia-Pino, A.; Hõrak, R.; Loris, R. A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT. Nat. Commun. 2019, 10, 972. [Google Scholar] [CrossRef]
- Wang, H.C.; Ko, T.; Wu, M.; Ku, S.; Wu, H.; Wang, A. Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor. Nucleic Acids Res. 2012, 40, 5718–5730. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.P.; Major, A.S.; Komarovsky, V.M.; Sullivan, J.T.; Dy, R.L.; Hynes, M.F.; Salmond, G.P.C.; Ronson, C.W. A widely conserved molecular switch controls quorum sensing and symbiosis island transfer in Mesorhizobium loti through expression of a novel antiactivator. Mol. Microbiol. 2013, 87, 1–13. [Google Scholar] [CrossRef]
- Ramsay, J.P.; Tester, L.G.L.; Major, A.S.; Sullivan, J.T.; Edgar, C.D.; Kleffmann, T.; Patterson-House, J.R.; Hall, D.A.; Tate, W.P.; Hynes, M.F.; et al. Ribosomal frameshifting and dual-target antiactivation restrict quorum-sensing-activated transfer of a mobile genetic element. Proc. Natl. Acad. Sci. USA 2015, 112, 4104–4109. [Google Scholar] [CrossRef]
- Kaneko, T.; Nakamura, Y.; Sato, S.; Minamisawa, K.; Uchiumi, T.; Sasamoto, S.; Watanabe, A.; Idesawa, K.; Iriguchi, M.; Kawashima, K.; et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002, 9, 189–197. [Google Scholar] [CrossRef]
- Shimada, T.; Hirao, K.; Kori, A.; Yamamoto, K.; Ishihama, A. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol. Microbiol. 2007, 66, 744–757. [Google Scholar] [CrossRef]
- Loh, K.D.; Hirao, K.; Kori, A.; Yamamoto, K.; Ishihama, A. A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 5114–5119. [Google Scholar] [CrossRef]
- Gogarten, J.; Townsend, J. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 2005, 3, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, J.; Chen, L.; Wang, J.; Cheng, Q.; Dixon, R.; Wang, Y. Using synthetic biology to distinguish and overcome regulatory and functional barriers related to nitrogen fixation. PLoS ONE 2013, 8, e68677. [Google Scholar] [CrossRef]
- McCloskey, D.; Xu, S.; Sandberg, T.E.; Brunk, E.; Hefner, Y.; Szubin, R.; Feist, A.M. Palsson Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism. Nat. Commun. 2018, 9, 3796. [Google Scholar] [CrossRef]
- Ryu, M.H.; Zhang, J.; Khokhani, T.T.D.; Geddes, B.A.; Mus, F.; Garcia-Costas, A.; Peters, J.W.; Poole, P.S.; Ané, J. Control of nitrogen fixation in bacteria that associate with cereals. Nat. Microbiol. 2020, 5, 314–330. [Google Scholar] [CrossRef]
- Doin de Moura, G.G.; Remigi, P.; Masson-Boivin, C. Delphine Capela Experimental Evolution of Legume Symbionts: What Have We Learnt? Genes 2020, 11, 339. [Google Scholar] [CrossRef]
- Inomura, K.; Bragg, J.; Follows, M.J. A quantitative analysis of the direct and indirect costs of nitrogen fixation: A model based on Azotobacter vinelandii. ISME J. 2017, 11, 166–175. [Google Scholar] [CrossRef]
- Taylor, B.N.; Menge, D.N.L. Light, nitrogen supply, and neighboring plants dictate costs and benefits of nitrogen fixation for seedlings of a tropical nitrogen-fixing tree. New Phytol. 2021, 231, 1758–1769. [Google Scholar] [CrossRef]
- Bañuelos-Vazquez, L.A.; Tejerizo, G.T.; la Luz, L.C.; Girard, L.; Romero, D.; Brom, S. Conjugative transfer between Rhizobium etli endosymbionts inside the root nodule. Environ. Microbiol. 2019, 21, 3430–3441. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, K.; Liu, Y.; Zou, D.; Wang, D.; Xie, Z. Effects of Calcium and Signal Sensing Systems on Azorhizobium caulinodans Biofilm Formation and Host Colonization. Front. Microbiol. 2020, 11, 563367. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Liu, W.; Li, Y.; Bai, X.; Wu, H.; Bian, L.; Hu, X. LuxR-Type Regulator AclR1 of Azorhizobium caulinodans Regulates Cyclic di-GMP and Numerous Phenotypes in Free-Living and Symbiotic States. Mol. Plant. Microbe Interact. 2020, 33, 528–538. [Google Scholar] [CrossRef]
- Mølbak, L.; Molin, S.; Kroer, N. Root growth and exudate production define the frequency of horizontal plasmid transfer in the Rhizosphere. FEMS Microbiol. Ecol. 2007, 59, 167–176. [Google Scholar] [CrossRef]
- Kroer, N.; Barkay, T.; Sørensen, S.; Weber, D. Effect of root exudates and bacterial metabolic activity on conjugal gene transfer in the rhizosphere of a marsh plant. FEMS Microbiol. Ecol. 1998, 25, 375–384. [Google Scholar] [CrossRef]
- Rashid, M.M.; Ikawa, Y.; Tsuge, S. GamR, the LysR-Type Galactose Metabolism Regulator, Regulates hrp Gene Expression via Transcriptional Activation of Two Key hrp Regulators, HrpG and HrpX, in Xanthomonas oryzae pv. oryzae. Appl. Environ. Microbiol. 2016, 82, 3947–3958. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).